Abstract
The goal of this study was to examine the efficacy of liver-targeted gene delivery by chitosan-DNA nanoparticles through retrograde intrabiliary infusion (RII). The transfection efficiency of chitosan-DNA nanoparticles, as compared with PEI-DNA nanoparticles or naked DNA, was evaluated in Wistar rats by infusion into the common bile duct, portal vein, or tail vein. Chitosan-DNA nanoparticles administrated through the portal vein or tail vein did not produce detectable luciferase expression. In contrast, rats that received chitosan-DNA nanoparticles showed more than 500 times higher luciferase expression in the liver 3 days after RII; and transgene expression levels decreased gradually over 14 days. Luciferase expression in the kidney, lung, spleen, and heart was negligible compared with that in the liver. RII of chitosan-DNA nanoparticles did not yield significant toxicity and damage to the liver and biliary tree as evidenced by liver function analysis and histopathological examination. Luciferase expression by RII of PEI-DNA nanoparticles was 17-fold lower than that of chitosan-DNA nanoparticles on day 3, but it increased slightly over time. These results suggest that RII is a promising routine to achieve liver-targeted gene delivery by non-viral nanoparticles; and both gene carrier characteristics and mode of administration significantly influence gene delivery efficiency.
Keywords: nanoparticles, gene delivery, liver-targeted, chitosan, retrograde intrabiliary infusion
Introduction
The liver represents one of the most important targets for therapeutic gene delivery because of the ready access of the transgene product to systemic circulation, and its susceptibility to many metabolic genetic disorders, viral infection and malignancies (Kren et al 2002; Prieto et al 2003; Nguyen and Ferry 2004). A critical barrier to realizing the potential of liver-targeted gene transfer is the development of a safe and efficient gene carrier in combination with a feasible and efficient administration route. Intravenous injection of naked plasmid DNA has showed no gene expression in the liver where the highest uptake is observed (Mahato et al 1995). High level of gene expression has been achievable in mouse and rat only by rapid injection of naked DNA in a large volume (∼8% body weight) (Liu et al 1999; Zhang et al 1999; Murakami et al 2001; Higuchi et al 2003). The concern is that such a procedure may cause significant mechanical damage, altering the physiological condition of the liver. Reducing the injection rate and volume to a physiologically acceptable range has been shown to abolish gene expression by lysosomal hydrolysis of the delivered DNA (Lecocq et al 2003). These factors highlight the need for a gene carrier that can efficiently protect DNA from degradation, facilitate its transport to the liver, and enhance its uptake by liver parenchymal cells (Nguyen and Ferry 2004).
Several gene carriers/vectors have been studied for liver-targeted gene delivery, including viral and non-viral vectors. Viral vectors, particularly adenovirus (Cristiano et al 1993; Nathwani et al 2002) and lentivirus (Cheung et al 2002), have demonstrated a high level of transgene expression. Viral vectors infect a wide variety of cell types in vivo (Jooss and Chirmule 2003); therefore, liver-targeted gene delivery can be achieved only by localized injection through the hepatic artery or portal vein (Nathwani et al 2002) or by conjugating the vectors to a polymer with hepatocyte-specific ligand (Cristiano et al 1993). However, significant concerns exist about clinical applications of viral vectors (Treco and Selden 1995; Cichon et al 1999) such as host immune response against the vectors (Jooss and Chirmule 2003), intrinsic toxicity of the viral proteins, and limited packaging size.
As an alternative, several non-viral carriers have been evaluated for liver-targeted gene delivery (Li and Huang 1997; Niidome and Huang 2002; Wu et al 2002). Liposomes and cationic polymers are the two most widely studied non-viral carriers. Ligand-conjugated lipoplexes (Kawakami et al 2002) and polymer-DNA complexes (Rogers et al 2000) have been constructed to achieve active targeting and enhance hepatocyte uptake through receptor-mediated endocytosis (Wu et al 2002). A significant problem of these lipoplexes and polyplexes for liver-targeted gene delivery is that these complexes aggregate in serum following intravenous injection. These aggregates then become trapped in lung vasculature (Li and Huang 1997) and are scavenged by macrophages. Even when injected through a more direct route, eg, intraportal injection, these complexes mediated higher gene expression in the lung than in the liver (Li and Huang 1997; Otsuka et al 2000; Zhang et al 2001).
Retrograde intrabiliary infusion (RII) has recently been explored as a liver-targeted gene delivery route (Uehara et al 1999; Otsuka et al 2000; Zhang et al 2001, 2003; Tominaga et al 2004). RII provides a direct delivery to parenchymal hepatocytes and avoids first contact with Kupffer cells (KCs). The large surface area and broadly distributed biliary system provides great access to nearly all the hepatocytes in liver parenchyma (Ludwig et al 1998). Because of these obvious advantages, delivery of gene vectors (adenovirus, retrovirus, lipoplexes, and polyplexes) to the liver through RII has been investigated in rats (Yang et al 1993; Terao et al 1998; De Godoy et al 1999; Uehara et al 1999; Kuhel et al 2000; Otsuka et al 2000; Tominaga et al 2004; Zhang et al 1997, 2001, 2003; Chen et al 2005), mice (Zhang et al 1997), pigs (Yang et al 1993; Otsuka et al 2000), and primates (Sullivan et al 1997). For example, Zhang et al (2001) showed that poly-L-lysine-molossin-DNA complexes (molossin is a polypeptide containing a 15-amino-acid integrin-binding domain), when directly infused to the portal vein of the rat liver, mediated a low level of transgene expressions, even with chloroquine-facilitated endolysosomal release of the complexes (Zhang et al 2003) On the other hand, the same complexes yielded about a 10-fold higher transgene expression when administered through RII. This was correlated with the prolonged presence of DNA in the liver after RII compared with intraportal infusion, where DNA was lost rapidly from the liver. Otsuka et al (2000) found that RII of multilamellar liposomes encapsulating DNA (0.3 mL/min) produced a 100-fold higher transgene expression than that given by intraportal vein infusion, which persisted for at least 6 days. These results suggest that RII is a promising administration route for liver-targeted gene delivery using non-viral vectors.
We have previously developed a nanoparticle gene delivery system based on a natural polymer, chitosan. Chitosan is a biodegradable and biocompatible polysaccharide derived from crustacean shells. Chitosan possesses several favorable characteristics for drug and gene delivery. Chitosan is soluble in acid solutions (pH<5.5) and can form complexes with anionic macromolecules to yield nanoparticles, microparticles, hydrogels, foams, and fibers (Borchard 2001; Liu and Yao 2002). In particular, we have prepared chitosan-DNA nanoparticles ranging from 150 to 300 nm in size. In addition, most of the positive charges in the chitosan polymer chain would be neutralized at physiological pH (pKa of the sidechain amino groups is 6.5), rendering chitosan molecule hydrophobic and less water soluble. This unique property ensures that chitosan-DNA nanoparticles formed at low pH remain stable at the physiological pH without chemical cross-linking. Its mucoadhesive property and ability to enhance transepithelial transport of macromolecules across tight junctions make chitosan a natural carrier for mucosal delivery (Schipper et al 1997; van der Merwe et al 2004; Prego et al 2005). We have shown in an oral DNA vaccine model that oral feeding of chitosan-DNA nanoparticles containing pCMVArah2 DNA encoding a major peanut allergen Arah2, elicited significant levels of secretory IgA and Th1 type T cell response (serum IgG2a) against Arah2 at week 4, while oral immunization with naked DNA failed (Roy et al 1999). This Arah2-specific immune response could protect the animals from challenge (anaphylactic). Besides the peanut allergy model, we have also demonstrated the efficiency of chitosan-DNA nanoparticles in an oral gene vaccine for dust mite allergen encoding Der p1 gene (Chew et al 2003) and in delivering Factor VIII gene orally (Bowman et al 2004). Successful transgene delivery has also been reported for intranasal and pulmonary administrations of chitosan-DNA nanoparticles (Koping-Hoggard et al 2001; Okamoto et al 2003; Zhang et al 2005).
This study examined the potential of liver-targeted gene delivery mediated by intrabiliary infusion of polymer-DNA nanoparticles. The gene delivery efficiency, as well as the toxicity and damage to the liver and biliary tree, of chitosan-DNA nanoparticles administered through RII, intraportal infusion or tail vein injection were characterized in comparison with PEI-DNA complexes and naked DNA.
