Abstract
Werner Syndrome (WS) is an inherited disease characterized by premature onset of aging, increased cancer incidence, and genomic instability. The WS gene encodes a 1,432-amino acid polypeptide (WRN) with a central domain homologous to the RecQ family of DNA helicases. Purified WRN unwinds DNA with 3′→5′ polarity, and also possesses 3′→5′ exonuclease activity. Elucidation of the physiologic function(s) of WRN may be aided by the identification of WRN-interacting proteins. We show here that WRN functionally interacts with DNA polymerase δ (pol δ), a eukaryotic polymerase required for DNA replication and DNA repair. WRN increases the rate of nucleotide incorporation by pol δ in the absence of proliferating cell nuclear antigen (PCNA) but does not stimulate the activity of eukaryotic DNA polymerases α or ɛ, or a variety of other DNA polymerases. Moreover, we show that functional interaction with WRN is mediated through the third subunit of pol δ: i.e., Pol32p of Saccharomyces cerevisae, corresponding to the recently identified p66 subunit of human pol δ. Absence of the third subunit abrogates stimulation by WRN, and stimulation is restored by reconstituting the three-subunit enzyme. Our findings suggest that WRN may facilitate pol δ-mediated DNA replication and/or DNA repair and that disruption of WRN-pol δ interaction in WS cells may contribute to the previously observed S-phase defects and/or the unusual sensitivity to a limited number of DNA damaging agents.
Werner Syndrome (WS) is an autosomal, recessive progeroid disorder characterized by genomic instability (1). Patients with WS prematurely exhibit age-related conditions such as atherosclerosis, osteoporosis, diabetes mellitus, and bilateral cataracts. Additionally, they show an increased incidence of cancers of non-epithelial cell lineage (2). Genetic instability in WS is manifested at the chromosomal level by breaks and rearrangements, and at the DNA level by multiple, large deletions (3–5).
The WS gene encodes a 1,432-amino acid polypeptide (WRN) containing a domain homologous to the RecQ family of DNA helicases (6). This family is represented by the prototypical Escherichia coli RecQ (7), Saccharomyces cerevisiae Sgs1 (8), Schizosaccharomyces pombe Rqh1 (9), Xenopus laevis FFA-1 (Focus forming activity) (10), and the human proteins RecQL (11), BLM (the product of the Bloom's syndrome gene) (12), RecQ4 (the product of the Rothmund-Thomson syndrome gene) and RecQ5 (13, 14).
Several RecQ family members, including WRN, have been purified and shown to possess 3′→5′ DNA unwinding activity in vitro (15–20). WRN helicase activity exhibits several features: First, unwinding of duplex DNA is ATP-dependent and typically requires a 3′ single-stranded tail (19, 21). Second, the activity is nonprocessive, limited to unwinding small stretches (<25 nt) of duplex DNA. Third, processivity is increased by ssDNA binding proteins, particularly by human replication protein A (22). In the presence of human replication protein A, WRN can unwind duplex DNA segments as long as 800 nt (23). Fourth, WRN can unwind alternate structures, such as tetraplex DNA assumed by CGG-rich DNA sequences (24).
In addition to unwinding DNA, WRN can also digest DNA exonucleolytically with 3′→5′ polarity (25, 26). WRN is the only RecQ-like helicase demonstrated to exhibit nuclease activity in vitro, although the X. laevis ortholog, FFA-1, potentially encodes a nuclease. WRN nuclease activity is nucleotide co-factor stimulated, shows a strong preference for duplex DNA with 3′-recessed termini containing either -OH or -PO4 groups, and preferentially digests DNA with a single 3′ terminal mismatch (27). In addition, unlike 3′→5′, proof-reading exonucleases, WRN nuclease does not hydrolyze single-stranded DNA (27, 28).
Although the DNA metabolic process/es in which WRN participates remain to be elucidated, WRN has been implicated in numerous DNA transactions. The postulation that WRN functions in DNA recombination arises by analogy with the involvement of E. coli RecQ in DNA recombination as well as from the hyperrecombination phenotype and elevated chromosomal instability of WS cells (3, 4, 29–31). The S-phase abnormalities of WS cells, including a decreased frequency of replicon initiation and a reduced rate of replication, implicate WRN in DNA replication (32, 33). Finally, a role for WRN in DNA repair is suggested by the observation that WS cells are uniquely sensitive to 4-nitroquinoline-1-oxide (34), an agent that generates base adducts as well as oxygen free radicals that produce a variety of DNA lesions, including double-strand DNA breaks. Most recently, WRN has been reported to be involved in RNA polymerase II-mediated transcription (35). Thus, the possible roles of WRN are multifaceted.
