Abstract
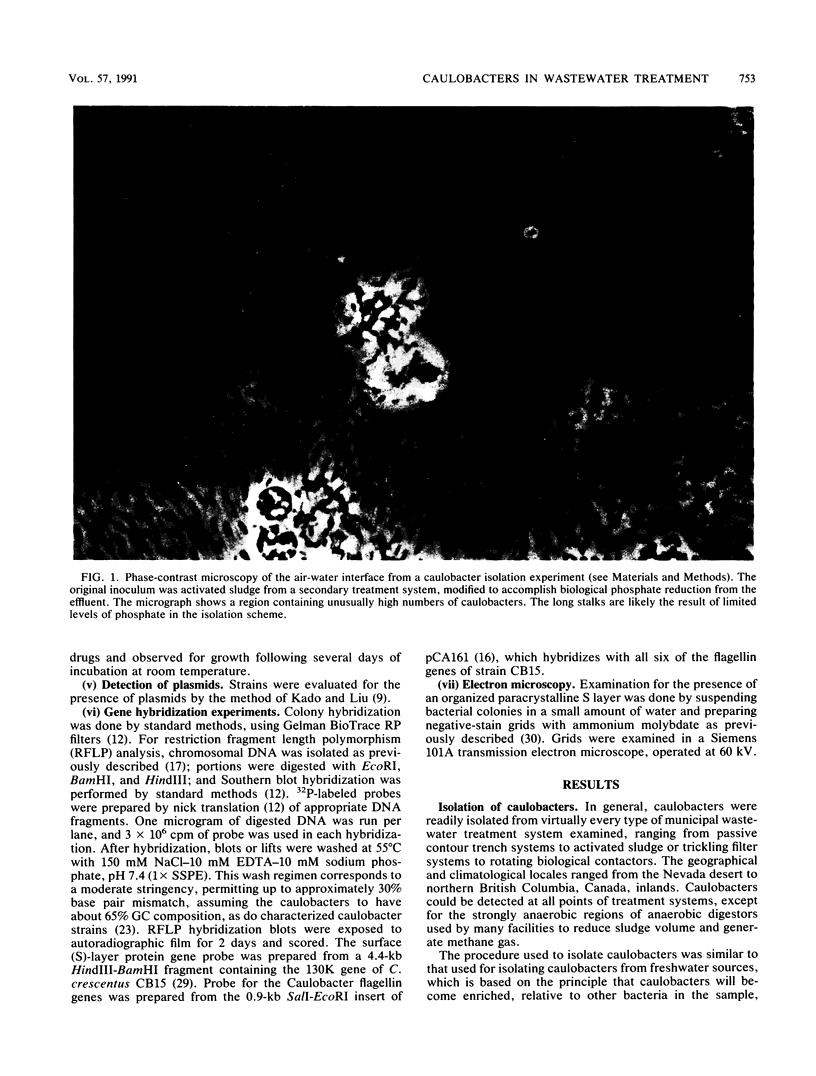
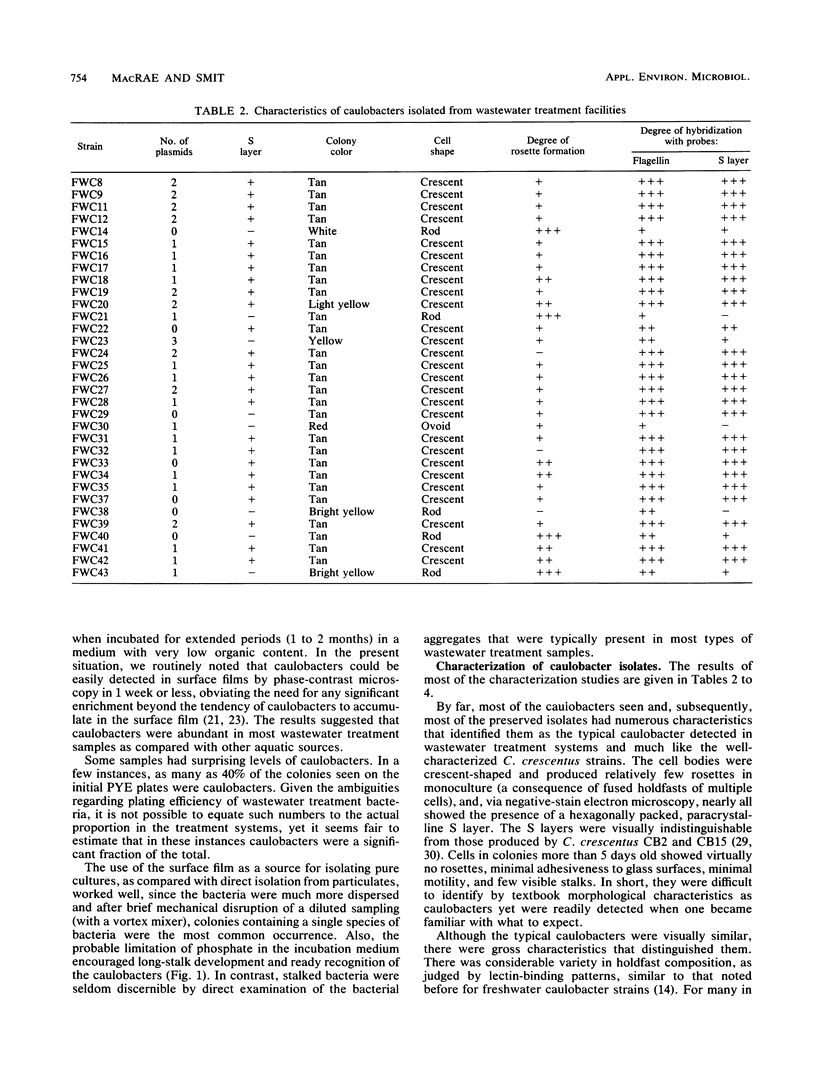
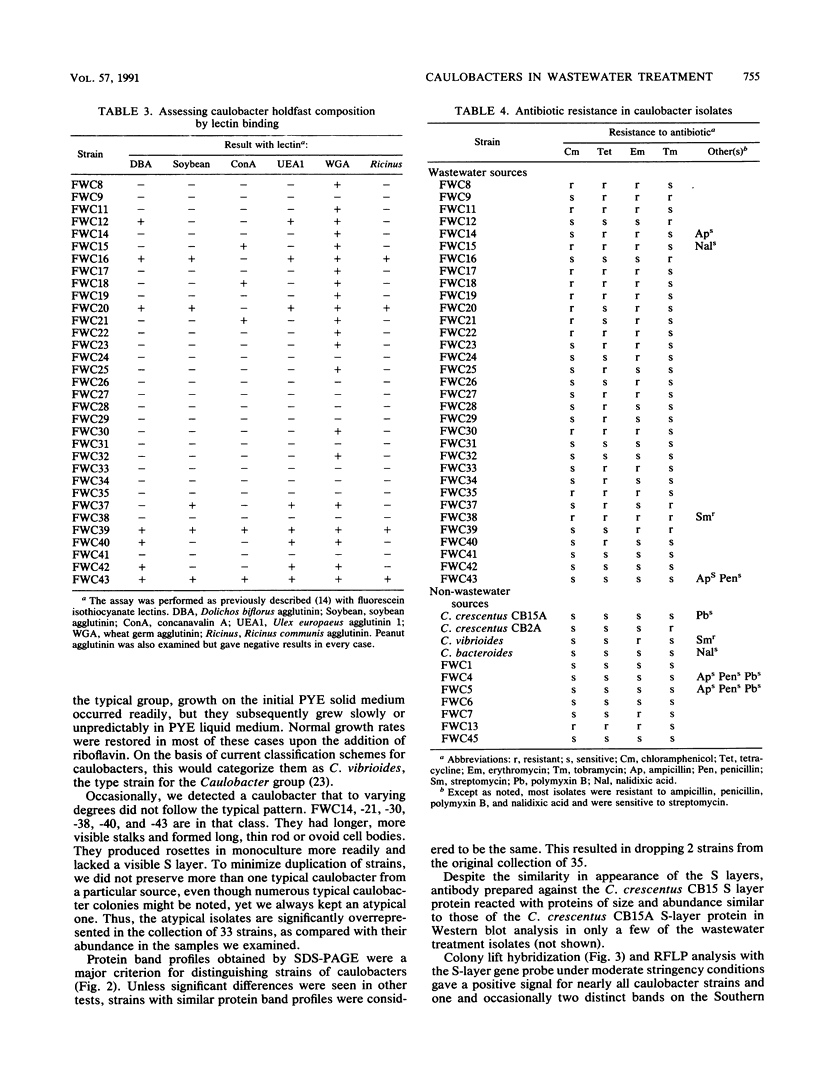
Caulobacters are generally assumed to be found only in environments of low organic content; however, we readily isolated strains from a variety of sewage treatment system designs and locations, and 33 distinct strains were characterized. Most were morphologically similar, having the crescent-shaped cell body, short stalk, and hexagonally packed, paracrystalline surface (S) layer characteristic of several Caulobacter crescentus laboratory strains. Upon closer examination, they were distinguishable on the basis of protein band profiles on polyacrylamide gel electrophoresis, gross colony characteristics, or holdfast composition or by DNA restriction fragment length polymorphism analysis with flagellin and S-layer gene probes. Most of the isolates contained one or more high-molecular-weight plasmids and were resistant to a number of antibiotics, characteristics generally not shared with caulobacters isolated from other sources. Six of the 33 strains were retained because they did not fit the typical isolate profile; these strains are overrepresented in our collection compared with their relative proportion in wastewater treatment systems. By colony hybridization and restriction fragment length polymorphism analysis, all of these and one typical isolate showed less homology than the others to the surface array gene of a laboratory strain (C. crescentus CB15), and three hybridized less strongly with the flagellin gene from the same strain. In sum, although the strains were distinguishable, caulobacters from the wastewater treatment systems we examined were relatively homogenous, were similar to characterized laboratory strains, and, with exceptions, could probably be reliably detected as a group by gene probes derived from C. crescentus strains.