Materials and methods
Reagents
Polyethylenimine (branched PEI, average molecular weight of 25 kDa), was purchased from Sigma-Aldrich (St Louis, MO, USA). PEI was purified by dialysis against water (dialysis tubing with MWCO 3500; Pierce, Rockford, IL, USA) for 3 days and lyophilization. Chitosan C390 (MW 390 kDa, deacetylation degree 83.5%) was kindly provided by Vanson Halosource (Redmond, WA, USA). Chitosan solution was prepared in 5 mM sodium acetate (NaAc) buffer at a concentration of 0.02 % (w/v). The pH of chitosan solution was adjusted to 5.5 and the solution was sterile-filtered through a 0.22 μm filter (Millipore, Billerica, MA, USA).
Amplification of plasmid DNA
Plasmid pVR1255 is a 6,413b pcDNA encoding luciferase driven by human CMV promoter (a gift from Dr Carl J Wheeler, Vical Inc., San Diego, CA, USA). The plasmid was amplified in Escherichia coli DH5α and purified by Qiagen Giga plasmid purification kit (endotoxin free, Qiagen, Hilden, Germany). Purified plasmids were dissolved in saline and kept in aliquots at a concentration of 1 to 2 mg/mL. Terrific Broth and ampicillin were purchased from Gibco BRL (Grand Island, NY, USA).
Preparation of chitosan-DNA and PEI-DNA nanoparticles
A chitosan solution (0.02% in 5 mM sodium acetate buffer, pH 5.5) and a DNA solution (100 μg/mL in 5 mM of NaAc solution) at an N/P ratio of 3 (N/P = molar ratio of chitosan nitrogen to DNA phosphate) were preheated to 50–55°C separately. An equal volume of both solutions were quickly mixed while vortexing. PEI-DNA nanoparticles were prepared by mixing PEI solution (133 μg/ml in 5% glucose) with equal volume of DNA solution (100 μg/ml in 5% glucose) at an N/P ratio of 10. Both chitosan-DNA and PEI-DNA nanoparticles were incubated at room temperature for 30 min before characterization and transfection. The particle size and zeta potential of chitosan-DNA and PEI-DNA nanoparticles were measured on a Zetasizer® 3000 (Malvern Instruments, UK).
Bile degradation
Bile was isolated from rat common bile ducts over 30- to 60-min periods and frozen at −80°C within a few hours. It was added to chitosan-DNA nanoparticles, PEI-DNA nanoparticles, and naked DNA solutions to reach the final concentrations of 10% and 50% (v/v), respectively. The solutions were then incubated at 37°C. At different time points, aliquots of samples were taken, quenched at 4°C before analysis by electrophoresis on a 1% agarose gel and ethidium bromide staining for visualization.
To investigate the degradation mechanism of bile, an aliquot of rat bile sample was pretreated by heating in a boiling water bath for 15 min and cooled on ice. It was added to chitosan-DNA nanoparticles and naked DNA solutions at a final concentration of 10%. In parallel, aliquots of chitosan-DNA nanoparticles and naked DNA were incubated with 10% untreated bile, 10% untreated bile with 50 mM of NAC, or 10 mM of hydrogen peroxide. The samples were incubated at 37°C. At 3-hour and 5-day time points, aliquots of samples were quenched at 4°C before analyzed by electrophoresis on a 1% agarose gel and ethidium bromide staining for visualization.
Animals
Six- to eight-week-old male Wistar rats were obtained and housed in National University of Singapore Animal Holding Unit. Rats were maintained on ad libitum rodent feed and water at room temperature, 40% humidity. All animal procedures were approved by the National University of Singapore Faculty of Medicine Animal Care and Use Committee.
Isolation and culture of hepatocytes, Kupffer cells, and normal rat cholangiocytes (NRC)
Hepatocytes were harvested from male Wistar rats weighting from 200 to 250 g by a two-step in situ collagenase perfusion as described previously (Zhang et al 2001). Hepatocytes were collected by centrifugation twice at 50×g for 3 min. Hepatocytes (pellet) were seeded on collagen coated substrates, and maintained in William’s E medium supplemented with 1 mg/mL BSA, 10 ng/mL of EGF, 0.5 μg/mL of insulin, 5 nM dexamethasone, 50 ng/mL linoleic acid, 100 U/mL penicillin, and 100 μg/mL streptomycin. The non-parenchymal cell fraction in the supernatant was washed with buffer and centrifuged at 650×g for 7 min at 4°C. Cell pellets were centrifuged on a density cushion of Percoll (25% and 50%) at 2500×g for 15 min at 4°C. The KCs fraction was then collected and seeded in a tissue culture flask, and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cell viability was determined using the Trypan Blue exclusion method. All cell culture media and supplements were from Gibco BRL (Grand Island, NY, USA) unless specified. The NRC cell line was kindly provided by Dr Nicholas F LaRusso (Mayo Clinic, Rochester, MN, USA) and were maintained on collagen-coated tissue culture inserts with polyester membrane (6.5 mm, Nalge Nunc, Rochester, NY, USA) in DMEM/Ham’s F-12 medium with supplements as described by Vroman and LaRusso (Vroman and LaRusso 1996).
Transfection of primary rat hepatocytes, KCs, and NRCs
Isolated hepatocytes and KCs were seeded 24 hours prior to transfection into a 24-well plates at an initial density of 3 × 105 cells per well. Before transfection, the medium in each well was replaced with 0.5 ml of fresh William’s E medium or RPMI 1640 for hepatocytes or KCs, respectively, with or without 10% bile. Nanoparticles containing 3 μg of DNA or 3 μg naked DNA was added to each well. The medium was refreshed after 4 h of incubation with cells. After 48 hours, cells were lysed with cell lysis buffer (Promega). Transfection of NRCs was conducted in transwell inserts (Nalge Nunc) in a 24-well tissue culture plate. When the NRCs reached 70% confluency, nanoparticles containing 3 μg of DNA or 3 μg naked DNA was added to each well. The medium was refreshed after 4 hours of incubation with cells. The analysis of luciferase expression was conducted according to the same procedure described above.
Luciferase activity in the cell extract was measured on a luminometer (LUMAT LB9507, Berthold, Germany) using a luciferase assay system from Promega (Madison, WI, USA). The relative light units (RLU) are normalized against protein concentration in the cell extracts using a micro-BCA protein assay kit (Pierce, Rockford, IL, USA).
Luciferase expression following RII, intraportal, and intravenous injections of nanoparticles and plasmid DNA
Wistar rats (male, 200–250 g) were randomly assigned to groups of 15–17 rats. Animals were laparotomized under general anesthesia and the liver was isolated from the surrounding tissue. A 33G needle was inserted into the common bile duct and a tie was used to secure the needle. Nanoparticles and naked DNA were administered at the dose equivalent to 200 μg of plasmid (∼0.8 mg/kg of body weight) in 4 mL of medium into the common bile duct over 20 min (0.2 ml/min) using a syringe pump. A tie was then placed around the bile duct between the liver and the point of infusion to prevent back flow, and the needle was withdrawn. After 30 min, all ties were removed. The needle hole in the bile duct might require stitches with 10-O nylon suture (Ethicon, Somerville, NJ, USA) to prevent bile leakage, whenever necessary. Rats were kept on normal diet. For portal vein injection using a syringe pump, surgical operation was performed as previously reported (Zhang et al 2001). Nanoparticles or naked DNA was infused at 0.2 mL/min using the same DNA concentration as described above. Tail vein injection was also performed using a syringe pump through a 30 G needle over 2 min.
On days 3, 7, and 14, 5 rats from each group were sacrificed. Major organs (liver, heart, lung, spleen, and kidney) were harvested and stored at −80°C for analysis. Each liver was divided into 4 sections composed of median, left, right, and caudate lobes. Two mL of lysis buffer (0.1% Triton X-100, 2 mM EDTA, and 0.1 M Tris-HCl, pH 7.8) per g of tissue was used for each sample, and the tissue was homogenized and subjected to 2 freeze-thaw cycles. The homogenates were centrifuged at 14000 rpm for 10 min. Luciferase activity in the homogenate was measured for 10 sec on a luminometer and converted to the mass of luciferase expressed per gram of tissue using a standard curve generated in parallel on the same luminometer. Student’s t-test was used to compare the difference between selected groups. Differences were considered statistically significant when p<0.05.
