Abstract
A major limitation of studies of the relevance of telomere length to cancer and age-related diseases in human populations and to the development of telomere-based therapies has been the lack of suitable high-throughput (HT) assays to measure telomere length. We have developed an automated HT quantitative telomere FISH platform, HT quantitative FISH (Q-FISH), which allows the quantification of telomere length as well as percentage of short telomeres in large human sample sets. We show here that this technique provides the accuracy and sensitivity to uncover associations between telomere length and human disease.
Keywords: human disease, quantitative FISH, telomeres
Telomeres are special structures at the ends of eukaryotic chromosomes that protect them from degradation and DNA repair activities (1). Telomerase is a reverse transcriptase that elongates telomeres in those cells where it is expressed, such as germ cells and stem cell populations (2, 3). Telomerase activity levels in adult tissues, however, are not sufficient to prevent telomere shortening associated to cell division, eventually leading to chromosomal instability and compromising tissue function (4–6). The notion that telomere shortening leads to loss of organismal viability is supported by premature aging phenotypes in late generation telomerase-deficient mice (5, 6) and by humans with decreased levels of telomerase and short telomeres, such as some cases of Dyskeratosis congenita and aplastic anemia (7, 8). In contrast, cancer cells maintain their telomeres by up-regulating telomerase or by activating alternative telomere lengthening mechanisms (9, 10). Despite the relevance of telomere length to cancer and age-related diseases, on a few studies validated telomere length as a predictor for organismal fitness in human populations (11–13). Similarly, only a few telomere-based anticancer therapies have been developed to date by screening large compound libraries (14). These important drawbacks in telomere research have been due, at least in part, to the lack of fast and reliable high-throughput (HT) quantitative platforms to measure telomere length in large sample sets.
Results
Design and Validation of an HT Telomere Length Quantification Method.
Quantitative fluorescence in situ hybridization (Q-FISH) of telomeres on metaphase spreads has been extensively used to obtain quantitative information on telomere length distributions (15–17). Also, the low detection limit of Q-FISH (<0.1 kb of telomere repeats) allows quantification of critically short telomeres (15–17), which is particularly relevant, because the frequency of critically short telomeres, rather than the mean telomere length, is determinant for telomere dysfunction (18, 19). An important drawback of the conventional Q-FISH technique on metaphases has been, however, that it is extremely laborious and time-consuming, thus precluding its application to human population studies and HT screenings. Here, we have developed an automated HT Q-FISH telomere-length analysis platform. Supporting information (SI) Fig. 5A shows the set-up and work-flow of this HT Q-FISH method, which builds on conventional Q-FISH (15–17) and combines labeling of telomeres in interphase nuclei, using a fluorescent peptide nucleic acid (PNA) probe against telomeric repeats, with automated HT microscopy in 96-well plates (see Materials and Methods). With this procedure, an entire 96-well plate can be processed in about 2 h.
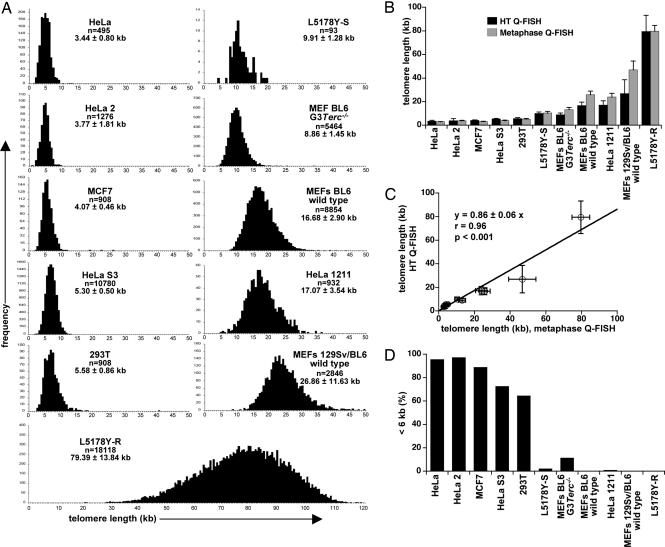
To validate automated HT Q-FISH, we chose a panel of mouse and human cell lines and determined their telomere lengths either by conventional telomere Q-FISH on metaphase spreads (SI Fig. 5B for representative metaphases) or HT Q-FISH (SI Fig. 5C for representative images of interphase nuclei, Fig. 1A for histograms showing the telomere length distribution of each cell line after HT Q-FISH). In all cases, telomere fluorescence values were converted into kb (see Materials and Methods) to compare HT Q-FISH results with those obtained by Q-FISH as well as other techniques (see below). Importantly, the mean telomere length values, as determined by HT Q-FISH, correlated excellently with the telomere length values that were obtained by using conventional Q-FISH analysis (highly significant, P < 0.001; Fig. 1 B and C), indicating that HT Q-FISH is as powerful and accurate as conventional Q-FISH for average telomere length determinations. Of notice, the larger standard deviation values of HT Q-FISH compared with Q-FISH on metaphases are likely to be because Q-FISH measures only a subpopulation of cells (mitotic cells), whereas HT Q-FISH measures all cells within the previously established size and shape limits of the DAPI-stained nucleus or the region of interest (ROI) (see Materials and Methods for details). Importantly, HT Q-FISH allows generation of telomere length frequency histograms and determination of the percentage of cells with critically short telomeres. In particular, we measured the percentage of nuclei with telomeres shorter than 6 kb (<6 kb) in all cell lines including third generation (G3) telomerase-deficient (Terc−/−) mouse embryonic fibroblasts (MEF), and the corresponding wild-type controls (Fig. 1D). As expected, human cell lines showed an increased percentage of nuclei with telomeres with a length <6 kb compared with mouse cell lines, and G3 Terc−/− MEF showed increased percentage of nuclei with telomeres <6 kb compared with the wild-type controls, in agreement with shorter telomeres in these mice (Fig. 1D). To further validate HT Q-FISH, we compared side-by-side HT Q-FISH results with those obtained with an independent Q-FISH technique based on flow cytometry, called “Flow-FISH” (20). We found a very good correlation between HT Q-FISH and Flow-FISH results (highly significant, P < 0.001) (SI Fig. 6). Finally, we also validated HT Q-FISH with an independent technique not based on fluorescence, terminal restriction fragment analysis, which is based on Southern blot analysis of MboI digested DNA (see Materials and Methods). Again, HT Q-FISH values nicely correlated with terminal restriction fragment values (highly significant, P < 0.001) (SI Fig. 7).