One approach to defining the role(s) of WRN is to identify the proteins with which it interacts. Currently, the proteins that have been shown to physically and functionally associate with WRN are human replication protein A and p53 (23, 36, 37). In addition, there is evidence that WRN interacts with proliferating cell nuclear antigen (PCNA) and topoisomerase I (38). We have pursued the identification of interacting proteins by screening for proteins that modulate WRN activity or vice versa, by monitoring the effect of WRN on the enzymatic activities of candidate proteins. Because cells from WS patients show DNA replication abnormalities, we have focused our search on replication proteins that influence WRN activity or are effected by WRN. We report here that WRN stimulates DNA synthesis by DNA polymerase δ (pol δ), an essential eukaryotic polymerase with central roles in DNA replication and DNA repair (39, 40). Moreover, WRN-mediated stimulation of pol δ activity requires Pol32p, the third subunit of S. cerevisiae pol δ (41) that is homologous to the recently discovered p66 subunit of human DNA polymerase δ (42).
Materials and Methods
Materials.
[γ-32P]ATP was purchased from NEN. High performance liquid chromatography-purified oligodeoxynucleotide primers and template used for primer extension assays were synthesized by Operon Technologies (Alameda, CA). Ultrapure deoxyribonucleoside triphosphates (dNTPs) were purchased from Promega. Bacteriophage T4 polynucleotide kinase was supplied by New England Biolabs.
WRN protein (>90% homogeneous) was purified by the protocol published by Shen et al. (26). Approximate protein concentration was determined from Coomassie stained SDS/polyacrylamide gels by using BSA as a standard. Homogeneous DNA pol δ and pol δ* from S. cerevisiae were also purified as described (43). The POL32 gene with a N-terminal (His)7 tag was overexpressed in the E. coli strain BL21(DE3) as described, except that the induction was performed at 17°C (43). Pol32p was purified from the lysate supernatant by successive phosphocellulose, Ni2+-agarose, and superose 12 chromatography. Pol δ was reconstituted in vitro by incubating purified pol δ* (50 μg, 250 pmol) and Pol32p (10 μg, 125 pmol dimer) for 1 h. Incubation was on ice in a total volume of 300 μl in buffer containing 40 mM Hepes-NaOH (pH 7.5), 10% glycerol, 1 mM EDTA, 0.02% Nonidet P-40, 0.2 M NaCl, 5 mM DTT, 5 mM sodium bisulfite, and 2 μM each of leupeptin and pepstatin A. After incubation, the reconstituted enzyme complex was isolated from the free subunits by chromatography through a Superose 6 gel filtration column. The reconstituted enzyme was stored at −80°C until use. Protein concentrations of pol δ preparations were determined spectrophotometrically at A280. Human DNA polymerase α-primase complex (pol α) was a kind gift of Teresa Wang (Stanford University), and human DNA polymerase ɛ (pol ɛ) was a generous gift of Hitomi Asahara and Stuart Linn (University of California at Berkeley).
Primer Extension Assays.
A 14-nt primer (5′ CGCGCCGAATTCCC 3′) was 5′-end labeled with 32P (44) and was boiled immediately after labeling to inactivate the kinase. The labeled oligomer was mixed with a 2-fold molar excess of the complementary unlabeled 46-nt oligonucleotide DNA template (5′G-CGCGGAAGCTTGGCTGCAGAATATTGCTAGCGG-GAATTCGGCGCG 3′) in 50 mM Tris·HCl buffer, pH 8.0, 10 mM MgCl2. The mixture was boiled for 5 min at 100°C and the denatured DNA strands were allowed to anneal by slow cooling to room temperature.
Reactions (10 μl) were carried out in buffer containing 40 mM Tris⋅HCl buffer (pH 7.4), 5 mM MgCl2, 5 mM DTT, 0.1 mg/ml BSA, and 100 μM each of dATP, dGTP, dCTP, and dTTP (unless indicated otherwise). Known amounts of DNA polymerase and WRN (specified in figure legends) were added, and the reactions were incubated at 37°C for 10 min. The molar amounts of polymerase were calculated based on pol δ being a dimer of a heterotrimer (≈500 kDa), and pol δ* being a heterodimer (≈180 kDa); the amount of WRN was calculated as moles of monomeric protein (≈165 kDa). The reactions were terminated by rapid cooling on ice and addition of an equal volume of denaturing loading buffer (44). The samples were boiled, and aliquots were electrophoresed through 14% polyacrylamide-urea gels. The gels were dried, and the extension products were visualized by autoradiography.
Single Nucleotide Extension Kinetics.