DNA distribution after RII of nanoparticles and naked DNA
To characterize DNA distribution, animals were injected with DNA nanoparticles containing 200 μg of Cy5-labeled plasmid DNA (pGeneGrip, Gene Therapy Systems, Inc.) through RII. Four hours after injection, major organs including liver, heart, spleen, lung, and kidney were resected, embedded in OCT medium, and frozen in liquid nitrogen as described above. For immunofluorescence staining, cryosections (μ8 μm) on microslides were fixed in cold acetone, and incubated with Hoechst 33258 to stain cell nuclei. Staining of endothelial cells was performed with a mouse anti-rat mAb against rat endothelial cell antigen (5 μg/mL, RECA-1, ab9774, Abcam, Cambridge, MA, USA) and a goat anti-mouse IgG2a HRP conjugate (1:100; Zymed, San Francisco, CA, USA), using a Tyramide Signal Amplification Fluorescein System (NEN, PerkinElmer, Wellesley, MA, USA). For staining of KCs, slides were incubated with mouse anti-rat macrophage F-6-J mAb [ab8173; Abcam; 10 μg/mL in I-Block (Tropix, Applied Biosystems, Foster City, CA, USA) and subsequently with a F(ab)2 goat anti-mouse IgG-FITC Ab (Abcam; 7 μg/mL in I-Block). Slides were mounted with Gel/Mounting medium (Abcam) and evaluated under a Nikon confocal microscope equipped with an imaging system (Nikon Singapore, Singapore).
Damage/toxicity to liver and biliary tree following RII
Blood samples were drawn on days 1, 2, 3, 7, and 14 from the tested rats. ALT, AST, ALP, and bilirubin levels in serum samples were analyzed using a multiparametric automatic analyzer in the Clinical Chemistry Laboratory at the National University Hospital in Singapore. Histopathological examination was performed on liver tissues collected at same time points. For collection of tissue, rats were anesthetized, perfused with 4% paraformaldehyde (PFA) in PBS, and euthanized. Extracted tissue will be further fixed in 4% PFA, and routinely processed for paraffin-section. Sections will be stained with hematoxylin-eosin.
Results
Characterization of PEI-DNA and chitosan-DNA nanoparticles
The chitosan-DNA nanoparticles were prepared by complex co-acervation between chitosan and DNA according to the procedure reported previously (Leong et al 1998; Mao et al 2001). Size and surface charges significantly affected the transfection and toxicity of the nanoparticles. We had identified that the optimal N/P ratios for PEI-DNA nanoparticles and chitosan-DNA nanoparticles were 10 and 3, respectively, for gene expression in the liver via RII in a pilot experiment (data not shown). These N/P ratios are consistent with other reported in vitro and in vivo gene transfer studies (Leong et al 1998; MacLaughlin et al 1998; Roy et al 1999; Mao et al 2001; Chew et al 2003; Tang et al 2003; Wang et al 2004). The following experiments are based on the PEI-DNA and chitosan-DNA nanoparticles prepared with these N/P ratios. At these N/P ratios, PEI-DNA nanoparticles displayed a high surface charge (+32 to +40 mV), with an average particle size of 170 nm, whereas chitosan-DNA nanoparticles exhibited a lower surface charge (∼+ 15 mV) and larger size (237 nm).
The stability of nanoparticles in bile or serum containing medium is one of the important factors that influence the particle transport and transfection efficiency. We observed that addition of 10% rat bile to the nanoparticles suspension induced significant aggregation (Table 1). Particles grew to a couple of microns in diameter within 15 min of incubation, yielded a suspension with a bimodal distribution. For example, light scattering plot of chitosan-DNA nanoparticles showed the majority (85–90% by number) of the particles in the range of 800–1200 nm and the remaining subset in the range of 2.5–3 μm. Particle size continued to grew until reaching a steady size after about 45 min of incubation. This aggregation behavior was likely due to the surface adsorption of negatively charged protein and bile salts present in the bile, leading to aggregation of positively charged nanoparticles. This is supported by the observation that zeta potential of DNA particles changed to highly negative following contact with 10% bile – the average zeta potentials of PEI-DNA and chitosan-DNA particles were −24 mV and −19 mV, respectively. Aggregation of particles was also observed when PEI-DNA and chitosan-DNA nanoparticles were incubated with serum containing medium (10%–50%), but at a slower rate compared with that in bile containing buffer (data not shown).
Table 1.
Size and surface charge of chitosan-DNA and PEI-DNA nanoparticles in preparation solutions and 10% bile-containing buffer
| Nanoparticles | Particle size (nm) | Zeta potential (mV) | ||
|---|---|---|---|---|
| in solutiona | in 10% bileb | in solutiona | in 10% bileb | |
| PEI-DNA (N/P=10) | 170 ± 13 | 3986 ± 117 | + (36 ± 4) | − (24.5 ± 2.5) |
| Chitosan-DNA (N/P=3) | 237 ± 32 | 3603 ± 155 | + (15 ± 3) | − (19 ± 4) |
Notes: Size and zeta potential measurements were carried out 30 min after preparation, with or without bile incubation. Data represents average ± standard deviation of 3 measurements.
The solution was 5% sucrose for PEI-DNA nanoparticles, and 50 mM sodium acetate buffer (pH 5.5) for chitosan-DNA nanoparticles.
Bile isolated from rat was added to the above mentioned solution to reach 10% final concentration (v/v). Size and zeta potential were measured after 60 min incubation with 10% bile.
Abbreviation: PEI, polyethylenimine.
Protection of plasmid DNA from bile degradation
Previously we have shown that chitosan-DNA nanoparticles render partial protection against nuclease degradation (Ludwig et al 1998). In order to characterize the potential protective effect during intrabiliary infusion, we incubated chitosan-DNA nanoparticles, PEI-DNA nanoparticles, or naked DNA with bile containing medium (10% and 50% bile in PBS, v/v) at 37°C, and examined the relative degrees of DNA degradation by gel electrophoresis. Degradation of naked DNA was apparent after incubating with 10% bile for 1 to 2 hours (Figure 1). Chitosan-DNA nanoparticles and PEI-DNA nanoparticles provided different degrees of protection to the encapsulated DNA. For chitosan-DNA nanoparticles, significant DNA degradation was observed after 6–12 hours incubation in the presence of 10% bile. The protective effect decreased as bile concentration increased, and degradation of plasmid was observed after incubating with 50% bile for only 1–2 hours. On the other hand, only slight plasmid degradation was apparent from PEI-DNA nanoparticles after 12 hours incubation under the same conditions. These data suggested that both nanoparticle formulations should render a significant level of protection to plasmid against bile degradation. The difference observed between chitosan and PEI carriers could be attributed to the degradation of chitosan in bile-containing medium.
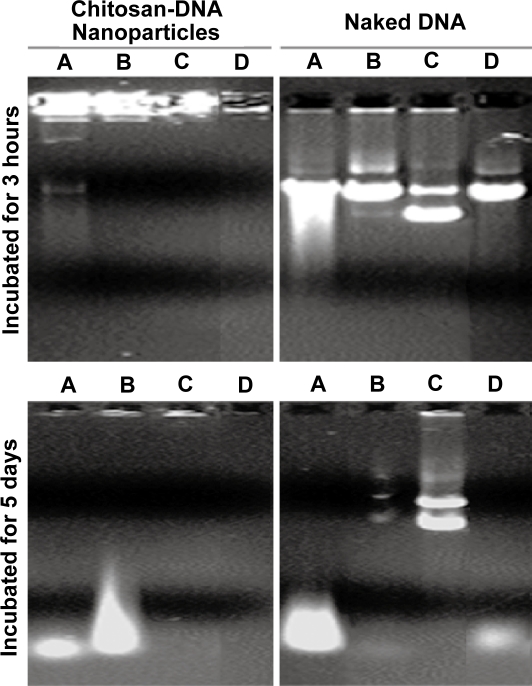
Figure 1.
Chitosan-DNA nanoparticles and PEI-DNA nanoparticles afford different degrees of protection to plasmid DNA in media containing 10% and 50% bile. To characterize the protection effect by nanoparticles in the presence of bile, chitosan-DNA nanoparticles, PEI-DNA nanoparticles and naked DNA were incubated with 10% or 50% bile diluted in PBS at 37°C. The relative degree of DNA degradation was analyzed by gel electrophoresis (0.8% agarose) and stained by ethidium bromide.
Abbreviation: PEI, polyethylenimine.
To determine whether bile-induced DNA damage is mediated by oxidative or enzymatic mechanisms, plasmid DNA was co-incubated for 3 hours at 37°C with 10% bile, 10% bile that was preheated at 100°C for 15 min, and 10% bile containing 50 mM of the anti-oxidant N-acetyl cysteine (NAC). As shown in Figure 2, the presence of NAC considerably preserved the super-coiled fraction of the plasmid, apparently preventing degradation and strand-breakage-induced linearization and nick-induced open coil formation in comparison to untreated and boiled bile incubations. Hydrogen peroxide (10 mM) treatment also mimicked the pattern of DNA fragmentation produced by bile treatment, whereas the DNA fragmentation was reduced in the group treated with boiled bile and absent with bile treatment in the presence of NAC. These data suggest that oxidative DNA damage accounted for the majority of strand scission observed; and it was significantly reduced by NAC treatment. Interestingly, neither DNA fragments nor intact/nicked plasmid was observed when chitosan-DNA particles were treated with bile in the presence of NAC, indicating that chitosan was not degraded under this condition in order to release DNA or its fragments. Compared with data on nanoparticles treated with bile and boiled bile, these results indicated that degradation of chitosan is partially dependent on the oxidative condition in the bile (Yoo et al 2005). It is worth noting that the exposure of plasmid to bile during RII is probably much less than the conditions tested in terms of bile concentration and exposure time, because of the large volume infused.
