Abstract
Apoptosis is an important event of the development of various organs. In this study, we used in situ terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) to visualize the temporal and spatial distribution of apoptosis in the developing medaka embryo, which is a useful model for developmental biology and genetics. Most of the apoptotic cells were distributed in the central nervous system and tailbud. In the brain and retina, most of the apoptosis occurred in the restricted period. In situ hybridization against caspase 3A and caspase 3B showed that these were distributed in the tailbud and the head, respectively. These results suggested that two types of caspase 3 were involved in apoptosis in different areas.
Keywords: apoptosis, caspase 3, TUNEL, medaka
I. Introduction
Developmental programmed cell death is a major event in the morphogenesis of almost all tissues and organs, along with cell proliferation, cell differentiation and coordinated cell migration [18, 20]. In development, excess cells are produced during morphogenesis and then removed. During this removal, dying cells reveal a highly conserved morphological process, showing cellular shrinkage, membrane blebbing, nuclear condensation and nuclear fragmentation sequentially. Such cell death is defined as apoptosis. Apoptosis is known to be under the control of a rigid regulated genetic program. In many animals, there are various signaling events for apoptosis. These signals, which may be derived from death receptors, oxidative stress or radiation damage, converge on caspase 3, which plays a direct terminator role. In medaka, two types of caspase 3 have been found [8, 9].
In the zebrafish, a useful model for developmental biology, many mutants have been reported, some of which exhibit abnormalities in developmental cell death [1, 5, 12]. Developmental apoptosis has been described in the zebrafish [2, 3]. However, there is a paucity of information on developmental cell death in this species.
The medaka is another useful model for studying developmental biology, neuroscience and genetics. The medaka embryo is highly transparent, so abnormalities of the abdominal organs are seen easily, compared with those in mice. An abundance of mutant medaka has been reported. Abnormalities of organs consist of abnormalities of growth, cell differentiation, and excess or lack of programmed cell death. Comparison of apoptosis in organogenesis between such mutants and wild-type medaka is important. However, the process of cell death in normal development has not been reported precisely in the medaka. In this study, the process of cell death is described quantitatively, and we describe the expression of each type of caspase 3, which is a key enzyme in apoptosis.
II. Materials and Methods
Fish strain and embryos
This study was performed using the Cab strain, a conventional wild-type strain of medaka (Oryzias latipes). Medaka eggs were collected from a laboratory colony. Embryos were incubated at 27°C after collection, and were staged according to Iwamatsu [7]. In order to compare the spontaneous cell death in development to the cell death under damage, we adopted ultraviolet (UV) light-exposed medaka embryos, in which apoptosis was induced by UV. For sampling of ultraviolet UV light-exposed medaka embryos, embryos at stage 17 were exposed to UV rays (UV-C; 254 nm) at an energy dose of 180 mJ/cm2 by a UV crosslinker (UV Stratalinker 2400, Stratagene, La Jolla, CA, USA). Twenty hours after UV-irradiation, the embryos were fixed as follows.
Tissue preparation
Embryos were fixed with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer for 4 hr at room temperature. After 3 hr of fixation, the embryos were dechorionated and the yolk sacs were removed in the PFA in phosphate buffer. The fixed embryos were washed twice in 0.1% Triton X-100 in phosphate-buffered saline (PBST) and gradually transferred to 100% methanol through a methanol series (25, 50, 75% methanol) for dehydration. Endogenous peroxidase activity was quenched with 0.3% H2O2 in 100% methanol for 30 min at room temperature.