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anast Nick, Smit John. Isolation and Characterization of Marine Caulobacters and Assessment of Their Potential for Genetic Experimentation. Appl Environ Microbiol. 1988 Mar;54(3):809–817. doi: 10.1128/aem.54.3.809-817.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- DIAS F. F., BHAT J. V. MICROBIAL ECOLOGY OF ACTIVATED SLUDGE. I. DOMINANT BACTERIA. Appl Microbiol. 1964 Sep;12:412–417. doi: 10.1128/am.12.5.412-417.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. E., Carr N. Examination of Genetic Relatedness of Marine Synechococcus spp. by Using Restriction Fragment Length Polymorphisms. Appl Environ Microbiol. 1988 Dec;54(12):3071–3078. doi: 10.1128/aem.54.12.3071-3078.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M. How do bacteria attach to solid surfaces? Microbiol Sci. 1987 May;4(5):133–136. [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Merker R. I., Smit J. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol. 1988 Aug;54(8):2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen M., Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983 Apr 14;302(5909):630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Mitchell D., Smit J. Identification of genes affecting production of the adhesion organelle of Caulobacter crescentus CB2. J Bacteriol. 1990 Sep;172(9):5425–5431. doi: 10.1128/jb.172.9.5425-5431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike E. B., Carrington E. G., Ashburner P. A. An evaluation of procedures for enumerating bacteria in activated sludge. J Appl Bacteriol. 1972 Jun;35(2):309–321. doi: 10.1111/j.1365-2672.1972.tb03703.x. [DOI] [PubMed] [Google Scholar]