Figure 2.
Bile-induced DNA damage is primarily mediated by oxidative cleavage. Plasmid DNA and chitosan-DNA nanoparticles were incubated with (A) 10% bile, (B) 10% preheated bile, (C) 10% bile with 50 mM of NAC, or (D) 10 mm H2O2solution at 37°C for 3 hours and 5 days. The relative DNA degradation was analyzed by gel electrophoresis (0.8% agarose) and stained by ethidium bromide for visualization.
Abbreviation: NAC, N-acetyl cysteine.
Transfection of primary rat hepatocytes, KCs, and normal rat cholangiocytes (NRCs)
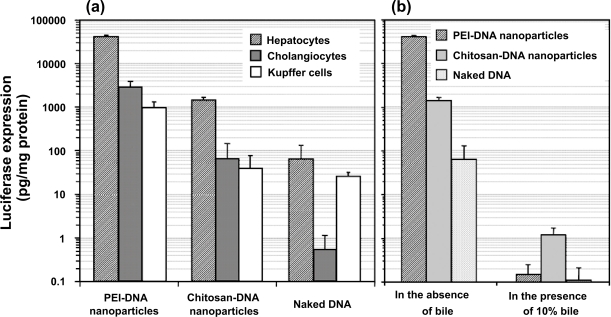
Previous reports have shown that the transfection efficiency of chitosan-DNA nanoparticles was generally lower than that of PEI-DNA nanoparticles, and it is cell type-dependent (Ludwig et al 1998). Figure 3a shows the difference in transfection efficiency of the two carriers in primary rat hepatocytes, KCs, and NRCs. Chitosan-DNA nanoparticles mediated a luciferase expression that was 4 orders of magnitude higher than background level; and transfections by chitosan-DNA nanoparticles were about 20 and 35 times less efficient in NRCs and KCs, respectively. PEI-DNA nanoparticles were more efficient, resulting in 26, 43, and 25 times higher luciferase expression than chitosan-DNA nanoparticles in transfecting hepatocytes, NRCs, and KCs, respectively. Interestingly, naked DNA alone effected more than 500 and 260 times higher luciferase activities than background level in hepatocytes and KCs, whereas it remained inefficient in NRCs.
Figure 3.
Transfection efficiency of PEI-DNA and chitosan-DNA nanoparticles and naked DNA in primary rat hepatocytes, Kupffer cells (KCs) and normal rat cholangiocytes (NRCs). (a). Transfection efficiency in three types of cells. Hepatocytes and KCs were transfected in 24-well dishes with the indicated complexes containing 3 μg VR1255 pcDNA for 4 hours. Luciferase expression was analyzed 48 hours after transfection. Relative light units were normalized to protein content and are the mean ± standard deviation of 2 independent experiments (n = 4). (b). Comparison of transfection efficiency in primary rat hepatocytes in the presence and absence of bile (10%). Cells were transfected in a medium containing 10% bile for 4 hours, and normal medium for 44 hours before luciferase analysis (n = 4).
Abbreviation: PEI, polyethylenimine.
We also investigated whether the exposure to bile would affect the transfection efficiency of nanoparticles. The transfection was carried out by exposing hepatocytes with nanoparticles or naked DNA in the absence or presence of 10% bile added in the medium. Bile containing medium was replaced with fresh medium after 4 hours of incubation with cells. The presence of bile almost completely abolished transgene expression mediated by PEI-DNA nanoparticles and naked DNA (Figure 3b). Chitosan-DNA nanoparticles retained a low level of transfection activity. This high sensitivity of nanoparticles and naked DNA to bile underscores the importance of developing strategies to stabilize the nanoparticles. The reduction of transfection efficiency by nanoparticles in bile containing medium could be due to a combination of nanoparticle aggregation and degradation of particles and DNA. In addition, gene expression in hepatocytes might also be dampened by the cytotoxicity of bile, which is evident from relative cell numbers at the end of transfection experiment (data not shown).
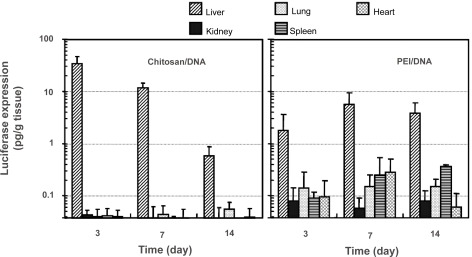
Luciferase expression in rat liver and other organs following RII, intraportal, and tail vein infusions of nanoparticles and naked DNA
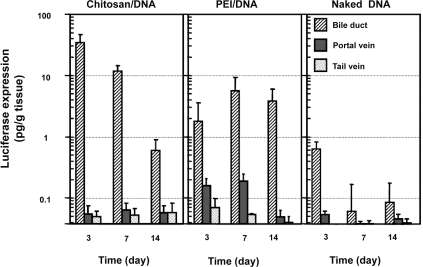
Transfection efficiency was analyzed using luciferase as a reporter gene. Nanoparticles were prepared using VR1255 plasmid. When given by tail vein infusion, chitosan-DNA nanoparticles, PEI-DNA nanoparticles and naked DNA groups all showed background level of luciferase expression on days 3, 7, and 14 (Figure 4). When given by intraportal vein infusion, PEI-DNA nanoparticle was the only group showing positive transgene expression (∼3 times higher than background level, p<0.05) during the first week. Chitosan-DNA nanoparticles and naked DNA both failed to show any detectable level of luciferase expression.
Figure 4.
Luciferase expression in rat liver following RII, intraportal infusion and tail vein injection of chitosan-DNA nanoparticles, PEI-DNA nanoparticles, and naked DNA. Each bar represents mean ± standard deviation (n = 5). Chitosan C390 and VR1255 plasmid were used in this experiment. Chitosan-DNA and PEI-DNA nanoparticles were prepared at N/P ratios of 3 and 10, respectively. Nanoparticles and naked DNA were infused to rats at a dose equivalent to 200 μg of plasmid per rat (∼0.8 mg/kg of body weight) in 4 mL of medium into the common bile duct (0.1 ml/min) or portal vein (1 ml/min), or tail vein (1 mL/min). On days 3, 7, and 14, 5 rats from each group were sacrificed, and livers were harvested and homogenized in lysis buffer and analyzed for luciferase activity. Rats receiving 4 mL of PBS infusions were included as the background control that defines the detection limit for this assay at 0.04 pg luciferase/g tissue.
Abbreviation: PEI, polyethylenimine.
RII, on the other hand, was a much more efficient delivery route. Three days after RII, rats that received chitosan-DNA nanoparticles showed a luciferase expression of 33 pg/g of tissue; this level is over 500 times greater than background signal in the luciferase expression assay observed in PBS controls (p<0.01). Gene expression level decreased over time to 0.6 pg/g of tissue on day 14. PEI-DNA nanoparticles were slightly less efficient than chitosan-DNA nanoparticles. Luciferase expression was about 17-fold lower than that by chitosan-DNA nanoparticles on day 3, but it increased by about 2 fold on day 7 and was maintained at the similar level for at least 2 weeks. Surprisingly, naked DNA given by RII also gave a transient luciferase expression (0.62 pg/g of tissue) on day 3 (p<0.05).
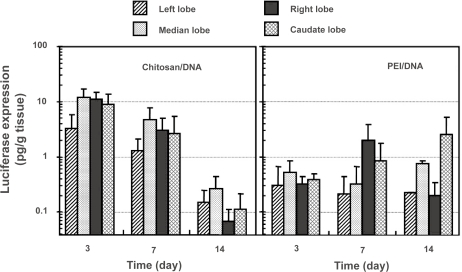
Lobular structure of the rat liver provided a convenient system to analyze the distribution of transgene product in the liver following RII. The distribution of transgene product in the liver was slightly different between chitosan-DNA and PEI-DNA nanoparticles (Figure 5). In chitosan-DNA nanoparticle-transfected rats, luciferase expression in the left lobe was consistently 3 to 4 times lower than other lobes of the liver (p<0.05) at the early time points. This might be a result of the nanoparticle aggregation in bile duct and canaliculi that limited the transport through the canaliculi to the left lobe of the liver. This difference was not significant on day 14. The expression levels among different lobes of the liver transfected with PEI-DNA nanoparticles were similar on day 3. Nevertheless, the right and caudate lobes, which are closer to the infusion point, seemed to have higher expression at later time points.