Detection of apoptotic cells in whole mounts
Fragmented nuclear DNA of apoptotic cells was detected by the TUNEL method (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling), using an in situ apoptosis detection kit (Takara, Shiga, Japan). Rehydrated embryos that had passed through a methanol series (75, 50, 25% methanol), were rinsed three times with PBST. The embryos were incubated with labeling solution, which contained terminal deoxynucleotide transferase and dUTP-conjugated FITC, for 90 min at 37°C. After three washes with PBST, these samples were reacted with anti-FITC antibody conjugated with horseradish peroxidase at 4°C overnight. Apoptotic cells were visualized with 3,3'-diaminobenzidine tetrahydrochloride (Dojin, Kumamoto, Japan) as a chromogen. Stained embryos were dehydrated with acetone and embedded in Technovit 8100 (Heraeus Kulzer, Wehrheim, Germany). Serial sections, 5 µm thick, were taken every three sections and stained with methylgreen (Wako, Osaka, Japan). The number of TUNEL-positive cells was counted in sections from four embryos for each stage group.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed using digoxigenin (DIG)-labeled RNA probes against caspase-3A mRNA and caspase 3B mRNA. We used EST clone MF01FFA032p01 and MF01FSA0333p01, which corresponded to caspase 3A and caspase 3B, respectively [8, 9]. Embryos were fixed for 3 hr in 4% PFA at room temperature. The fixed embryos were dechorionated, washed twice in 0.1% Tween-20 in phosphate-buffered saline (PBSTw), and gradually transferred to 100% methanol through a methanol series (25, 50, 75% methanol) for dehydration. The dehydrated embryos were then rehydrated by transferring to PBST through a reversed methanol series (75, 50, 25% methanol). After washing twice in PBSTw, the embryos were treated with proteinase K (Roche, Basel, Switzerland, 10 µg/ml in PBSTw) for 5–10 min and refixed for 20 min in 4% PFA at room temperature. Following several washes with PBSTw, the embryos were prehybridized for 4 hr in hybridization buffer (50% formamide, 0.1% Tween-20, 50 µg/ml tRNA, 50 µg/ml heparin, 2×SSC) at 70°C, and then hybridized overnight with DIG-labeled sense or antisense RNA probe (100–200 ng/ml) in the hybridization buffer at 70°C. The hybridized embryos were washed four times in 50% formamide/2×SSC for 30 min each time at 70°C, twice in 2×SSC for 15 min each time at 70°C, twice in 0.2×SSC for 20 min each time at 70°C, and three times in PBSTw for 10 min each time at room temperature. After the embryos were blocked for 1 hr by incubating with 1% blocking agent (Roche) in PBSTw at room temperature, they were incubated overnight at 4°C with 1/5000-diluted anti-DIG antibody conjugated with alkaline phosphatase (Roche) in PBSTw. Following six washes in PBSTw for 30 min each time at room temperature, the embryos were further washed three times in NTMT (100 mM NaCl, 100 mM Tris-HCl pH 9.5, 50 mM MgCl2, 0.1% Tween-20) for 10 min each time at room temperature. Finally, the embryos were incubated in NTMT containing 3.5 µl NBT (4-nitro blue tetrazolium chloride, Roche, 100 mg/ml) and 3.5 µl BCIP (5-bromo-4-chloro-3-indolyl phosphate, Roche, 50 mg/ml) per 1 ml, in the dark. The colored embryos were washed several time in PBSTw and photographed in 2% methylcellulose.
III. Results
Detection of apoptotic cell death by TUNEL method
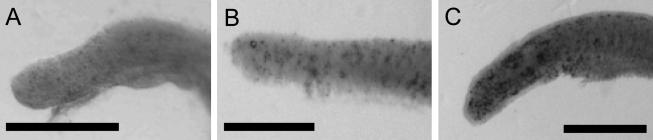
TUNEL-positive cells were detected at all stages at which embryos were sampled. Especially in the head area at stage 24 and tail bud at stage 27, many dark brown signals were observed by binocular microscope. In the head, relatively large particles were found in the brain (Fig. 1A). In contrast, in embryos exposed to UV light, TUNEL signals were detected in the whole embryo as many small dots. Large particles, as detected in normal development at stage 24, were not detected. (Fig. 1B). In the tailbud, TUNEL-positive cells increased from stage 23 to stage 27. TUNEL signals were detected in the whole embryo as many small dots (Fig. 2A, B, C).
