Abstract
Two-thirds of the 54 proteins of the Escherichia coli ribosome interact directly with the rRNAs, but the rRNA binding sites of only a very few proteins are known. We present a method (selection of random RNA fragments; SERF) that can identify the minimal binding region for proteins within ribonucleo-protein complexes such as the ribosome. The power of the method is exemplified with the ribosomal proteins L4 and L6. Binding sequences are identified for both proteins and characterized by phosphorothioate footprinting. Surprisingly, the binding region of L4, a 53-nt rRNA fragment of domain I of 23S rRNA, can simultaneously and independently bind L24, one of the two assembly initiator proteins of the large subunit.
Identifying the minimal length of RNA sequences that bind specifically to a protein is a challenge for structural research. The classical approach of digesting an RNA-protein complex with RNase usually does not give the minimal binding region, because the protein(s) within the complex might hinder the access of an RNase. Alternatively, the RNase can cut the RNA within the complex into short sequences that lose the binding capacity. Crosslinking approaches and protection experiments with base modifying reagents indicate vicinity but not necessarily the binding sequence. We decided to exploit the enormous power of in vitro selection methods that can identify the best fitting RNA sequences from about 1015 variants (SELEX, systematic evolution of ligands by exponential enrichment; ref. 1). However, when randomized sequences are used as starting conditions, affinity selection methods usually do not lead to the naturally occurring binding site for a target as such.
Short random fragments from genomic DNA were used to select the DNA binding site of transcription factor IIIA in vitro (2), and a similar idea has been put forward for the selection of protein–nucleic acid interactions (3). In vivo small DNA fragments randomly obtained from 14 genes were identified that stimulate expression of the lacZ gene (4). Expression libraries made of short rRNA sequences have been used to create resistance against antibiotics in vivo (5, 6).
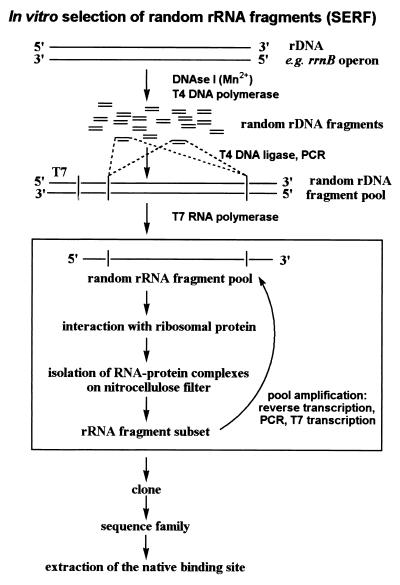
The basic idea presented here is that the use of a pool of random fragments from large RNAs directly reveals the native binding site when selected for affinity to a certain protein in vitro. The SELEX variant is abbreviated SERF for selection of random RNA fragments. The principle is shown in Fig. 1. A pool of random rDNA fragments is generated by a random cutting of rDNA. The rDNA fragments are transcribed into RNA in a second step. The advantages of the SERF method are: (i) the selection does not work on just any RNA-protein interactions but rather on the native ones; (ii) the size of the selected rRNA can be chosen and a series of overlapping fragments can reveal the minimal binding site; and (iii) the selection should be fast and efficient because the variability of the pool is low compared with the starting pool used in classical affinity selection experiments.
Figure 1.
Outline of the experimental strategy for SERF: rDNA is digested with DNase I in a manganese-dependent manner producing random rDNA fragments. The fragments are blunt-ended and ligated into a vector, thereby introducing constant primer regions. An rDNA fragment pool is amplified by PCR, and the pool of random rRNA fragments is prepared by T7 in vitro transcription. A certain size distribution of the fragments results from the area of RNA cut out from a denaturing acrylamide gel during purification of the RNA. The rRNA–protein complexes are formed and collected on nitrocellulose filters. Bound rRNA is recovered from the filter, reverse-transcribed to cDNA, and amplified in a PCR. The PCR products are the template for further T7 in vitro transcription to produce rRNA fragments for the next round of selection. After a significant increase in binding, the fragments are cloned and sequenced. The SERF technique leads directly to the native binding site for the protein.
More than two-thirds of the 54 ribosomal proteins from Escherichia coli are known to interact with rRNAs (7), but only a very few of these interactions have been characterized. Apart from the L5-L25-L18–5SrRNA complex, the rRNA binding sites for only six proteins have been narrowed to less than 100 nt: S8, S15, L1, L2, L10, and L11 (for reviews see refs. 8 and 9). Mapping the rRNA binding sites of ribosomal proteins is a prerequisite for the structural analysis of RNA–protein complexes, which might be as important for the determination of the three-dimensional (3D) structure of the ribosome as the 3D structures of isolated proteins (10, 11) and rRNA fragments (12). The latter were fitted into the ribosomal structure and played a prominent role for the interpretation of ribosomal 3D maps determined by cryo-electron microscopy or x-ray analysis (13–17).