- Poindexter J. S. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev. 1981 Mar;45(1):123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasam T. B., Dondero N. C. Aerobic heterotrophic bacterial populations of sewage and activated sludge. II. Method of characterization of activated sludge bacteria. Appl Microbiol. 1967 Sep;15(5):1122–1127. doi: 10.1128/am.15.5.1122-1127.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SKINNER F. A., JONES P. C., MOLLISON J. E. A comparison of a direct- and a plate counting technique for the quantitative estimation of soil micro-organisms. J Gen Microbiol. 1952 May;6(3-4):261–271. doi: 10.1099/00221287-6-3-4-261. [DOI] [PubMed] [Google Scholar]
- Schmidt J. M., Stanier R. Y. The development of cellular stalks in bacteria. J Cell Biol. 1966 Mar;28(3):423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenlein P. V., Ely B. Plasmids and bacteriocins in Caulobacter species. J Bacteriol. 1983 Feb;153(2):1092–1094. doi: 10.1128/jb.153.2.1092-1094.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J Cell Biol. 1982 Oct;95(1):41–49. doi: 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J Bacteriol. 1984 Dec;160(3):1137–1145. doi: 10.1128/jb.160.3.1137-1145.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Grano D. A., Glaeser R. M., Agabian N. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J Bacteriol. 1981 Jun;146(3):1135–1150. doi: 10.1128/jb.146.3.1135-1150.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D., Lagenaur C., Agabian N. Isolation and characterization of Caulobacter crecentus bacteriophage phi Cd1. J Virol. 1976 Feb;17(2):568–575. doi: 10.1128/jvi.17.2.568-575.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoman S., Stephenson T., Lester J. N., Perry R. Biotechnology for phosphorus removal during wastewater treatment. Biotechnol Adv. 1986;4(1):13–26. doi: 10.1016/0734-9750(86)90003-0. [DOI] [PubMed] [Google Scholar]