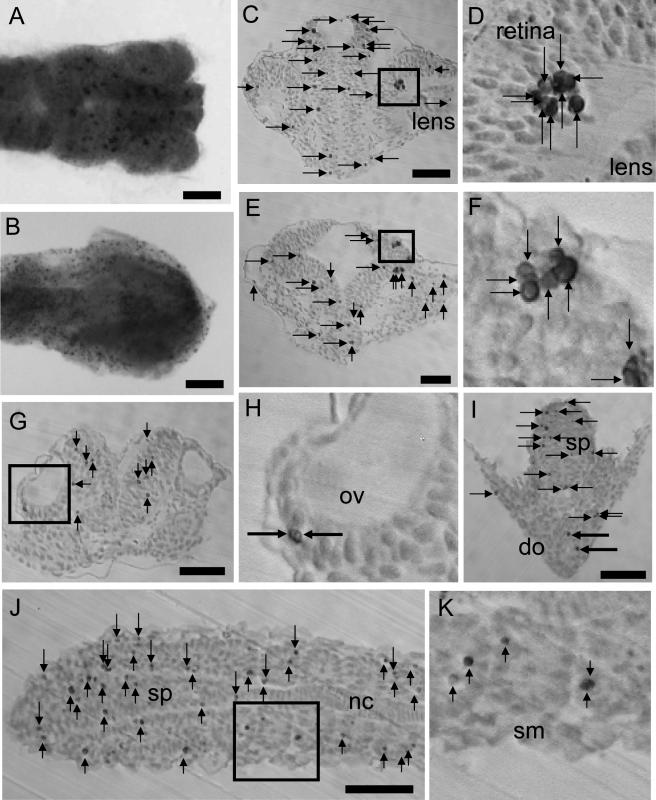
Fig. 1.
Visualization of apoptosis by TUNEL in the head. A: Stage 24 embryos observed from the dorsal view. B: TUNEL-positive cells after 20 hr of UV light exposure. C: Coronal section of forebrain containing eye. D: The square in C is magnified in this photograph. E: Coronal section of midbrain. F: The square in E is magnified in this photograph. G: Coronal section of hindbrain containing otic vesicle. H: The square in G is magnified in this photograph. I: Coronal section of spinal cord and digestive organ. J: Horizontal section of spinal cord and somite. K: The square in J is magnified in this photograph. ov, otic vesicle; do, digestive organ; sp, spinal cord; nc, notochord; sm, somite. Arrows indicate apoptosis counted. Large arrows in I indicate apoptosis counted in the digestive organ. Bar=50 µm.
Fig. 2.
Visualization of apoptosis by TUNEL in the tailbud. A: Embryo were observed from the lateral view at stage 22 (A), stage 24 (B) and stage 27 (C). Bar=500 µm.
Microscopic observation showed that TUNEL-positive cells had a rounded and or shrunken shape. Large number of TUNEL-positive cells were found in the central nervous system compared with other tissues. Especially at stage 24, apoptotic cells formed clusters in the brain and retina (Fig. 1D, F). The size of the contiguous clusters ranged from two to ten cells. In other areas containing the spinal cord and tailbud, clusters of apoptosis were not found. Most of TUNEL-positive cells were localized in the brain and spinal cord. A moderate number of apoptotic cells were detected in the retina and somites or undivided myotomes (Fig. 1J, K). Smaller number of apoptotic cells were detected in the otic vesicle (Fig. 1G, H), digestive organ (Fig. 1I), epidermal cells of yolk (data not shown), lens (Fig. 1C) and notochord (data not shown).