Ribosomal protein L11 binds a 58-nt-long fragment of 23S rRNA. This complex is one of the best-characterized RNA–protein structures of the ribosome. The 3D structure of this complex was reported recently (18, 19). This well-defined L11-rRNA interaction was used as a model to elaborate the SERF method. A series of binding fragments derived from 23S rRNA were isolated in three independent SERF experiments. The common overlap of the various rRNA fragments indicated the minimal binding sequence for L11 (58 nt; data not shown).
Here, we report the identification of rRNA binding sites involving the ribosomal proteins L4 and L6; the corresponding fragments contain 53 and 35 nt, respectively. L4 is essential for early events of the 50S assembly (20) and seems to be adjacent to the peptidyl-transferase center (21). L6 is one of the most conserved proteins and is present in ribosomes of all organisms (22) at or near the elongation-factor binding site (15). Furthermore, preliminary experiments demonstrated a low unspecific binding of these proteins to an rRNA fragment pool. Low unspecific binding facilitates the selection of specific high-affinity binding sites. In addition, we present data about the rRNA interactions with the assembly-initiator protein L24.
Materials and Methods
Construction of a Random rRNA Fragment Pool from the rrnB Operon.
DNase I digestion on the Plasmid ptac-1 (23) was performed in 50 mM Tris⋅HCl (pH 8.0 at 25°C), 0.01 mM MnCl2 (freshly prepared) at 16°C for 15–50 min. A total of 35 ng/μl DNA was digested with 0.3 μg/μg DNA DNase I (RNase-free). The reaction was stopped by the addition of EDTA, and the mixture was phenol-extracted twice. Blunt ends of the fragments were generated by T4 DNA polymerase and the Klenow fragment under standard conditions (24). The rDNA fragments were ligated into the SmaI site of pGem 3Z- (Amersham Pharmacia, AC#65307) with T4 DNA ligase. PCR (150 μl) was carried out in the presence of 0.5 μM of each of the T7–5′(+) (5′-TAATACGACTCACTATAGGGCGAATTCGAGCTCG-3′) and 3′(−) (5′-GTCGACTCTAGAGGATCC-3′) primers. A 100-μl T7 in vitro transcription (25) using the purified PCR product (Qiagen PCR purification kit; Chatsworth, CA) as template yielded 2.5 A260 units of rRNA fragments after gel purification.
SERF Procedure.
The ribosomal proteins were purified from total proteins of the 50S ribosomal subunit as described (26) and were essentially pure as judged from Coomassie-stained two-dimensional gels. For in vitro selection from randomly fragmented rRNA, rRNA fragments were incubated for 3 min at 70°C in 20 mM Hepes-KOH (pH 7.5), 4 mM MgCl2, 400 mM NH4Cl, and 6 mM β-mercaptoethanol (Rec-4 buffer) and cooled to 37°C; proteins were incubated 15 min at 37°C in the same buffer before mixing with the rRNA fragments. The RNA/protein ratio was subsequently increased during the following selection rounds from 0.67 to 8 at μM concentrations: from 0.6 μM RNA and 0.9 μM protein to 0.8 μM RNA and 0.1 μM protein. After incubation for 10 min at 37°C and 10 min on ice, the mixture was directly filtrated (0.45-μm nitrocellulose filter, prewetted and degassed for 30 min in Rec-4 buffer). The filter was washed with 500 μl of ice-cold Rec-4 buffer, cut into pieces, and extracted with 400 μl phenol-Tris⋅HCl (pH 7.8) and 200 μl 7 M urea (freshly prepared) for 60 min at room temperature. RNA was recovered by EtOH precipitation in the presence of 1 μg/ml RNase-free glycogen. For reverse transcription, the selected RNA was dissolved in 10 μl MQ-water containing 1.5 pmol/μl primer-3′(−), heated at 70°C for 5 min, and cooled to room temperature for 5 min. Reverse transcription reaction with avian myeloblastosis virus reverse transcriptase was done in 30 μl at 42°C; for amplification, 11.3 μl of the reaction was directly used in a 150-μl PCR.