Assays to measure the Km and Vmax values for the incorporation of the initiating nucleotide by pol δ were carried out by using a protocol modified from that of Boosalis et al. (45). After determining the requisite conditions for kinetic assays, extension was monitored as a function of dGTP concentration, keeping the template and enzyme concentrations and reaction time constant. The reactions were carried out at 37°C and were processed as described above. Amounts of extension products were quantitated by PhosphorImager analysis (Molecular Dynamics); the kinetic constants of pol δ −/+ WRN were calculated from Hanes-Woolf plots.
Pol δ Holoenzyme Assays.
The assays were carried out as described (43). Reaction mixtures (45 μl) contained 40 mM Tris⋅HCl (pH 7.8), 8 mM magnesium acetate, 75 mM NaCl, 0.2 mg/ml BSA, 1 mM DTT, 100 μM each of dATP, dCTP, and dGTP, 10 μM [α-32P] dTTP, 1 mM ATP, 300 ng of singly primed single-stranded mp18 DNA, 2.5 μg of E. coli single-stranded DNA binding protein, 125 fmol of replication factor C, and 500 fmol of PCNA (as trimers). After a 1-min incubation at 13°C (to load PCNA), pol δ with or without WRN was added, and the incubation was continued at 13°C for the indicated times. Aliquots were analyzed on a 1% alkaline agarose gel.
Results
As a first step in identifying WRN partners, we have surveyed the ability of candidate proteins to functionally interact with WRN. Because WS cells show defects in DNA replication, we have searched for interactions between WRN and DNA replication proteins, in particular, DNA polymerases.
WRN Increases Primer Extension by DNA Polymerase δ.
We examined the ability of WRN to modulate the polymerization activity of several eukaryotic DNA polymerases. Polymerization activity was monitored by visualizing extension of a 5′-end labeled 14-mer primer on a 46-mer DNA template. Hybridization of the primer to the 3′ end of the template created a blunt-ended terminus that was resistant to the 3′→5′unwinding activity of WRN. Reactions contained limiting amounts of purified DNA polymerase α, δ, or ɛ such that less than 20% of the primer was extended to full length product.
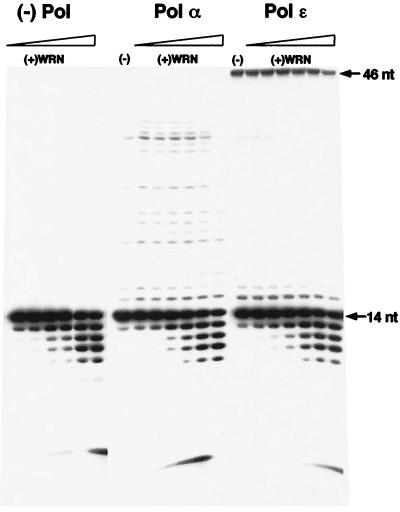
Under the assay conditions used, each polymerase exhibited a characteristic extension profile. Addition of increasing amounts of WRN to reactions containing human pol α or human pol ɛ did not significantly alter this profile, and neither did it increase the yield of full-length product (Fig. 1). In fact, at the highest concentration of WRN, a slight inhibition of extension was observed in some cases. Presumably, this is a result of competition between WRN and DNA polymerase for binding at the 3′-primer terminus, as reflected in the increasing amount of degradation products observed with increasing amounts of WRN. Thus, to a first approximation, WRN does not modulate the activities of DNA polymerases α or ɛ on a short synthetic oligonucleotide DNA substrate. Likewise, addition of varying concentrations of DNA polymerase α or ɛ had no effect on the helicase/exonuclease activities of WRN (data not shown; Fig. 1), again suggesting a lack of functional interaction.
Figure 1.
WRN does not increase primer extension by DNA polymerase α or ɛ. A 5′-end labeled 14-nt DNA primer was hybridized to a 46-nt DNA template. The primer (0.1 pmol) was extended by DNA polymerase α or ɛ in the absence or presence of increasing concentrations of WRN (1.2–60 fmol). The reactions were incubated at 37°C for 10 min and were terminated by the addition of an equal volume of denaturing loading buffer. Aliquots were electrophoresed through 14% polyacrylamide-urea gels, and extension products were visualized by autoradiography. The panel labeled “(−) Pol” is a control of increasing concentrations of WRN incubated with the primer/template in the absence of DNA polymerase to demonstrate the products of the 3′→5′ exonucleolytic activity of WRN.