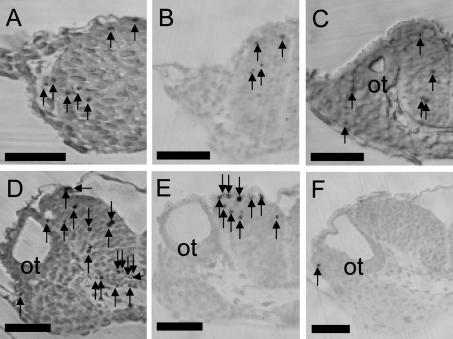
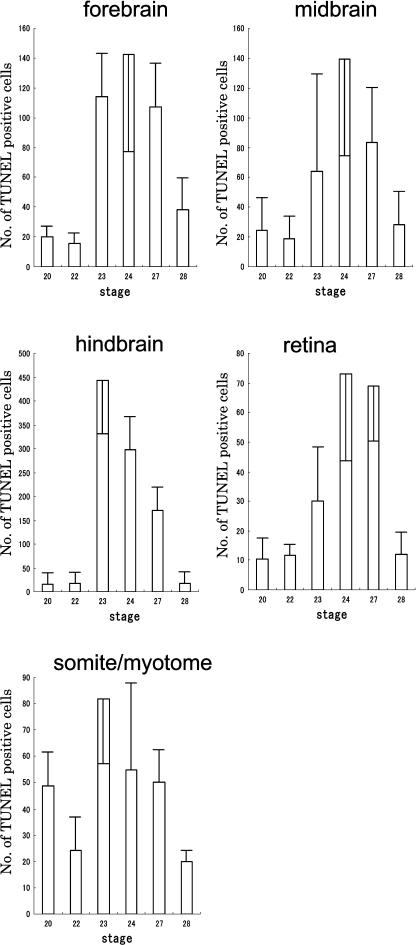
In the hindbrain, in which a large number of apoptotic cells were detected, the dynamics of apoptosis were observed in development. At stage 24, strongly TUNEL-positive cells were detected (Fig. 3). A quantitative study of TUNEL-positive cells showed that apoptotic cells located in the tissues, which were derived from neural tube, constituted 50–80% of the total TUNEL-positive cells in individual embryos. At stages 20 and 22, the numbers of TUNEL-positive cells in the forebrain, midbrain, hindbrain and retina were small; they increased at stage 23 or 24 to a peak, and then decreased (Fig. 4). In the central nervous system, along dorsal-ventral axis, distinct differences in the numbers of cell death was not detected. No distinct differences in the number of cell death was found between the area around the ventricle, in which neuronal progenitor cells were located, and other areas. In the somites and undivided myotome, a relative large number of apoptotic cells was observed at stage 23 (Fig. 4).
Fig. 3.
Apoptosis in the hindbrain. Coronal sections of hindbrain containing otic vesicle at stage 20 (A), 22 (B), 23 (C), 24 (D), 27 (E) and 28 (F). Arrows indicate apoptosis counted. Bar=50 µm.
Fig. 4.
Dynamics of apoptosis in each area. Apoptotic cells were counted on sections in each area separately at stage 20, 22, 23, 24, 27 and 28. Data are presented as mean profile±standard deviation, of four embryos at each stage.
Localization of caspase 3A mRNA and caspase 3B mRNA
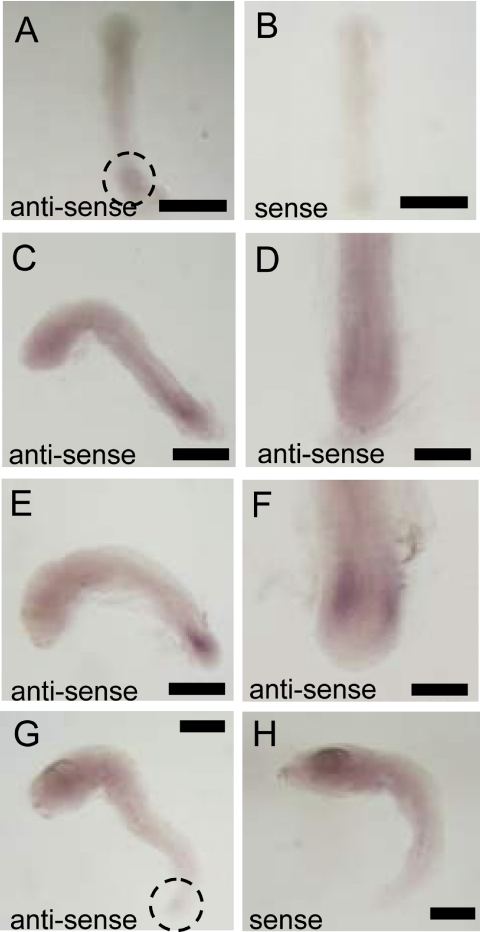
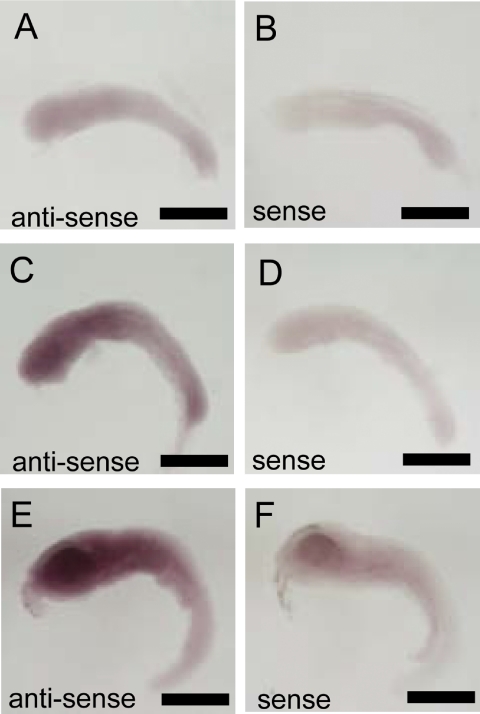
Caspase 3A mRNA was localized in the tailbud. It was detected weakly at stage 20 (Fig. 5A). Expression of caspase 3A mRNA was strongest at stage 24 (16-somite stage; Fig. 5E, F). At stages 27, expression of caspase 3A mRNA was weaker than that at stage 24 (Fig. 5G). In contrast, caspase 3B mRNA was widely expressed in the head area, and its expression was not restricted to particular organs or tissues. The posterior portion of the body expressed caspase 3B mRNA more weakly. Expression of caspase 3B mRNA increased from stage 20 to stage 27 (Fig. 6A, C, E).
Fig. 5.