Iodine cleavage experiments were done in Rec-4 buffer (omitting β-mercaptoethanol) with 0.1 μM 32P-labeled and phosphorothioated RNA. The protein concentration was 0.3 μM for L4 and L24, 0.4 μM for L6, keeping the protein concentration below saturation during complex formation. Iodine was added in EtOH to a final concentration of 1 mM (2% EtOH) for 1 min at 0°C, and the cleavage reaction was stopped by the addition of DTT. In interference experiments, the complex was collected on nitrocellulose filters and the RNA was cleaved with iodine after extraction. In the protection experiments, iodine was added to the complex in solution, the complex was then collected on a nitrocellulose filter, and the extracted RNA was analyzed on a sequencing gel. The cleavage products are separated on a 13% polyacrylamide-7.5 M urea gel and visualized with the help of a PhosphorImager system. In a first processing step, each band was scanned (imagequant program) and its intensity was normalized to that of a band of the same lane that did not show an interference or protection effect. In a second step, the normalized intensity of a band was compared with that of the corresponding control band of the RNA in solution. Control experiments carried out with nonphosphorothioated RNA or without addition of iodine did not result in cleavages. The experiments were repeated 2–6 times.
Results and Discussion
The Binding Site of L4.
L4 is an early assembly protein of the large ribosomal subunit and is essential for the formation of an active peptidyl-transferase center (7). It is a regulatory multitalent in that it is involved in both transcriptional and translational control of its own polycistronic mRNA (27).
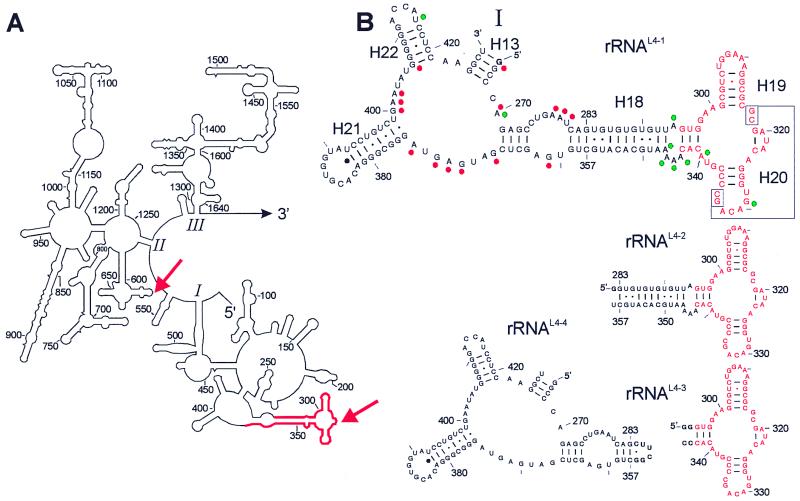
Following the scheme in Fig. 1, the rDNA of the rrnB operon was digested with DNase I in the presence of 0.01 mM Mn2+ ions. DNase I usually produces single-strand cuts, but in the presence of Mn2+ the enzyme makes double-strand cuts and slows the digestion process (28), thus allowing a convenient adjustment of the fragment length to about 70–200 nt. The random rDNA fragments were inserted into an appropriate plasmid, thereby introducing constant primer regions. To increase the specificity of selection, the molar ratio of rRNA fragments over L4 was increased from 0.67 to 8 during subsequent rounds of selection (29). A total of 5% of the rRNA fragment input was bound nonspecifically by L4 in the first round of selection, binding increased to about 14% in the seventh round as determined in control filter-binding experiments with stoichiometric molar ratios of fragments and L4. A subset of rRNA fragments was cloned and sequenced. A total of 26 of 71 cloned rRNA fragments contained the A282–U369 (88 nt) fragment of domain I of 23S rRNA as a consensus (red in Fig. 2A). All other fragments were derived from plasmid DNA, except four antisense 23S sequences. Subsequently, the fragments rRNAL4-1 to rRNAL4-3 were synthesized, and their binding to L4 was assessed (Fig. 2B). All fragments bind to L4 with about the same affinity of Ka = 8.3 × 106 M−1, i.e., the smallest fragment rRNAL4-3 (53 nt) still contains the binding site (red in Fig. 2B).
Figure 2.