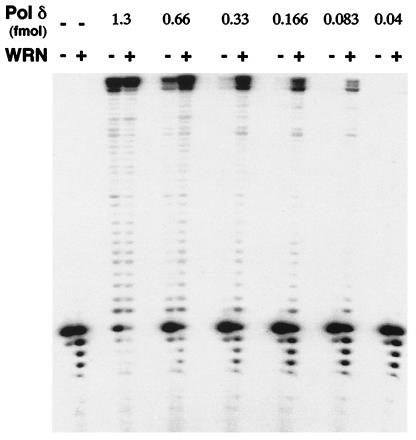
In contrast to the above findings, WRN had a dramatic effect on DNA synthesis by yeast pol δ (Fig. 2). Addition of WRN to reactions containing pol δ increased both the amount of primer extended and the yield of the full length 46-nt product. Notably, in the presence of higher concentrations of pol δ [as in reactions with either 0.66 or 1.3 fmol (Fig. 2)], exonucleolytic degradation, presumably catalyzed by WRN, was markedly diminished. Furthermore, the ability of WRN to increase the extent of DNA synthesis by pol δ was apparent even at concentrations of polymerase at which little or no primer was extended by pol δ alone (Fig. 2, lanes with 0.083 and 0.166 fmol pol δ). As also observed with pol α and pol ɛ, pol δ did not affect the helicase activity of WRN (not shown).
Figure 2.
WRN increases primer extension by DNA polymerase δ. Indicated amounts of pol δ were mixed with 0.1 pmol of the 14/46 primer/template in the absence or presence of a fixed concentration of WRN (≈6 fmol/10 μl reaction). Reactions were incubated at 37°C for 10 min and were processed as described in the legend to Fig. 1. Lanes: 1, substrate alone; 2, primer/template incubated with ≈6 fmol WRN in the absence of pol δ.
WRN Increases the Rate of Initiation of DNA Synthesis by Polymerase δ.
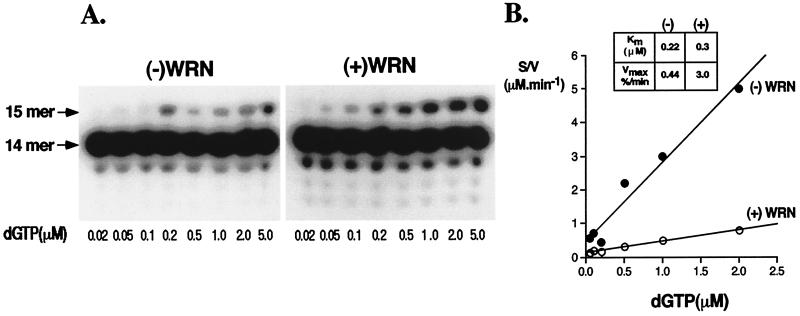
To determine whether WRN has any effect on incorporation of the first/initiating nucleotide by pol δ, we carried out a systematic kinetic analysis of extension by a single nucleotide in the absence or presence of WRN. Quantitation of the results presented in Fig. 3 A and B revealed that addition of WRN had an insignificant effect on altering the Km of pol δ for dGTP, 0.3 μM (+) WRN versus 0.22 μM (−) WRN. On the other hand, it had a more dramatic effect on the Vmax value, increasing the rate of incorporation of dGTP by ≈6-fold, 0.44% extension/min (−) WRN versus 3% extension/min (+) WRN.
Figure 3.
WRN stimulates the rate of incorporation of the initiating nucleotide by DNA pol δ. 0.5 pmol of the 14/46 primer/template was incubated with ≈0.3 fmol of pol δ in the absence (−) or presence (+) of 24 fmol of WRN. dGTP was included at concentrations ranging from 0.02–5 μM, and the reactions were incubated for either 5 min [(−)WRN] or 3 min [(+)WRN] at 37°C. The reactions were quenched by the addition of denaturing loading buffer and were electrophoresed through 14% polyacrylamide-urea gels as described in Materials and Methods. The extension products were visualized by autoradiography (A), and the amounts of 15-mer generated were quantitated by PhosphorImager analysis. The kinetic constants were derived from Hanes-Woolf plots of the data (B).
WRN Does Not Increase the Activity of DNA Polymerase δ Holoenzyme.