In situ hybridization against caspase 3A mRNA. A: Using antisense probe in stage 20 embryo. B: Using sense probe in stage 20 embryo. C: Using antisense probe in stage 22 embryo from lateral view. D: Using antisense probe in stage 22 embryo from dorsal view. E: Using antisense probe in stage 24 embryo from lateral view. F: Using antisense probe in stage 24 embryo from dorsal view. G: Using antisense probe in stage 27 embryo. H: Using sense probe in stage 27 embryo. Bars=500 µm (A, B, C, E, G and H), 200 µm (D and F).
Fig. 6.
In situ hybridization against caspase 3B. A: Using antisense probe in stage 22 embryo. B: Using sense probe in stage 22 embryo. C: Using antisense probe in stage 24 embryo. D: Using sense probe at stage 24 embryo. E: Using antisense probe at stage 27 embryo. F: Using sense probe in stage 27 embryo. Bar=500 µm.
IV. Discussion
In this study, we first obtained an overview of TUNEL-positive cell death in medaka embryos. Two markedly TUNEL-stained areas were found, the head and the tailbud. Counting TUNEL-positive cell showed the dynamics of apoptotic cell death in the head area. In the forebrain, midbrain, hindbrain and retina, which are derived from neuronal tube, marked peaks of TUNEL-positive cells were detected. At stage 23 (12-somite stage) and stage 24 (16-somite stage), the number of apoptotic cells increased rapidly and reached a peak. Most of the apoptotic cell deaths in the head area were located in the brain. In the tailbud, a large amount of apoptosis was detected at stage 27. These results were similar to reports of apoptosis in the zebrafish embryo [3]. Clusters of apoptotic cells have also been reported in zebrafish embryos and other animals [3, 10]. In these reports, they are speculated to be caused by gap junctions, which might coordinate the activities of transient neuronal circuits before innervation of their axons [15, 16]. Such junctions might determine the fate of cells located near dying cells in the medaka embryo. It is thought that signals involved in cell death might be transferred from dying cells to neighboring cells though gap junctions. In the embryos exposed to UV light, in which cell death was not physiological, clusters of apoptotic cells were not found. While this raises the possibility that the mechanism of jointing cell fate might not have developed among apoptotic cells under UV irradiation, the reason for the presence of such clusters in restricted developmental periods is not precisely understood.