Secondary structures of a part of 23S rRNA and some fragments analyzed for binding to the ribosomal protein L4. (A) 5′ part of the 23S rRNA with the domains I to III (1–1646, E. coli numbering; ref. 48). The rRNA fragment selected in the SERF procedure with protein L4 is highlighted in red. Crosslinks to the protein are indicated by red arrows (31, 32). (B) rRNAL4-1 is a subdomain of domain I with 163 nt comprising the sequence G-G266-U427. The minimal L4-binding site is in red. Relevant structural probing data for proteins L4 (red dots; ref. 33) and L24 (green dots; ref. 35) are indicated (RNase-digestion data have identified the same region and are not integrated). Phylogenetic studies suggest that the identified binding site contains a pseudoknot by means of the interaction of G317–C318 with G333–C334 (enboxed; ref. 39). Other rRNA fragments used in this study are rRNAL4-2 (G-G283-U358; 77 nt), rRNAL4-3 (GG-G295-C343-CC; 53 nt), and for control rRNAL4-4 [G-(G266-G283)-CUUCG-(G356-U427); 96 nt]. Nucleotides in bold at 5′ and 3′ ends indicate deviations from the E. coli 23S rRNA sequence.
Evidence was reported previously that some nucleotides within this rRNA sequence are of importance for the binding of L4. Free L4 causes a transcription pause during the mRNA synthesis of the S10 operon. A large part of domain I of 23S rRNA (48–386; 339 nt) competes well with the pausing event and even a 122-nt-long sequence (265–386) showed a lower but significant competition; a deletion of nucleotides 321–325 (within the binding site identified here) abolished the competition (30). Furthermore, L4 could be crosslinked to nucleotides 320–325, whereas the second crosslink site to nucleotides 613–617 (31, 32) is not identified as an L4 binding site in the approach presented here (Fig. 2A, arrows).
L4 protects a number of bases, outside but close to the binding sequence of L4, against modifying reagents (red dots in Fig. 2B; ref. 33). To test whether the corresponding structure left of the base pair G283–C357 (Fig. 2B) binds L4 independently of the binding site rRNAL4-3, we closed this base pair by the tetraloop 5′-CUUCGG-3′, thus shortening helix H18 (rRNAL4-4 in Fig. 2B). This rRNA fragment does not bind to ribosomal protein L4 (data not shown).
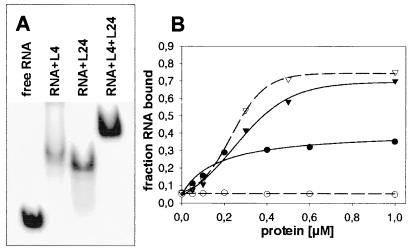
The ribosomal protein L24 initiates the ribosomal assembly process together with L3 (34). L24 protects bases directly adjacent to the L4 binding site (green dots in Fig. 2B; ref. 35). The internal loop bordered by the helices H18, H19, and H20 was considered one of two putative binding sites of L24. Therefore, we tested the binding of L24 to the same fragment. Surprisingly, L24 binds alone (Ka = 6.5 × 106 M−1) as well as simultaneously with L4 to the same rRNA fragment as indicated by gel shift experiments, where L4 and L24 individually induce slightly different gel shifts that are well separated from the complex simultaneously containing both proteins (Fig. 3A). Interestingly, a single mutation G298U was observed in 60% of the high-affinity rRNA fragments picked in the L4-dependent selection. The presence of this mutation slightly improves the interaction with L4, but completely abolishes L24 binding (see the binding experiments in Fig. 3B). This result underscores the specificity of the rRNA–protein interactions in the ternary complex of L4-rRNA-L24 and suggests that both proteins recognize distinct features of the fragment.
Figure 3.
Binding experiments with L4 and L24. (A) Band-shift assay with RNAL4-2 in the presence of the ribosomal proteins L4, L24, and L4 + L24, respectively: 7.5 μM RNA, 15 μM protein(s). The ionic conditions were 10 mM Hepes (pH 7.4), 4 mM MgCl2, 100 mM NH4Cl, and 4 mM β-mercaptoethanol. An equivalent experiment was performed with the shorter RNAL4-3 (53 nt), where the L4-rRNAL4-3 and L4-rRNAL4-3-L24 complexes migrated at similar positions. The band with the putative L4-rRNAL4-3-L24 complex was excised from the gel and checked for protein content by SDS-gel electrophoresis. Both L4 and L24 were detected, thus verifying the formation of the ternary complex. (B) Nitrocellulose filtration: Binding of L4 (triangles) and L24 (circles) to the fragment rRNAL4-3 (solid lines) and to the corresponding mutant G289U (broken lines).
The rRNAL4-3 fragment complexed with L4 and L24, respectively, was further characterized by the phosphorothioate technique (36, 37). Statistically, one phosphate group per molecule containing a sulfur atom instead of a nonbridging oxygen is randomly incorporated 5′ to A, U, G, or C into the RNA by T7 in vitro transcription. The resulting RNA grossly maintains its biological activity as demonstrated in a comprehensive functional analysis with tRNAs (38). Adding the small inert iodine molecule I2 induces a cleavage at the modified phosphate group, provided that the sulfur modification does not prevent complex formation and that iodine has access within the complex. These two aspects of cleavage inhibition are explored in interference and protection analysis, respectively.