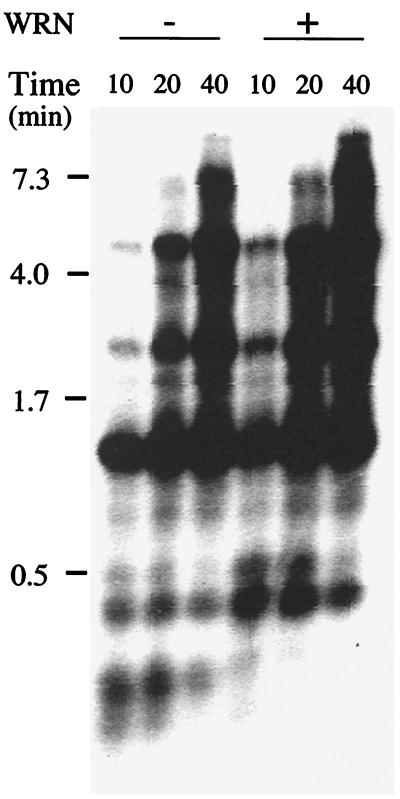
Efficient replication of natural DNA templates by pol δ requires the processivity factor, PCNA. To determine whether WRN has any effect on DNA synthesis by the pol δ-PCNA complex, we monitored replication of a singly primed natural DNA template. PCNA was loaded onto primed single-stranded mp18 DNA by replication factor C and ATP. The reaction was initiated by the addition of pol δ in the absence or presence of added WRN, and synthesis was followed over several minutes. As shown in Fig. 4, the pol δ-PCNA complex carried out processive DNA synthesis; progressively longer products accumulated over time, with full length product (7.25 kb) being generated after a 40-min incubation at 13°C. Notably, in contrast to the stimulatory effect of WRN on the oligonucleotide primer-template, WRN had a minimal incremental effect on the rate of accumulation and/or the amount of extension products synthesized by pol δ holoenzyme. The stimulatory effect of WRN can be attributed to displacement of template secondary structure by the helicase activity of WRN; no stimulation was observed when the reactions were carried out at 30°C, a temperature at which secondary structures are expected to be less stable (data not shown).
Figure 4.
WRN does not increase the activity of DNA polymerase δ holoenzyme. 0.125 pmol of singly primed single-stranded mp18 DNA was coated with E. coli single-stranded DNA binding protein and was loaded with PCNA (0.5 pmol) by replication factor C (0.125 pmol). DNA synthesis was initiated by the addition of pol δ (0.125 pmol) in the absence or presence of WRN (0.25 pmol). Reactions were incubated at 13°C; aliquots (12 μl) were removed at 10, 20, and 40 min after incubation and were electrophoresed through alkaline agarose gels. Extension products were visualized by autoradiography of the dried gel. Positions of migration of size markers, expressed in kilobases, are indicated on the left.
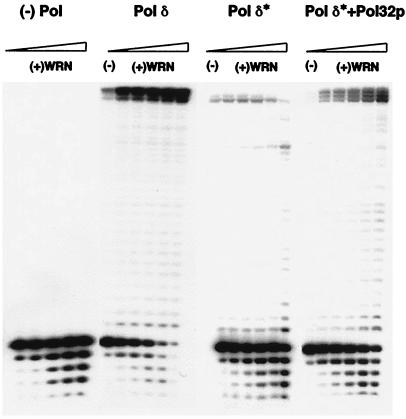
The Interaction of WRN with Yeast pol δ Requires the Third Subunit, Pol32p.
Yeast pol δ is comprised of three subunits, designated Pol3p (125 kDa), Pol31p (58 kDa), and Pol32p (55 kDa) (41). The former two subunits, corresponding to the p125 and p48/p50 subunits, respectively, of mammalian pol δ, form a heterodimeric complex called pol δ*. The third subunit, Pol32p, forms a complex with pol δ* and induces dimerization of the heterotrimer. In addition, through its interaction with PCNA, Pol32p increases the processivity of pol δ relative to that of pol δ* (41, 43).
To determine which subunit of pol δ is necessary for the functional interaction with WRN, we compared the ability of WRN to modulate the activity of the two-subunit and three-subunit enzyme complexes. Concentrations of pol δ* and pol δ that yielded comparable activities, measured as percent of primer extended, were used. As presented in Fig. 5, the extension profile of pol δ* is distinct from that of pol δ; the predominant product of synthesis was 44 nt instead of 46 nt, the full-length product generated by pol δ. However, whereas the addition of WRN had a dramatic effect on primer extension by pol δ, it had little or no effect on the extent of DNA synthesized by pol δ*. Even at the maximal concentration of WRN, there was no increase in full length product. In fact, there was a decrease in the yield of the 44-mer and an increase in the amount of some of the smaller extension products. This inhibition of extension is attributable to interference from a component of the WRN storage buffer (data not shown). The lack of a functional interaction between pol δ* and WRN is also evidenced by the more extensive exonucleolytic degradation, presumably catalyzed by WRN, in reactions with pol δ* versus those with pol δ.
Figure 5.
Functional interaction with WRN requires the Pol32p subunit of S. cerevisiae DNA pol δ. Pol δ (≈0.3 fmol), pol δ* (≈0.9 fmol), or in vitro reconstituted pol δ [Pol δ*+Pol32p] (≈0.6 fmol)] were incubated with the 14/46 primer/template such that comparable amounts of primer were extended in each case. Each preparation of polymerase was incubated with increasing amounts of WRN (2.4–60 fmol); assays were carried out and processed as described in the legend to Fig. 1.