We also visualized medaka caspase 3 mRNA (caspase 3A mRNA and caspase 3B mRNA). Caspase 3A and caspase 3B were expressed in the tailbud and the head area, respectively. In general, caspase 3 is a key enzyme for apoptotic cell death, and is recognized as a terminator for cells to be killed. Interestingly, caspase 3-dominant areas coincided with the areas in which a large amount of apoptosis occurred. Thus, it is thought that cell death in medaka development depends on caspase 3A and caspase 3B. In the zebrafish, caspase 3 was expressed throughout the embryo at every developmental stage [21]. When caspase 3 was over-expressed by introducing its cDNA into zebrafish eggs, extensive apoptosis was induced [21]. It is speculated that, in the medaka as in the zebrafish, the developmental expression patterns of the caspase 3 gene regulate the spatial distribution of apoptotic cells in the head area and tailbud. However, the largest number of TUNEL positive-cells appeared at stage 24 in the head, with the peak of expression of caspase 3B in the head occurring at stage 28. Hence, the amount of caspase 3 mRNA alone did not strictly determine the frequency of cell death in development.
In caspase 3 knock-out mice, excess neuronal cells were produced to close the third ventricle [10, 17, 22]. This suggests that neuronal cell death in the developing forebrain depends on caspase 3. The excess cells were neuronal progenitor cells [4]. However, in this study, the area around the ventricle, in which many neuronal progenitor cells were located, did not contain more apoptotic cells than other areas in the brain.
In the developing nervous system, the machinery of cell death has not been elucidated completely. Regarding the triggers of apoptosis, bone morphogenic protein, which is produced from the roof plate of the neural tube, and sonic hedgehog, which is produced from the floor plate of the neural tube and from the notochord, have been reported to be a death signal and a survival signal, respectively [6, 11, 13, 19]. Cell-to-cell communication mediated by Notch was reported to function as a survival signal [14]. In the medaka, the contribution of these factors to neural cell death is unknown. Future studies can be expected to elucidate neural cell death in the medaka brain.
V. Acknowledgments
We are grateful to Dr K. Naruse (Tokyo University) for kindly providing the EST clone DNA of caspase 3A and caspase 3B, and we thank K. Oka for his help in laboratory work. This research was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
VI. References
- 1.Abdelilah S., Mountcastle-Shah E., Harvey M., Solnica-Krezel L., Schier A. F., Stemple D. L., Malicki J., Neuhauss S. C., Zwartkruis F., Stainier D. Y., Rangini Z., Driever W. Mutations affecting neural survival in the zebrafish. Danio rerio. Development. 1996;123:217–227. doi: 10.1242/dev.123.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Biehlmaier O., Neuhauss S. C., Kohler K. Onset and time course of apoptosis in the developing zebrafish retina. Cell Tissue Res. 2001;306:199–207. doi: 10.1007/s004410100447. [DOI] [PubMed] [Google Scholar]
- 3.Cole L. K., Ross L. S. Apoptosis in developing Zebrafish embryo. Dev. Biol. 2001;240:123–142. doi: 10.1006/dbio.2001.0432. [DOI] [PubMed] [Google Scholar]
- 4.D’Sa-Eipper C., Roth K. A. Caspase regulation of neuronal progenitor cell apoptosis. Dev. Neurosci. 2000;22:116–124. doi: 10.1159/000017433. [DOI] [PubMed] [Google Scholar]
- 5.Furutani-Seiki M., Jiang Y. J., Brand M., Heisenberg C. P., Houart C., Beuchle D., van Eeden F. J., Granato M., Haffter P., Hammerschmidt M., Kane D. A., Kelsh R. N., Mullins M. C., Odenthal J., Nusslein-Volhard C. Neural degeneration mutants in the zebrafish. Danio rerio. Development. 1996;123:229–239. doi: 10.1242/dev.123.1.229. [DOI] [PubMed] [Google Scholar]