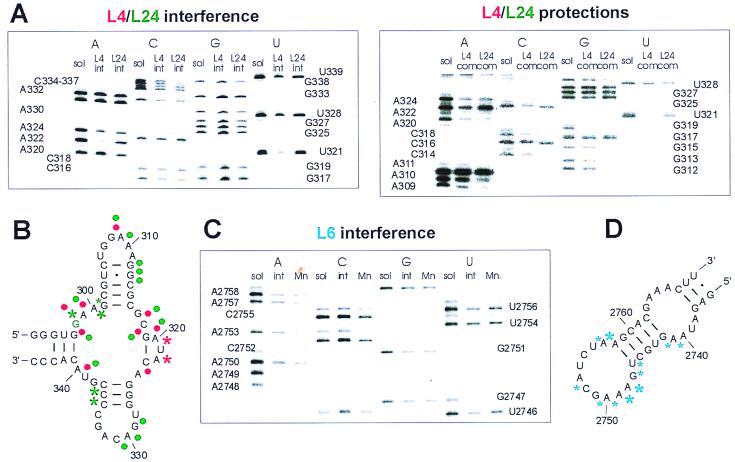
To assess a possible interference of the thio-modified phosphate groups with complex formation, the protein–rRNAL4-3 complex was formed and purified from the nonbound rRNA. The rRNA fragment was cleaved with iodine after the extraction from the complex, and the intensities of the iodine-induced cleavages were compared with those of the fragment in solution. If a distinct modified phosphate group interferes with complex formation, the corresponding band in the sequence gel is weakened or absent. Two sulfur substitutions in the backbone 5′ of U321 and A322 strongly interfere with binding of L4 to rRNAL4-3. Strong interference with L24 binding was observed at positions A299, G301, C336, and C337, weaker for C302 (see, for example, U321 and A322 for interference with L4 binding and C336 and C337 for interference with L24 binding in Fig. 4A). The clear differences in the interference patterns of both proteins support the above conclusion that L4 and L24 recognize different features of this short rRNA fragment.
Figure 4.
Iodine cleavage experiments of phosphorothioated rRNA fragments. (A and B) Results with rRNAL4-3 in complexes with the proteins L4 and L24. (A) Sections of sequencing gels from interference and protection experiments, respectively. sol, rRNA in solution; int, rRNA isolated from the complexes before iodine cleavage (interference); com, rRNA isolated from the complexes after iodine cleavage (protection). (B) Superposition of the results obtained with L4 (red symbols) and L24 (green symbols). Stars indicate interference positions, dots protections. G298 essential for L24 binding is highlighted green. Weak interference, the normalized intensity of a band was >0.33 and <0.66 of that of the corresponding band from the rRNA in solution; strong effects, the intensity ratio was <0.33. Because most of the positions were protected, we only show the strongly protected positions. (C and D) Results with rRNAL6-2 in a complex with L6. (C) Gel section from an interference experiment. (D) Strong and weak interferences are indicated by large and small blue stars, respectively. Mn, interference experiment in the presence of 3 mM Mn2+.
In contrast to the interference experiment, the protection patterns are assessed by adding iodine to the complex in solution, i.e., before isolation of the bound rRNA fragment. Thus, strong interference sites cannot be judged for protection effects. A surprising exception of this rule is seen in the position A322. As mentioned above, a thio-phosphate at this nucleotide prevents binding of this fragment to L4, whereas we see a relatively strong band in L4-protection experiments (see Fig. 4A). A possible explanation is that cleavage in solution at this position of an rRNA fragment allows binding, in contrast to the intact fragment that is thioated at this position. The other positions protected from iodine cleavage within the complexes are spread over the whole rRNA fragment: G298, A300, A309, C318, G319, A320, and A340 are protected by either of the proteins. Protections only seen with one protein are A324 with L4 and A310, G312, G313, C314, G329, A330, and C331 with L24 (compare gel section in Fig. 4A).
The large number of protections common to both proteins indicates that reduced access of I2 at these positions is caused by tight folding of the rRNA and not by the proteins themselves. The interpretation is supported by the fact that the same positions (in addition to others) also show reduced cleavage efficiency in free rRNA when compared with denatured rRNA in the presence of 7.5 M urea (not shown). The L24 interferences and protections (Fig. 4B) suggest that L24 folds the fragment by bringing together the loops around 310 and 330. This view agrees with both the phylogenetically proposed pseudoknot (39) and the role of L24 as an assembly initiator protein and organizer of rRNA folding (34, 35). L4 interacts mainly with the bulge between helices H19 and helix H20 around U321. The identified rRNA fragment rRNAL4-3 within the 23S rRNA together with L24 and L4 probably play a key role for the folding and organization of the 23S rRNA during early assembly events.