To further demonstrate that the Pol32p subunit mediates the functional interaction with WRN, we reconstituted the three-subunit complex in vitro with pol δ* and Pol32p. The reconstituted enzyme generated an extension profile slightly different from that exhibited by pol δ purified from yeast cells co-expressing all three subunits (Fig. 5, Pol δ*+Pol32p lanes). Rather than synthesizing the 46-mer as the predominant product, the reconstituted pol δ enzyme generated 44-, 45-, and 46-nt-long products in equivalent amounts. However, in contrast to the results with pol δ*, addition of WRN increased the yield of this triplet product, with the highest amount of WRN preferentially elevating the amount of the 44-mer and 45-mer. Thus, the addition of Pol32p to pol δ* restored the functional interaction with WRN. Taken together, the foregoing results suggest that the Pol32p subunit is required for the stimulatory effect of WRN.
Discussion
We demonstrate in this report that there is a functional interaction between the WS protein, WRN, and DNA polymerase δ, a key enzyme in DNA replication and DNA repair (39, 40). By using limiting concentrations of three major eukaryotic replicative DNA polymerases, polymerases α, δ, and ɛ, we show that WRN dramatically and selectively stimulates the polymerization activity of DNA polymerase δ (compare Figs. 1 and Fig. 2). WRN does not affect DNA synthesis carried out by eukaryotic DNA polymerase β, the Klenow fragment of E. coli DNA polymerase I, murine Moloney leukemia viral reverse transcriptase, or the thermostable Thermus aquaticus (Taq) DNA polymerase (data not shown), suggesting that the functional association of WRN with DNA polymerases may be limited to pol δ.
By using an oligonucleotide primer-template substrate, we show that WRN increases the amount of primer extended by pol δ and the yield of full length product (Fig. 2). Assuming the functional unit of WRN, like that of the closely related DNA helicase, BLM, is predominately a hexamer (46), stimulation of polymerization activity is observed at molar ratios of pol δ:WRN ranging from 1:1 to 1:30. The increase in the turnover number of pol δ can be accounted for, at least in part, by the ability of WRN to stimulate (6-fold higher Vmax) the rate of incorporation of the first nucleotide (Fig. 3B). Analysis of the Km values of pol δ for dGTP, however, indicates that WRN does not increase the affinity of pol δ for the initiating nucleotide.
Data presented in Figs. 2 and 3 suggest that the helicase activity of WRN may be dispensable for stimulation of pol δ activity, at least under our assay conditions. First, the increase in primer extension, particularly with a single dNTP (Fig. 3), is observed in the absence of ATP/dATP, the nucleotide co-factor essential for helicase activity of WRN. dGTP concentrations used in the kinetic studies (0.02–5 μM) do not support unwinding by WRN (22) (our unpublished results). Second, the oligonucleotide primer/template lacks a 3′ tail, required for WRN helicase activity (19, 21). In addition, because the rate of polymerization exceeds the rate of exonucleolytic degradation of primer DNA, the exonuclease activity of WRN may also be dispensable for stimulation of pol δ activity. Notably, however, it is possible that the helicase and/or exonuclease activities of WRN are used in vivo to create a substrate for concomitant WRN-stimulated nucleotide incorporation by pol δ.
In contrast to the stimulatory effect of WRN on polymerization activity of pol δ in the absence of PCNA, WRN does not significantly increase the activity of pol δ holoenzyme, i.e., pol δ-PCNA complex, on natural DNA (Fig. 4). These results suggest that WRN may not function in normal, processive DNA synthesis reactions in which the pol δ-PCNA complex is required. Instead, our current hypothesis is that WRN, like RecQ, may function in replication restart at forks blocked by DNA damage, or stalled/collapsed by unusual secondary structures from which the normal replication machinery (including pol δ and PCNA) have dissociated. Our data suggest that an alternative DNA synthetic system involving a PCNA-independent activity of pol δ in combination with WRN may rescue replication. Several alternate DNA replication systems have been identified in E. coli (47). Most recently, work from the Leach laboratory reports that replicative bypass of secondary structures (hairpins) can occur by a RecQ helicase-dependent pathway (48). This is consistent with the studies of Courcelle and Hanawalt (49) that demonstrate that RecQ and RecJ are required to process nascent DNA at blocked replication forks before replication can resume.