Figure 5.
Luciferase expression in different lobes of the rat liver following intrabiliary injection of chitosan-DNA nanoparticles or PEI-DNA nanoparticles containing 200 μg of DNA. Each bar represents mean ± standard deviation (n = 5). Experimental condition were the same as described in Figure 3.
Abbreviation: PEI, polyethylenimine.
Transgene distribution among different organs was different for gene transfers mediated by chitosan-DNA nanoparticles and PEI-DNA-nanoparticles (Figure 6). This information is important not only in evaluating the liver-targeting effect by the carriers, but also in understanding particle transport mechanism. In rats receiving intrabiliary infusion of chitosan-DNA nanoparticles, transgene expression levels in the kidney, lung, spleen, and heart were negligible (p<0.01). However, in PEI-DNA nanoparticle-transfected rats, low level of luciferase expressions were detected in the lung, spleen, and heart, even though the expression in the liver still accounted for more than 95% of the total transgene expression.
Figure 6.
Luciferase expression in major organs of the rats after receiving intrabiliary injection of chitosan-DNA nanoparticles or PEI-DNA nanoparticles containing 200 μg of DNA. Each bar represents mean ± standard deviation (n = 5). Experimental conditions are the same as described in Figure 3.
Abbreviation: PEI, polyethylenimine.
Distribution of DNA following RII of nanoparticles and naked DNA
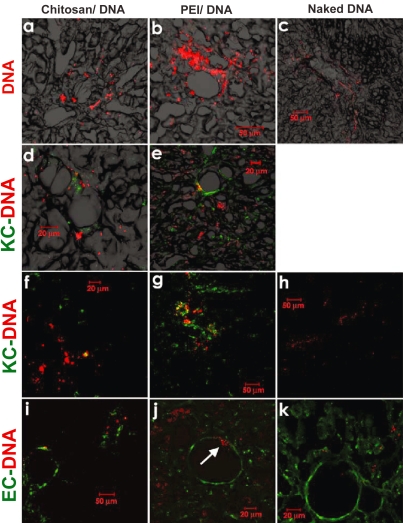
To characterize the distribution of nanoparticles and naked DNA following RII, plasmid DNA was labeled with Cy5 using pGeneGrip technology before nanoparticle preparation. Tissues from major organs were harvested 4 hours after infusion, and cryo-sectioned for the examination of Cy5-labelled plasmid. For animals that received nanoparticles, the reporter gene was detected throughout the liver, although the pattern was rather heterogeneous; occasionally clusters of Cy5 fluorescence were found in close proximity to vessel structure or portal triads (Figures 7a, b). Only a low level of reporter gene was found in naked DNA-transfected rats, highlighting the importance of gene carriers. Comparing the two nanoparticle groups, more fluorescence was observed for the PEI-DNA nanoparticle group than the chitosan-DNA nanoparticle group. The relative levels of reporter gene observed in the tissue, however, seemed to contradict the relative levels of transgene expression in the liver: chitosan-DNA nanoparticles showed a lower level of presence in the liver, but yielded higher level of luciferase expression, in contrast to PEI-DNA nanoparticles. This may be due to the fact that chitosan is degradable, therefore resulting in higher level of DNA release following cell uptake.
Figure 7.
Confocal fluorescence images indicating DNA distribution in the right lobe of the liver that received naked DNA and nanoparticles via RII. Liver samples were collected 4 hours after intrabiliary infusion of chitosan-DNA or PEI-DNA nanoparticles containing 200 μg of Cy5 labeled DNA (pGeneGrip) or 200 μg of naked Cy5-DNA. (a–c): distribution of DNA (red); (d–h): co-localization of DNA (red) and KCs (green). KCs were stained with FITC-labeled mouse anti-rat macrophage F-6-J mAb. (i–k): co-localization of DNA (red) and endothelial cells (ECs, green). Endothelial cells were identified by immunostaining with FITC-labeled mouse anti-rat RECA-1 mAb.
Abbreviation: PEI, polyethylenimine; EC, endothelial cell; KC, Kupffer cell.
Another possibility is that more PEI-DNA nanoparticles were taken up by KCs than chitosan-DNA nanoparticles. This is supported by additional immunofluorescence staining for KCs using an antibody specific for rat macrophages (Figures 6d–h). Even though the majority of cells that took up reporter gene (or particles) were hepatocytes, we observed a significantly higher level of DNA co-localization with KCs in the PEI-group (Figures 6e, g), compared with chitosan-group and naked DNA group. It is also interesting to note that there was a substantially higher number of KCs observed in the PEI-groups than the other two groups. Relatively lower frequency of KCs found in the chitosan-group might be related to the anti-inflammatory effect of chitosan (Chou et al 2003; Seo et al 2003; Kim et al 2004). We have observed a very low degree of co-localization between reporter gene and endothelial cells (Figures 6i–k), indicating a lower level of uptake of nanoparticles by endothelial cells. On the other hand, we did observe the passage of PEI-DNA particles through the endothelium occasionally (arrow in Figure 7j).
Despite the fact that low levels of gene expression were observed in heart, spleen, lung, and kidney in rats receiving PEI-DNA nanoparticles, we failed to observe Cy5-fluorescence (reporter gene) in these organs. This is likely due to low sensitivity of this assay method.
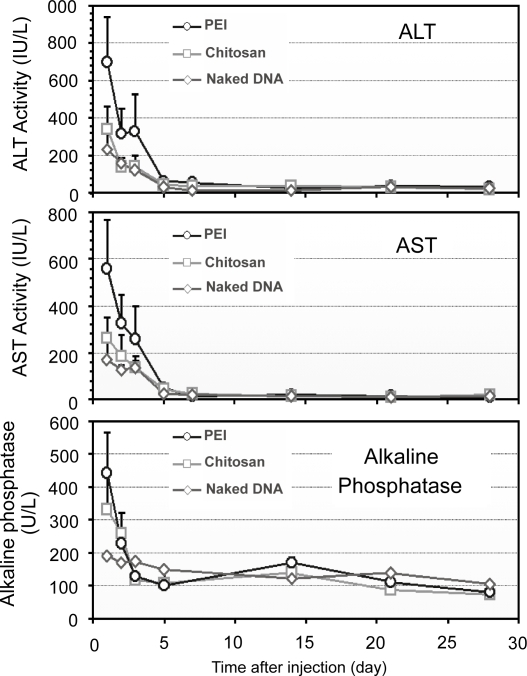
Toxicity/damage to the liver and biliary tree by RII of nanoparticles and naked DNA
The acute liver damage and toxicity following RII of nanoparticles and naked DNA were assessed by analyzing serum aspartate transaminase (AST) and alanine transaminase (ALT) levels during the experimental period, in comparison with the infusions of PEI-DNA nanoparticles (Figure 8). A slight increase in serum AST and ALT activities was observed 1 day after the administration of naked DNA, followed by a rapid decrease to the normal level by day 3. Chitosan-DNA nanoparticles mediated only a moderate increase of both ALT and AST activities, slightly higher than that of naked DNA group. In contrast, significant elevation of both AST and ALT in the PEI-DNA nanoparticle group was observed (∼1150 IU/L for ALT and ∼900 IU/L for AST). The levels of alkaline phosphatases (ALP) were monitored to reflect potential damage to biliary tree. The ALP levels followed the same trends for all three groups (Figure 8). The transient increase of ALP level indicated that mild damage to biliary tree occurred in response to the infusion pressure, but this damage was transient. Nanoparticle infusions caused higher levels of ALP during the first two days. Throughout the experimental period, bilirubin levels for all groups were in the normal range (5–30 IU/L), indicating that there was no obstructive jaundice caused by the injection procedure (data not shown).
Figure 8.
Serum ALT, AST, and ALP levels in rats that received nanoparticles and naked DNA through RII (n = 3).
Abbreviation: ALT, alanine transaminase; AST, aspartate transaminase; ALP, alkaline phosphatase; PEI, polyethylenimine.