The Binding Site of L6.
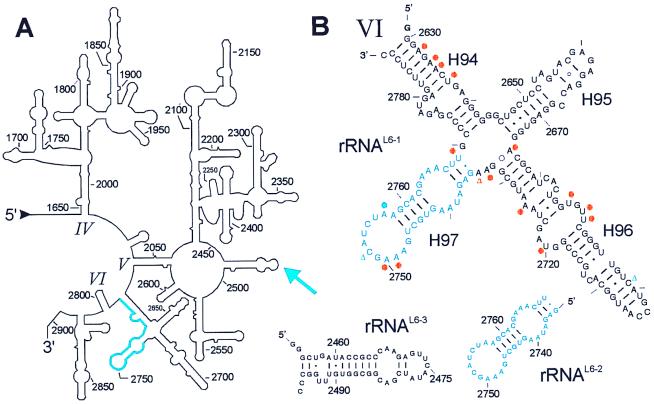
Ribosomal protein L6 is one of the most highly conserved ribosomal proteins located at the factor binding site of the large subunit (15, 22). The SERF technique identified a consensus sequence U2739–C2789 of domain VI of 23S as the binding site of L6 after four rounds of selection (blue in Fig. 5A). A total of 22 of 43 clones contained the 2739–2789 (51 nt) fragment of domain VI of 23S rRNA. Other 23S rRNA fragments contained sequences from domain V of 23S rRNA: 2098–2204 (107 nt, found twice), 2111–2197 (87 nt, found once), and 2131–2196 (66 nt, found once). These sequences contain the L1 binding site (40) and do not bind the protein L6 in filter binding assays (data not shown). The rest were antisense rRNA sequences found only once, or originated from the plasmid ptac-1 [with the exception of one from 16S rRNA 778–830 (53 nt)]. The fragments rRNAL6-1 and rRNAL6-2 were synthesized and tested for binding to L6. Both fragments bound the protein with the same affinity of Ka = 2 × 106 M−1, revealing a short sequence with only 35 nt from G2735 to U2789 as the binding site for this ribosomal protein (blue in Fig. 5B).
Figure 5.
Secondary structures of a part of the 23S rRNA and some fragments analyzed for binding to the ribosomal protein L6. (A) Secondary structure of the 3′ region of 23S rRNA with domain IV-VI (1647–2904; E. coli numbering; ref. 48). The rRNA fragment selected in the L6-SERF experiment is highlighted in blue. The blue arrow indicates a crosslink between L6 and helix H89 of domain V in 23S rRNA (31, 42). (B) rRNAL6-1 is a subdomain of domain VI with 161 nt comprising the sequence G-G2630-C2789 and including the functionally important sarcin/ricin stem loop (H95). The minimal L6-binding site is in blue. L6 induced changes of base reactivities against dimethylsulphate are indicated with a blue dot (protection) and blue triangles (enhancements) (41). Some bases are protected against base-specific chemical probing by protein L3 within rRNAL6-1 (orange dots, protection; triangle, enhancement; refs. 41 and 47). This fragment binds both L3 and L6. Other fragments used in this study are RNAL6-2 (G2735-U2769, 35 nt), the minimal fragment for L6 binding, and rRNAL6-3 (G-G2455-C2496-CC, 46 nt; helix H89) that originates from domain V of 23S rRNA and contains the crosslink site of L6. This fragment does not bind the protein.
Two observations agree well with the binding site reported here. (i) The binding affinity determined for L6 compares well with that reported for the rRNA sequence 2640–2774 (Ka = 3 × 106 M−1; ref. 41), i.e., the rRNAL6-1 fragment. (ii) L6 changed the extent of base modifications with dimethylsulphate at three positions (41), and two of them are located within the binding site identified here (blue dots and triangles in Fig. 5B). In addition, Uchiumi et al. (41) showed that a mutation of the protected A2757 within the identified sequence abolishes L6 binding.
However, a crosslink between L6 and the loop of helix 89 around nucleotide 2475 (31, 42) is not covered by the L6 binding site reported here (see arrow in Fig. 5A). Therefore, we examined whether helix H89 is an L6 binding site that has escaped the selection process of the L6-SERF experiment. To this end, we synthesized helix H89 and tested it for binding to L6, with negative results (rRNAL6-3; Fig. 5B). This helix might be in the vicinity of L6 within the ribosome, but is not an L6 binding site. We found no hints of two rRNA binding sites as proposed in the presentation of the 3D structure of L6 (43, 44).