We have also established that Pol32p, the third subunit of S. cerevisiae pol δ, is required for WRN-mediated stimulation of DNA synthesis. Two independent lines of evidence support this conclusion (Fig. 5). First, the two-subunit complex lacking Pol32p (pol δ*) is refractory to the stimulatory effect of WRN, and, second, reconstituting pol δ from purified pol δ* and Pol32p subunits restores stimulation. Consistent with the hypothesis that the third subunit of pol δ is necessary to mediate the functional interaction with WRN, we have observed that WRN yields variable stimulation of the activity of purified human and calf thymus pol δ (data not shown). Mammalian pol δ has been purified from tissue as a two-subunit complex lacking the Pol32p homolog (50, 51). The enzyme preparation-dependent variability in stimulation by WRN could arise from a variation in the amount of this subunit that co-purifies with the two-subunit form. Hughes et al. (42) have recently cloned and sequenced the human subunit, p66, that corresponds to the S. cerevisiae Pol32p subunit (41) and the S. pombe Cdc27 subunit (52). Once this subunit can be expressed and co-purified with the two-subunit enzyme, it will be feasible to directly test our hypothesis with human pol δ.
A growing literature implicates the RecQ family of DNA helicases in DNA replication. Courcelle and Hanawalt (49) have shown that E. coli RecQ is required to process blocked replication forks before resumption of replication in UV-irradiated cells. Lee et al. (53) have recently demonstrated that SGS1 is required for DNA replication in S. cerevisiae. In the fission yeast S. pombe, Rqh1p is necessary for recovery from hydroxy urea-induced S-phase arrest, suggesting that it may facilitate replication reinitiation after DNA damage (9, 54, 55). Analyses of WS and Bloom's syndrome cells also suggest the involvement of WRN and BLM in DNA replication. WS cells exhibit a reduced rate of DNA replication, a prolonged S-phase, and a decreased frequency of replication initiation (32, 33, 56, 57). Bloom's syndrome cells show retarded replication fork progression, accumulation of abnormal replication intermediates, and an elevated rate of sister-chromatid exchanges arising in S-phase in response to 5′-bromodeoxyuridine (58). In addition, both WS and Bloom's syndrome cells are sensitive to the S-phase specific topoisomerase I inhibitor, camptothecin (58, 59). Furthermore, the X. laevis WRN homolog, FFA-1, has been shown to be present in replication foci (10). The demonstration of a functional interaction between a replicative DNA polymerase, pol δ, and WRN may thus provide a molecular link toward understanding the basis of some of the S-phase defects of WS cells. For example, during DNA replication, the replication fork can encounter DNA lesions and/or secondary structure. Analogous to the situation in E. coli, replication fork progression would arrest, and the replisome containing DNA pol δ would disassemble. If WRN were involved in replication restart under these conditions, then its absence in WS cells (60) may interfere with the ability of pol δ to resume replication and delay S-phase progression.
DNA polymerase δ also functions in cellular DNA repair pathways (40). Thus, the functional interaction between pol δ and WRN may be restricted in vivo to specific repair processes. The selective sensitivity of WS cells to camptothecin (59) and 4-nitroquinoline-1-oxide (34) implicates WRN in the repair/resolution of DNA damaged by these agents. Further, the ability of WRN to unwind G4 tetraplex DNA (24) in vitro suggests that WRN may be able to process unusual structures that arise during replication/recombination. Therefore, it is conceivable that the helicase/exonuclease activities of WRN are required to first process DNA damage and/or alternate DNA structures before pol δ can initiate repair synthesis, the rate of which could also be markedly regulated by WRN.
Mutations in a single gene, WRN, can result in a wide spectrum of physiological and pathological alterations that characterize aging. Our finding that WRN functionally associates with pol δ suggests that subtle mutations in pol δ might also be associated with aging. Furthermore, polymorphic differences in WRN and pol δ may contribute to differences in aging among various populations and families. Considering the central role of pol δ in a number of DNA transactions, it will also be important to analyze spontaneous tumors of the types observed in WS patients (e.g., osteosarcomas) for mutations in pol δ and to study their effects on genetic stability. Of particular interest would be mutations in the human p66 pol δ subunit.
Acknowledgments
We thank Ann Blank for critical reading of the manuscript, Jue Zhang for assistance with graphics, and members of our laboratories for useful discussions. This work was supported by National Cancer Institute Grant CA77852 and National Institutes of Health Grant AG01751 to L.A.L., National Institutes of Health Grant GM58534 to P.M.J.B., and by a fellowship from Cancerfonden to E.J.