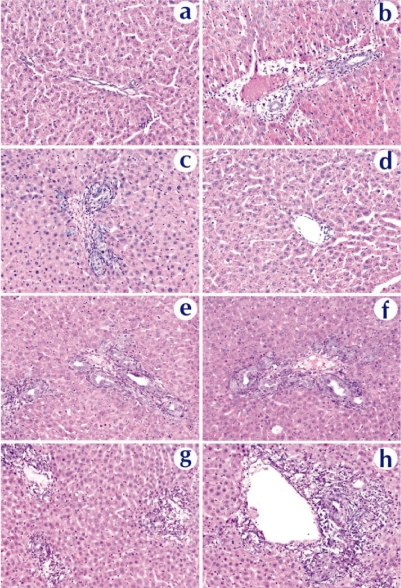
Histopathological examination confirmed these conclusions. Figure 9 showed tissue reactions in the liver 3 days after nanoparticle and naked DNA infusions. Liver sections from a naked DNA-infused rat showed minimal changes with patchy mild bile ductular proliferation and mild reactive changes in the biliary epithelium (Figure 9c). The reactive bile ducts/ductules appeared regenerative with increased mitoses, decreased basophilic cytoplasm, and increased nuclear size with open chromatin. Most of portal tracks are essentially normal (Figure 9d). The focal and limited proliferation of hepatocytes and ductular cells is in good agreement with the result by Polimeno et al (1995) showing that stimulation of a significant proliferative response requires obstruction of the bile duct for longer than 24 hours. Rats that received chitosan-DNA nanoparticles showed some mild biliary changes with a higher level of diffused but mild bile ductular proliferation and biliary epithelial reactive/regenerative changes (Figures 9e, f); other portal tracks were normal. PEI-DNA nanoparticles induced a stronger biliary tract change than chitosan-DNA nanoparticles. In addition to bile ductular proliferation and reactive epithelial changes, some sections showed portal edema and significant portal inflammation (Figures 9g, h). No animals showed evidence for lobular hepatitis, vascular flow abnormalities, lobular cholestasis, or portal fibrosis. Long-term morphological damages to the liver and biliary tree remain to be characterized. Biochemical analyses discussed above suggest that these damages were transient.
Figure 9.
Histopathological examination of liver tissue extracted on day 3 from rats received naked DNA (c and d), chitosan-DNA nanoparticles (e and f), or PEI-DNA nanoparticles (g and h), in comparison with the naïve rat (a and b). Liver samples were collected on day 14, cryosectioned (8 μm thickness), and stained with H&E.
Abbreviation: PEI, polyethylenimine.
Discussion
Retrograde intrabiliary infusion has several advantages that make it attractive for improving liver-targeted gene transfer efficiency. The biliary system is distensible with a large bile duct volume (∼29 mL for human liver). It consists of 7–10 orders of cholangiographically visible bile ducts spanning the expanse of the hepatic parenchyma (Saxena et al 2003). This large surface area (eg, the ultrastructural surface of an entire normal biliary tree for a human liver would be around 3000 cm2) and broadly distributed biliary system provides great access to large number of hepatocytes in liver parenchyma through bile canaliculi (Ludwig et al 1998). RII achieves a more direct delivery to parenchymal hepatocytes than intraportal vein infusion and tail vein injection; and avoids first contact with serum and KCs lining endothelium, even though leakage through tight junction to Space of Disse and hepatic sinusoid likely occurs under this infusion condition. RII of adenovirus (Ad5) encoding LacZ gene yielded transfection of similar expression levels (∼30%) and duration in hepatocytes as that delivered through intraportal infusion, however, re-administration of Ad by RII on day 35 and 70 induced similar level of re-expression of transgene in the liver, despite the fact that the first administration of Ad5 also generated neutralizing antibody in the serum. This was in contrast to that by intraportal infusion, which failed to show gene expression upon re-administration (Tominaga et al 2004). No cholangiocytes were stained positive. The distinct outcome of these two delivery routes highlights the advantage of RII as a more efficient route for liver-targeted gene delivery.
One particular concern for RII delivery of plasmid DNA is the degradation and damage to DNA by contact with bile. Bile has long been known to cause DNA damage and strand breakage, having been implicated as a mutagen in colon cancer (Cheah and Bernstein 1990) and biliary tract carcinoma (Masamune et al 1997). In particular, strand breakage has been shown to occur through the production of hydroxyl radicals and other reactive oxygen species by the bilirubin-Cu(II) complex (Asad et al 1999, 2002). The decreased fragmentation in the presence of boiled bile indicates that a native protein, such as bilirubin, might be involved, although significant nuclease activity is effectively precluded by the marked protective effect of NAC treatment (Figure 2). NAC is commonly administered as an antioxidant to treat paracetamol poisoning of the liver. It could thus be readily added to the solution after nanoparticle formation or used as a pre-treatment prior to nanoparticle delivery to further enhance the delivery efficiency.
Compared with literature-reported RII procedures (Otsuka et al 2000; Zhang et al 2001, 2003), we made two modifications in our procedure: (1) the infusion was carried out with a syringe pump in order to achieve a consistent flow and reproducible results. Understandably, this minimizes the potential hepatic and biliary damage as the intrabiliary pressure can be maintained steady during infusion; (2) the intrabiliary infusion was given through the common bile duct to the whole liver, as opposed to infusion only to one of the liver lobes (Otsuka et al 2000; Zhang et al 2001, 2003) This is apparently a more practical procedure to be adapted in the clinical setting.
In the literature, infusion rate ranging from 0.02 to 2.7 mL/min have been tested for RII in various experiments in rats, including non-transfection related studies (Coleman et al 1989; De Godoy et al 1999; Zhang et al 2001; Chen et al 2005). Although no systematic study has been reported on examining the effect of infusion rate and volume on transfection efficiency, it is assumed that there is a balanced condition that correlates with high transfection and acceptable hepatic injury and toxicity (De Godoy et al 1999; Zhang et al 2001; Chen et al 2005). It has been shown that rats do not tolerate high infusion rate (0.4–2.25 mL/min) and high infusion volume (>4 mL) well (Coleman et al 1989; Chen et al 2005). A pilot study suggested that using our protocol, an infusion rate up to 0.25 min/mL and a total infusion volume of 2–4 mL could be well tolerated by Wistar rats. For the purpose of comparing the delivery efficiency of different gene carriers, we fixed the infusion rate at 0.2 mL/min and infusion volume at 4 mL for intrabiliary infusion in this study.
Our study demonstrated that the transgene expression is both administration route-dependent and carrier dependent. Among the three administration routes tested, RII showed the highest transgene expression in the liver for all both nanoparticle groups and naked DNA. Even naked DNA delivered by RII yielded about 10-times higher transient gene expression than background level; whereas intraportal and tail vein infusion both failed to show any expression. Gene expressions by PEI-DNA and chitosan-DNA nanoparticles showed very different kinetics. Chitosan was 17 times more efficient than PEI under the test condition, but the expression was more transient than PEI. PEI-DNA nanoparticles showed a more persistent level of gene expression, even though the maximum level of gene expression was 6-fold lower than that of chitosan-DNA nanoparticles. These data suggested that polymeric carrier improves transgene expression delivered through RII, but the level and kinetics of transgene expression is dependent on the nature of gene carrier. In addition, the difference in transgene expression between the two types of carriers in vivo (Figures 4 and 6) does not correlate with that in vitro (Figure 3). These differences could be related to the degradation of chitosan in vivo, even though chitosan degradation is not appreciable in typical cell culture medium. Macrophages present in the liver (Kupffer cells) and other tissues may also play a role in modulating chitosan degradation and nanoparticle clearance, therefore influence the transgene expression in vivo (Chellat et al 2005).
Both nanoparticles mediated less transgene expression in the left lobe, a distance lobe from the infusion point, suggesting that particle aggregation could be barrier to achieving uniform transfection throughout the liver. This is also corroborated by the observation that both nanoparticles aggregate upon contact with 10% bile or medium. Further improvement of the colloidal stability of the nanoparticles will likely increase the transfection efficiency and the accessibility of nanoparticles to the whole liver tissue.
Immunofluorescence microscopy also suggested that DNA uptake was restricted to focal areas; and DNA was associated with hepatocytes and to a lesser extent with macrophages. It is also of importance to investigate whether biliary epithelial cells (cholangiocytes) can be transfected under this RII condition. No significant level of DNA was found in the portal triad region in the fluorescence microscopy study; neither did we observe β-galactosidase expression in the portal triad region in a transfection experiment using LacZ as the reporter gene (data not shown). This is consistent with literature reports showing very limited transfection in cholangiocytes with liposomes (Otsuka et al 2000), PLL-DNA complexes (Zhang et al 2001, 2003), and adenovirus (Tominaga et al 2004) using RII protocols. Detailed localization of gene expression with cholangiocytes is currently under investigation.