We also tested the short binding fragment rRNAL6-2 by phosphorothioate analysis. The results were strikingly different to those obtained with L4. In the L4–rRNA complex, only two of 53 sulfur substitutions interfere with binding, whereas modification of 11 phosphates within a short sequence of 18 nt (2741–2758) impaired the binding of L6, three of which abolished binding (A2748, A2749, and A2762; see Fig. 4 C and D). The accumulation of interfering thio-substitutions made assessment of a protection pattern prohibitively difficult, so that a contact pattern could not be established as in the case of the L4- and L24–rRNA complexes. One reason for an interference of a thio-phosphate with the binding of the rRNA fragment could be the involvement of a coordinating Mg2+ ion, because Mg2+ can coordinate with oxygen but not with sulfur. Mn2+ can coordinate with both ions (45, 46); replacement of Mg2+ ions by Mn2+ therefore should reestablish the binding. A control experiment in the presence of up to 3 mM Mn2+ did not change the interference pattern (“Mn” lanes in Fig. 4C), indicating that Mg2+ ions are not directly involved in coordinating contacts of L6 with these phosphate groups. The massive presence of phosphate groups that are not allowed to be modified indicates a tight binding pocket within the tertiary structure of the sugar-phosphate backbone of this rRNA sequence for L6. The L6 binding site identified with SERF comprises only 35 nt, which is one of the shortest rRNA fragments identified so far that shows specific binding to a ribosomal protein.
It has been reported that, in and around the L6 binding site identified here, a number of bases were protected against chemical probing on binding of ribosomal protein L3 (orange dots in Fig. 5B, refs. 41 and 47), the second initiator protein of the 50S assembly after L24 (34). We tested the binding of the two fragments rRNAL6-1 and rRNAL6-2 (Fig. 5B); only the fragment rRNAL6-1 binds L3 (not shown).
In summary, the SERF technique can identify short protein binding sites within large RNAs in a straightforward manner. The resulting short sequences together with the high affinity for proteins have a promising potential for NMR and crystallographic studies. The SERF method is not restricted to substructures of the ribosome and might be applied to other RNA–protein complexes, such as the spliceosome, or to mapping studies of protein-binding sites within large mRNAs.
Acknowledgments
We thank Drs. Richard Brimacombe and Sean Connell for help and discussions.
Abbreviations
- SERF
selection of random RNA fragments
- 3D
three-dimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090009297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090009297
References
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K W, Vogelstein B. Nucleic Acids Res. 1989;17:3645–3653. doi: 10.1093/nar/17.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer B S, Shtatland T, Brown D, Gold L. Nucleic Acids Res. 1997;25:781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreyfus M. J Mol Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 5.Tenson T, Deblasio A, Mankin A. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thom G, Prescott C D. Bioorg Med Chem. 1997;5:1081–1086. doi: 10.1016/s0968-0896(97)00060-6. [DOI] [PubMed] [Google Scholar]
- 7.Nierhaus K H. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 8.Ehresmann B, Ehresmann C, Romby P, Mougel M, Baudin F, Westhof E, Ebel J-P. In: The Ribosome: Structure, Function and Evolution. Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warners J R, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 148–159. [Google Scholar]
- 9.Draper D E. In: Ribosomal RNA–Structure, Evolution, Processing and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 171–197. [Google Scholar]
- 10.Ramakrishnan V, White S W. Trends Biochem Sci. 1998;23:208–212. doi: 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 11.Nikonov S V, Nevskaya N A, Fedorov R V, Khairullina A R, Tishchenko S V, Nikulin A D, Garber M B. Biol Chem. 1998;379:795–805. doi: 10.1515/bchm.1998.379.7.795. [DOI] [PubMed] [Google Scholar]