- 6.Israsena N., Kessler J. A. Msx2 and p21(CIP1/WAF1) mediate the proapoptotic effects of bone morphogenetic protein-4 on ventricular zone progenitor cells. J. Neurosci. Res. 2002;69:803–809. doi: 10.1002/jnr.10362. [DOI] [PubMed] [Google Scholar]
- 7.Iwamatsu T. Stages of normal development in the medaka. Oryzias latipes. Zool. Sci. 1994;11:825–839. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Kohara Y., Shin-i T., Kimura T., Narita T., Jindo T., Takeda H. BJ724549 MF01FFA cDNA Oryzias latipes cDNA clone MF01FFA032p09 3', mRNA sequence. Available at: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=45265641 2001
- 9.Kohara Y., Shin-i T., Kimura T., Narita T., Jindo T., Takeda H. BJ724864 MF01FFA cDNA Oryzias latipes cDNA clone MF01FFA033p01 3', mRNA sequence. Available at: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=45265956 2001
- 10.Kuan C. Y., Roth K. A., Flavell R. A., Rakic P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Helms A. W., Johnson J. E. Distinct activities of Msx1 and Msx3 in dorsal neural tube development. Development. 2004;131:1017–1028. doi: 10.1242/dev.00994. [DOI] [PubMed] [Google Scholar]
- 12.Malicki J., Neuhauss S. C., Schier A. F., Solnica-Krezel L., Stemple D. L., Stainier D. Y., Abdelilah S., Zwartkruis F., Rangini Z., Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- 13.Miao N., Wang M., Ott J. A., D’Alessandro J. S., Woolf T. M., Bumcrot D. A., Mahanthappa N. K., Pang K. Sonic hedgehog promotes the survival of specific CNS neuron populations and protects these cells from toxic insult in vitro. J. Neurosci. 1997;17:5891–5899. doi: 10.1523/JNEUROSCI.17-15-05891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi K., Kamakura S., Isazawa Y., Yoshimatsu T., Kuida K., Nakafuku M., Masuyama N., Gotoh Y. Notch promotes survival of precursor cells via mechanisms distinct from those regulating neurogenesis. Dev. Biol. 2004;276:172–184. doi: 10.1016/j.ydbio.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 15.Patenostro M. A., Reyher C. K., Brunjes P. C. Intercellular junctions of Lucifer yellow into lightly fixed mitral cells reveal neuronal dye-coupling in the developing rat olfactory bulb. Brain Res. Dev. Brain Res. 1995;84:1–10. doi: 10.1016/0165-3806(94)00146-q. [DOI] [PubMed] [Google Scholar]
- 16.Peinado A., Yuste R., Katz L. C. Gap junctional communication and the development of local circuits in neocortex. Cereb. Cortex. 1993;3:488–498. doi: 10.1093/cercor/3.5.488. [DOI] [PubMed] [Google Scholar]
- 17.Roth K. A., Kuan C., Haydar T. F., D’Sa-Eipper C., Shindler K. S., Zheng T. S., Kuida K., Flavell R. A., Rakic P. Epistatic and independent functions of caspase-3 and Bcl-X(L) in developmental programmed cell death. Proc. Natl. Acad. Sci. U S A. 2000;97:466–471. doi: 10.1073/pnas.97.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders E. J., Wride M. A. Programmed cell death in development. Int. Rev. Cytol. 1995;163:105–173. doi: 10.1016/s0074-7696(08)62210-x. [DOI] [PubMed] [Google Scholar]
- 19.Solloway M. J., Robertson E. J. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 20.Thomaidou D., Mione M. C., Cavanagh J. F., Parnavelas J. G. Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J. Neurosci. 1997;17:1075–1085. doi: 10.1523/JNEUROSCI.17-03-01075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabu T., Kishi S., Ozaki T., Yamashita M. Characterization of zebrafish caspase-3 and induction of apoptosis through ceramide generation in fish fathered minnow tailbud cells and zebrafish embryo. Biochem. J. 2001;360:39–47. doi: 10.1042/0264-6021:3600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida H., Kong Y. Y., Yoshida R., Elia A. J., Hakem A., Hakem R., Penninger J. M., Mak T. W. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]