Of particular note is the different pattern of distribution in transgene expression among major organs mediated by chitosan-DNA nanoparticles and PEI-DNA nanoparticles following RII. No significant gene expression was detected in organs other than the liver in the chitosan group, in contrast to significant luciferase expression observed in lung, spleen and heart for PEI-DNA nanoparticles on days 3 to 14. Positive gene expression in other organs suggested that transport of nanoparticles from the bile canaliculi to hepatic sinusoid and hepatic vein occurred, and a higher degree of “leakage” occurred for PEI-DNA nanoparticles, compared with chitosan-DNA nanoparticles. Nevertheless, it is also possible that chitosan-DNA nanoparticles were transported to other organs at a similar degree but failed to transfect those tissues. Considering the acute disruption of the tight junctions under mildly elevated pressure, RII actually can be viewed as an alternative route to deliver genes to hepatic sinusoid, although detailed transport mechanism remains to be characterized.
Our RII protocol mounted a mild level of hepatic toxicity/damage as evidenced by the transient and slight elevation of the AST, ALT, and ALP levels following naked DNA delivery. Understandably, the degree of liver and biliary tree damage should be dependent on the infusion rate and volume (Roy et al 1999; Mao et al 2001; Chew et al 2003). Chitosan and PEI increased the AST, ALT, and ALP levels significantly at early time points, but PEI showed much higher toxicity/damage than chitosan (ALT and AST levels in PEI group were 3 times higher than that of chitosan group on the first day). This suggests that gene carrier does contribute to and is probably the most significant factor in the acute toxicity/damage by this route of delivery.
Efforts in optimizing non-viral vector-mediated gene transfer have been centered on the design and modification of gene vectors and constructs. More studies are needed to systematically optimize the non-viral gene vector properties in concert with the administration route. This study demonstrates that the optimal characteristics of a gene carrier are closely dependent upon the administration route; and establishes the feasibility of RII for liver-targeted gene delivery using biodegradable and biocompatible nanoparticles. It is important to note that in clinical settings RII can be achieved via endoscopic retrograde cholangiopancreatography (ERCP), a routine bile duct canulation procedure (Chia et al 2000). This study laid the foundation for further optimization of nanoparticles to enhance liver-targeted gene delivery and for systematic study on the mechanism of nanoparticle transport to liver parenchymal cells through different routes.
Acknowledgments
This study is supported in part by Agency for Science, Technology and Research (A*STAR) of Singapore through a grant to the Division of Biomedical Sciences, Johns Hopkins in Singapore, the Office of Life Sciences, National University of Singapore (WBS 152-000-600-712, H.Q. Mao), and National Institutes of Health (EB002849, KW Leong; and DK068399, HQ Mao). The authors thank Dr Pengchi Zhang, Roy Lee, and Peggy Tang for technical assistance.
References
- Asad SF, Singh S, Ahmad A, et al. Bilirubin-Cu(II) complex degrades DNA. Biochim Biophys Acta. 1999;1428:201–8. doi: 10.1016/s0304-4165(99)00075-6. [DOI] [PubMed] [Google Scholar]
- Asad SF, Singh S, Ahmad A, et al. Bilirubin/biliverdin-Cu(II) induced DNA breakage; reaction mechanism and biological significance. Toxicol Lett. 2002;131:181–9. doi: 10.1016/s0378-4274(02)00031-0. [DOI] [PubMed] [Google Scholar]
- Borchard G. Chitosans for gene delivery. Adv Drug Deliv Rev. 2001;52:145–50. doi: 10.1016/s0169-409x(01)00198-3. [DOI] [PubMed] [Google Scholar]
- Bowman K, Sarkar R, Wang XL, et al. Evaluation of non-viral vectors for gene therapy of hemophilia A. Mol Ther. 2004;9:S314. [Google Scholar]
- Cheah PY, Bernstein H. Modification of DNA by bile acids: a possible factor in the etiology of colon cancer. Cancer Lett. 1990;49:207–210. doi: 10.1016/0304-3835(90)90160-y. [DOI] [PubMed] [Google Scholar]
- Chellat F, Grandjean-Laquerriere A, Le Naour R, et al. Metalloproteinase and cytokine production by THP-1 macrophages following exposure to chitosan-DNA nanoparticles. Biomaterials. 2005;26:961–70. doi: 10.1016/j.biomaterials.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Chen CY, Liu HS, Lin XZ. Hydrodynamics-based gene delivery to the liver by bile duct injection of plasmid DNA – the impact of lasting biliary obstruction and injection volume. Hepatogastroenterology. 2005;52:25–8. [PubMed] [Google Scholar]
- Cheung ST, Tsui TY, Wang WL, et al. Liver as an ideal target for gene therapy: expression of CTLA4Ig by retroviral gene transfer. J Gastroenterol Hepatol. 2002;17:1008–14. doi: 10.1046/j.1440-1746.2002.02784.x. [DOI] [PubMed] [Google Scholar]
- Chew JL, Wolfowicz CB, Mao HQ, et al. Chitosan nanoparticles containing plasmid DNA encoding house dust mite allergen, Der p 1 for oral vaccination in mice. Vaccine. 2003;21:2720–9. doi: 10.1016/s0264-410x(03)00228-7. [DOI] [PubMed] [Google Scholar]
- Chia SM, Leong KW, Li J, et al. Hepatocyte encapsulation for enhanced cellular functions. Tissue Eng. 2000;6:481–95. doi: 10.1089/107632700750022134. [DOI] [PubMed] [Google Scholar]
- Chou TC, Fu E, Shen EC. Chitosan inhibits prostaglandin E2 formation and cyclooxygenase-2 induction in lipopolysaccharide-treated RAW 264.7 macrophages. Biochem Biophys Res Commun. 2003;308:403–7. doi: 10.1016/s0006-291x(03)01407-4. [DOI] [PubMed] [Google Scholar]
- Cichon G, Schmidt HH, Benhidjeb T, et al. Intravenous administration of recombinant adenoviruses causes thrombocytopenia, anemia and erythroblastosis in rabbits. J Gene Med. 1999;1:360–71. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<360::AID-JGM54>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Coleman R, Rahman K, Kan KS, et al. Retrograde intrabiliary injection of amphipathic materials causes phospholipid secretion into bile. Taurocholate causes phosphatidylcholine secretion, 3-[(3-cholamidopr opyl)dimethylammonio]-propane-1-sulphonate (CHAPS) causes mixed phospholipid secretion. Biochem J. 1989;258:17–22. doi: 10.1042/bj2580017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano RJ, Smith LC, Kay MA, et al. Hepatic gene therapy: efficient gene delivery and expression in primary hepatocytes utilizing a conjugated adenovirus-DNA complex. Proc Natl Acad Sci U S A. 1993;90:11548–52. doi: 10.1073/pnas.90.24.11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Godoy JL, Malafosse R, Fabre M, et al. In vivo hepatocyte retrovirus-mediated gene transfer through the rat biliary tract. Hum Gene Ther. 1999;10:249–57. doi: 10.1089/10430349950019039. [DOI] [PubMed] [Google Scholar]
- Higuchi N, Maruyama H, Kuroda T, et al. Hydrodynamics-based delivery of the viral interleukin-10 gene suppresses experimental crescentic glomerulonephritis in Wistar-Kyoto rats. Gene Ther. 2003;10:1297–310. doi: 10.1038/sj.gt.3301988. [DOI] [PubMed] [Google Scholar]
- Jooss K, Chirmule N. Immunity to adenovirus and adeno-associated viral vectors: implications for gene therapy. Gene Ther. 2003;10:955–63. doi: 10.1038/sj.gt.3302037. [DOI] [PubMed] [Google Scholar]
- Kawakami S, Yamashita F, Nishida K, et al. Glycosylated cationic liposomes for cell-selective gene delivery. Crit Rev Ther Drug Carrier Syst. 2002;19:171–90. doi: 10.1615/critrevtherdrugcarriersyst.v19.i2.30. [DOI] [PubMed] [Google Scholar]
- Kim MS, You HJ, You MK, et al. Inhibitory effect of water-soluble chitosan on TNF-alpha and IL-8 secretion from HMC-1 cells. Immunopharmacol Immunotoxicol. 2004;26:401–9. doi: 10.1081/iph-200026887. [DOI] [PubMed] [Google Scholar]
- Koping-Hoggard M, Tubulekas I, Guan H, et al. Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 2001;8:1108–21. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- Kren BT, Chowdhury NR, Chowdhury JR, et al. Gene therapy as an alternative to liver transplantation. Liver Transpl. 2002;8:1089–1108. doi: 10.1053/jlts.2002.36844. [DOI] [PubMed] [Google Scholar]
- Kuhel DG, Zheng S, Tso P, et al. Adenovirus-mediated human pancreatic lipase gene transfer to rat bile: gene therapy of fat malabsorption. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1031–6. doi: 10.1152/ajpgi.2000.279.5.G1031. [DOI] [PubMed] [Google Scholar]