- 12.Moore P B. Annu Rev Biophys Biomol Struct. 1998;27:35–58. doi: 10.1146/annurev.biophys.27.1.35. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra A, Penczek P, Agrawal R K, Gabashvili I S, Grassucci R A, Jünemann R, Burkhardt N, Nierhaus K H, Frank J. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 14.Clemons W M J, May J L, Wimberly B T, McCutcheon J P, Capel M S, Ramakrishnan V. Nature (London) 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 15.Ban N, Nissen P, Hansen J, Capel M, Moore P B, Steitz T A. Nature (London) 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 16.Cate J H, Yusupov M M, Yusupova G Z, Earnest T N, Noller H F. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 17.Tocilj A, Schlünzen F, Janell D, Glühmann M, Hansen H A S, Harms J, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Proc Natl Acad Sci USA. 1999;96:14252–14257. doi: 10.1073/pnas.96.25.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conn G L, Draper D E, Lattman E E, Gittis A G. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 19.Wimberly B T, Guzmon R, McCutcheon J P, White S W, Ramakrishnan V. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 20.Spillmann S, Dohme F, Nierhaus K H. J Mol Biol. 1977;115:513–523. doi: 10.1016/0022-2836(77)90168-1. [DOI] [PubMed] [Google Scholar]
- 21.Dohme F, Fahnestock S R. J Mol Biol. 1979;129:63–81. doi: 10.1016/0022-2836(79)90060-3. [DOI] [PubMed] [Google Scholar]
- 22.Müller E C, Wittmann-Liebold B. Cell Mol Life Sci. 1997;53:34–50. doi: 10.1007/PL00000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewicki B T U, Margus T, Remme J, Nierhaus K H. J Mol Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Triana-Alonso F J, Dabrowski M, Wadzack J, Nierhaus K H. J Biol Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 26.Diedrich G, Burkhardt N, Nierhaus K H. Protein Expression Purif. 1997;10:42–50. doi: 10.1006/prep.1996.0702. [DOI] [PubMed] [Google Scholar]
- 27.Zengel J M, Lindahl L. Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 28.Melgar E, Goldthwait D A. J Biol Chem. 1968;243:4409–4416. [PubMed] [Google Scholar]
- 29.Irvine D, Tuerk C, Gold L. J Mol Biol. 1991;222:739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- 30.Zengel J M, Lindahl L. Nucleic Acids Res. 1993;21:2429–2435. doi: 10.1093/nar/21.10.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osswald M, Greuer B, Brimacombe R. Nucleic Acids Res. 1990;18:6755–6760. doi: 10.1093/nar/18.23.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiede B, Urlaub H, Neubauer H, Grelle G, Wittmann-Liebold B. Biochem J. 1998;334:39–42. doi: 10.1042/bj3340039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostergaard P, Phan H, Johansen L B, Egebjerg J, Ostergaard L, Porse B T, Garrett R A. J Mol Biol. 1998;284:227–240. doi: 10.1006/jmbi.1998.2185. [DOI] [PubMed] [Google Scholar]
- 34.Nowotny V, Nierhaus K H. Proc Natl Acad Sci USA. 1982;79:7238–7242. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egebjerg J, Leffers H, Christensen A, Andersen H, Garrett R A. J Mol Biol. 1987;196:125–136. doi: 10.1016/0022-2836(87)90515-8. [DOI] [PubMed] [Google Scholar]
- 36.Verma S, Eckstein F. Annu Rev Biochem. 1998;67:99–134. doi: 10.1146/annurev.biochem.67.1.99. [DOI] [PubMed] [Google Scholar]
- 37.Strobel S A. Curr Opin Struct Biol. 1999;9:346–352. doi: 10.1016/S0959-440X(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 38.Dabrowski M, Spahn C M T, Nierhaus K H. EMBO J. 1995;14:4872–4882. doi: 10.1002/j.1460-2075.1995.tb00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutell R R, Woese C R. Proc Natl Acad Sci USA. 1990;87:663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egebjerg J, Christiansen J, Garrett R A. J Mol Biol. 1991;222:251–264. doi: 10.1016/0022-2836(91)90210-w. [DOI] [PubMed] [Google Scholar]
- 41.Uchiumi T, Sato N, Wada A, Hachimori A. J Biol Chem. 1999;274:681–686. doi: 10.1074/jbc.274.2.681. [DOI] [PubMed] [Google Scholar]
- 42.Urlaub H, Kruft V, Bischof O, Mueller E C, Wittmann-Liebold B. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golden B L, Ramakrishnan V, White S W. EMBO J. 1993;12:4901–4908. doi: 10.1002/j.1460-2075.1993.tb06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies C, Bussiere D E, Golden B L, Porter S J, Ramakrishnan V, White S W. J Mol Biol. 1998;279:873–888. doi: 10.1006/jmbi.1998.1780. [DOI] [PubMed] [Google Scholar]
- 45.Christian E L, Yarus M. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 46.Cate J H, Hanna R L, Doudna J A. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 47.Leffers H, Egebjerg J, Andersen A, Christensen T, Garrett R A. J Mol Biol. 1988;204:507–522. doi: 10.1016/0022-2836(88)90351-8. [DOI] [PubMed] [Google Scholar]
- 48.Gutell R R. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]