Abbreviations
- WS
Werner Syndrome
- pol δ
DNA polymerase δ
- PCNA
proliferating cell nuclear antigen
References
- 1.Epstein C J, Martin G M, Schultz A L, Motulsky A G. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Goto M, Miller R W, Ishikawa Y, Sugano H. Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 3.Salk D, Au K, Hoehn H, Martin G M. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 4.Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht K W, Jonas J B. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 5.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 8.Watt P M, Hickson I D, Borts R H, Louis E J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, Chen C Y, Kobayashi R, Newport J. Nat Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- 11.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 12.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 13.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 14.Kitao S, Shimamoto A, Goto M, Miller R W, Smithson W A, Lindor N M, Furuichi Y. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 15.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Nature (London) 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 17.Bennett R J, Sharp J A, Wang J C. J Biol Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 18.Tada S, Yanagisawa J, Sonoyama T, Miyajima A, Seki M, Ui M, Enomoto T. Cell Struct Funct. 1996;21:123–132. doi: 10.1247/csf.21.123. [DOI] [PubMed] [Google Scholar]
- 19.Gray M D, Shen J-C, Kamath-Loeb A S, Blank A, Martin G M, Oshima J, Loeb L A. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 20.Karow J D, Chakraverty R K, Hickson I D. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J-C, Gray M D, Oshima J, Loeb L A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosh R M, Jr, Orren D K, Nehlin J O, Ravn P H, Kenny M K, Machwe A, Bohr V A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 24.Fry M, Loeb L A. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Li B, Gray M D, Oshima J, Mian I S, Campisi J. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J-C, Gray M D, Oshima J, Kamath-Loeb A S, Fry M, Loeb L A. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 27.Kamath-Loeb A S, Shen J-C, Loeb L A, Fry M. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki N, Shiratori M, Goto M, Furuichi Y. Nucleic Acids Res. 1999;27:2361–2368. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng R Z, Murano S, Kurz B, Shmookler-Reis R J. Mutat Res. 1990;237:259–269. doi: 10.1016/0921-8734(90)90008-f. [DOI] [PubMed] [Google Scholar]
- 31.Stefanini M, Scappaticci S, Lagomarsini P, Borroni G, Beradesca E, Nuzzo F. Mutat Res. 1989;219:179–185. doi: 10.1016/0921-8734(89)90013-1. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi F, Hanaoka F, Goto M, Akaoka I, Hori T, Yamada M, Miyamoto T. Hum Genet. 1982;60:365–368. doi: 10.1007/BF00569220. [DOI] [PubMed] [Google Scholar]
- 33.Fujiwara Y, Higashikawa T, Tatsumi M. J Cell Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- 34.Ogburn C E, Oshima J, Poot M, Chen R, Hunt K E, Gollahon K A, Rabinovitch P S, Martin G M. Hum Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 35.Balajee A S, Machwe A, May A, Gray M D, Oshima J, Martin G M, Nehlin J O, Brosh R, Orren D K, Bohr V A. Mol Biol Cell. 1999;10:2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spillare E A, Robles A I, Wang X W, Shen J C, Yu C E, Schellenberg G D, Harris C C. Genes Dev. 1999;13:1355–1360. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blander G, Kipnis J, Leal J F, Yu C E, Schellenberg G D, Oren M. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 38.Lebel M, Spillare E A, Harris C C, Leder P. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 39.Hindges R, Hubscher U. Biol Chem. 1997;378:345–362. doi: 10.1515/bchm.1997.378.5.345. [DOI] [PubMed] [Google Scholar]
- 40.Burgers P M. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 41.Gerik K J, Li X, Pautz A, Burgers P M. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 42.Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G. Nucleic Acids Res. 1999;27:2108–2114. doi: 10.1093/nar/27.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgers P M, Gerik K J. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Reference Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 45.Boosalis M S, Petruska J, Goodman M F. J Biol Chem. 1987;262:14689–14699. [PubMed] [Google Scholar]
- 46.Karow J K, Newman R H, Freemont P S, Hickson I D. Curr Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 47.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cromie G A, Millar C B, Schmidt K H, Leach D R. Genetics. 2000;154:513–522. doi: 10.1093/genetics/154.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y, Zhang S J, Wu S M, Lee M Y. Arch Biochem Biophys. 1995;320:297–304. doi: 10.1016/0003-9861(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J Q, He H, Tan C K, Downey K M, So A G. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo S, Gibbs E, Kelman Z, Wang T S, O'Donnell M, MacNeill S A, Hurwitz J. Proc Natl Acad Sci USA. 1997;94:11244–11249. doi: 10.1073/pnas.94.21.11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S K, Johnson R E, Yu S L, Prakash L, Prakash S. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 54.Murray J M, Lindsay H D, Munday C A, Carr A M. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davey S, Han C S, Ramer S A, Klassen J C, Jacobson A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Exp Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- 57.Poot M, Hoehn H, Runger T M, Martin G M. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 58.Chakraverty R K, Hickson I D. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 59.Poot M, Gollahon K A, Rabinovitch P S. Hum Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- 60.Moser M J, Kamath-Loeb A S, Jacob J E, Bennett S E, Oshima J, Monnat R J., Jr Nucleic Acids Res. 2000;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]