- Lecocq M, Andrianaivo F, Warnier MT, et al. Uptake by mouse liver and intracellular fate of plasmid DNA after a rapid tail vein injection of a small or a large volume. J Gene Med. 2003;5:142–56. doi: 10.1002/jgm.328. [DOI] [PubMed] [Google Scholar]
- Leong KW, Mao HQ, Truong L, et al. DNA-polycation nanospheres as non-viral gene delivery vehicles. J Control Release. 1998;53:183–93. doi: 10.1016/s0168-3659(97)00252-6. [DOI] [PubMed] [Google Scholar]
- Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Liu WG, Yao KD. Chitosan and its derivatives – a promising non-viral vector for gene transfection. J Control Release. 2002;83:1–11. doi: 10.1016/s0168-3659(02)00144-x. [DOI] [PubMed] [Google Scholar]
- Ludwig J, Ritman EL, LaRusso NF, et al. Anatomy of the human biliary system studied by quantitative computer-aided three-dimensional imaging techniques. Hepatology. 1998;27:893–9. doi: 10.1002/hep.510270401. [DOI] [PubMed] [Google Scholar]
- MacLaughlin FC, Mumper RJ, Wang J, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56:259–72. doi: 10.1016/s0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- Mahato RI, Kawabata K, Takakura Y, et al. In vivo disposition characteristics of plasmid DNA complexed with cationic liposomes. J Drug Target. 1995;3:149–57. doi: 10.3109/10611869509059214. [DOI] [PubMed] [Google Scholar]
- Mao HQ, Roy K, Troung L, et al. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- Masamune K, Kunitomo K, Sasaki K, et al. Bile-induced DNA strand breaks and biochemical analysis of bile acids in an experimental model of anomalous arrangement of the pancreaticobiliary ducts. J Med Invest. 1997;44:47–51. [PubMed] [Google Scholar]
- Murakami T, Sato H, Nakatani S, et al. Biliary tract of the rat as observed by scanning electron microscopy of cast samples. Arch Histol Cytol. 2001;64:439–47. doi: 10.1679/aohc.64.439. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Davidoff AM, Hanawa H, et al. Sustained high-level expression of human factor IX (hFIX) after liver-targeted delivery of recombinant adeno-associated virus encoding the hFIX gene in rhesus macaques. Blood. 2002;100:1662–9. doi: 10.1182/blood-2002-02-0589. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Ferry N. Liver gene therapy: advances and hurdles. Gene Ther. 2004;11(Suppl 1):S76–84. doi: 10.1038/sj.gt.3302373. [DOI] [PubMed] [Google Scholar]
- Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647–52. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Nishida S, Todo H, et al. Pulmonary gene delivery by chitosan-pDNA complex powder prepared by a supercritical carbon dioxide process. J Pharm Sci. 2003;92:371–80. doi: 10.1002/jps.10285. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Baru M, Delriviere L, et al. In vivo liver-directed gene transfer in rats and pigs with large anionic multilamellar liposomes: routes of administration and effects of surgical manipulations on transfection efficiency. J Drug Target. 2000;8:267–79. doi: 10.3109/10611860008997905. [DOI] [PubMed] [Google Scholar]
- Polimeno L, Azzarone A, Zeng QH, et al. Cell proliferation and oncogene expression after bile duct ligation in the rat: evidence of a specific growth effect on bile duct cells. Hepatology. 1995;21:1070–8. [PMC free article] [PubMed] [Google Scholar]
- Prego C, Garcia M, Torres D, et al. Transmucosal macromolecular drug delivery. J Control Release. 2005;101:151–62. doi: 10.1016/j.jconrel.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Prieto J, Herraiz M, Sangro B, et al. The promise of gene therapy in gastrointestinal and liver diseases. Gut. 2003;52:ii49–54. doi: 10.1136/gut.52.suppl_2.ii49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JV, Bigley NJ, Chiou HC, et al. Targeted delivery of DNA encoding herpes simplex virus type-1 glycoprotein D enhances the cellular response to primary viral challenge. Arch Dermatol Res. 2000;292:542–9. doi: 10.1007/s004030000181. [DOI] [PubMed] [Google Scholar]
- Roy K, Mao HQ, Huang SK, et al. Oral gene delivery with chitosan – DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat Med. 1999;5:387–91. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- Saxena R, Zucker SD, Crawford JM. Anatomy and physiology of the liver. In: Zakim D, Boyer TD, editors. Hepatology, a textbook of liver disease. 4th ed. Philadelphia: Saunders; 2003. pp. 3–30. [Google Scholar]
- Schipper NG, Olsson S, Hoogstraate JA, et al. Chitosans as absorption enhancers for poorly absorbable drugs 2: mechanism of absorption enhancement. Pharm Res. 1997;14:923–9. doi: 10.1023/a:1012160102740. [DOI] [PubMed] [Google Scholar]
- Seo SB, Jeong HJ, Chung HS, et al. Inhibitory effect of high molecular weight water-soluble chitosan on hypoxia-induced inflammatory cytokine production. Biol Pharm Bull. 2003;26:717–21. doi: 10.1248/bpb.26.717. [DOI] [PubMed] [Google Scholar]
- Sullivan DE, Dash S, Du H, et al. Liver-directed gene transfer in non-human primates. Hum Gene Ther. 1997;8:1195–206. doi: 10.1089/hum.1997.8.10-1195. [DOI] [PubMed] [Google Scholar]
- Tang Y, Shah K, Messerli SM, et al. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14:1247–54. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- Terao R, Honda K, Hatano E, et al. Suppression of proliferative cholangitis in a rat model with direct adenovirus-mediated retinoblastoma gene transfer to the biliary tract. Hepatology. 1998;28:605–12. doi: 10.1002/hep.510280301. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Kuriyama S, Yoshiji H, et al. Repeated adenoviral administration into the biliary tract can induce repeated expression of the original gene construct in rat livers without immunosuppressive strategies. Gut. 2004;53:1167–73. doi: 10.1136/gut.2003.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco DA, Selden RF. Non-viral gene therapy. Mol Med Today. 1995;1:314–21. doi: 10.1016/s1357-4310(95)80030-1. [DOI] [PubMed] [Google Scholar]
- Uehara T, Honda K, Hatano E, et al. Gene transfer to the rat biliary tract with the HVJ-cationic liposome method. J Hepatol. 1999;30:836–42. doi: 10.1016/s0168-8278(99)80137-8. [DOI] [PubMed] [Google Scholar]
- van der Merwe SM, Verhoef JC, Verheijden JH, et al. Trimethylated chitosan as polymeric absorption enhancer for improved peroral delivery of peptide drugs. Eur J Pharm Biopharm. 2004;58:225–35. doi: 10.1016/j.ejpb.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Vroman B, LaRusso NF. Development and characterization of polarized primary cultures of rat intrahepatic bile duct epithelial cells. Lab Invest. 1996;74:303–13. [PubMed] [Google Scholar]
- Wang J, Gao SJ, Zhang PC, et al. Polyphosphoramidate gene carriers: effect of charge group on gene transfer efficiency. Gene Ther. 2004;11:1001–10. doi: 10.1038/sj.gt.3302248. [DOI] [PubMed] [Google Scholar]
- Wu J, Nantz MH, Zern MA. Targeting hepatocytes for drug and gene delivery: emerging novel approaches and applications. Front Biosci. 2002;7:717–25. doi: 10.2741/A806. [DOI] [PubMed] [Google Scholar]
- Yang Y, Raper SE, Cohn JA, et al. An approach for treating the hepatobiliary disease of cystic fibrosis by somatic gene transfer. Proc Natl Acad Sci U S A. 1993;90:4601–5. doi: 10.1073/pnas.90.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Lee JS, Park SY, et al. Effects of selective oxidation of chitosan on physical and biological properties. Int J Biol Macromol. 2005;35:27–31. doi: 10.1016/j.ijbiomac.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–7. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Zhang G, Vargo D, Budker V, et al. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum Gene Ther. 1997;8:1763–72. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- Zhang X, Collins L, Sawyer GJ, et al. In vivo gene delivery via portal vein and bile duct to individual lobes of the rat liver using a polylysine-based nonviral DNA vector in combination with chloroquine. Hum Gene Ther. 2001;12:2179–90. doi: 10.1089/10430340152710522. [DOI] [PubMed] [Google Scholar]
- Zhang X, Sawyer GJ, Dong X, et al. The in vivo use of chloroquine to promote non-viral gene delivery to the liver via the portal vein and bile duct. J Gene Med. 2003;5:209–18. doi: 10.1002/jgm.340. [DOI] [PubMed] [Google Scholar]