Fig. 1.
Validation of the HT Q-FISH technique. (A) HT Q-FISH histograms of telomere length distributions of the indicated cell lines. Telomere length values are given as mean ± SD, and the number of nuclei analyzed per sample (n) is indicated. (B) Mean telomere length values ± SD of the indicated cells, determined either by conventional metaphase Q-FISH (gray bars) or HT Q-FISH analysis of interphase nuclei (black bars). (C) Linear regression analysis of the correlation between mean telomere length values ± SD obtained with HT Q-FISH and metaphase Q-FISH. (D) Analysis of percentage of nuclei with short (<6 kb) telomeres by HT Q-FISH analysis of interphase nuclei. Note that G3 Terc−/− MEF show an increased percentage of nuclei with short telomeres (<6 kb) compared with wild type controls.
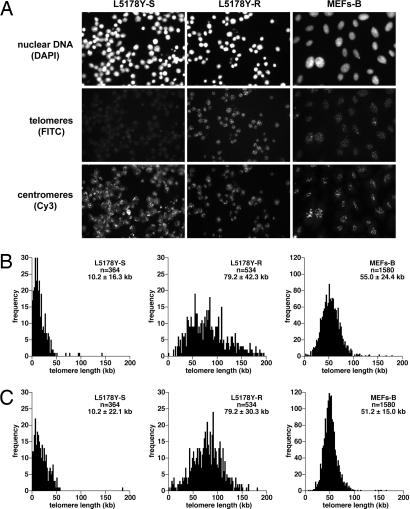
We have further developed HT Q-FISH to simultaneously capture and analyze telomere and centromere (major satellite) fluorescence on an individual cell-to-cell basis (see Materials and Methods), which allows normalization of average telomere fluorescence to the centromeric major satellite fluorescence for every individual cell. As shown in Fig. 2, both average telomere length as well as telomere length frequency histograms were very similar in different mouse cell lines [MEF-B and lymphocyte cell lines L5178Y-S (10.2 kb) and L5178Y-R (79.7 kb)] when considering either the telomere fluorescence values alone or after their normalization by mouse major satellite fluorescence in the same cell.
Fig. 2.
Normalization of HT Q-FISH. (A) Representative HT microscopy images of interphase nuclei stained for nuclear DNA, mouse major satellite repeats, and telomeres with DAPI-, FITC-, and Cy3-labeled PNA oligonucleotides, respectively. (B) HT Q-FISH histograms of telomere length distributions of interphase nuclei from murine lymphocytes with short (L5178Y-S cells) and long (L5178Y-R cells) telomeres and of mouse MEFs-B (MEFs 129Sv/BL6 B). Telomere length was calculated without normalization with major satellite signals. (C) Telomere length histograms after normalization with major satellite fluorescence in each nuclei. Telomere length values are given as mean ± SD, and the number of nuclei analyzed per sample (n) is indicated. Very similar distributions were obtained with or without centromeric normalization.
All together, these results obtained with a large panel of human and mouse cell lines validate the HT Q-FISH method as a reliable and robust tool for HT telomere length determinations.
Impact of Age, Gender, and Geographic Factors on Human Telomere Length.
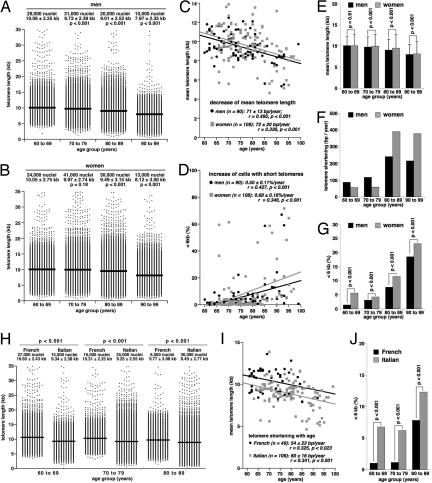
To validate HT Q-FISH directly on human samples, we analyzed peripheral blood lymphocytes from 198 healthy donors (90 men and 108 women) ranging between 60 and 99 years of age (see Materials and Methods). Human peripheral blood lymphocytes are generally quiescent (G0 phase of the cell cycle); therefore, the HT Q-FISH values obtained represent telomere length in cells with a G0 DNA content. To normalize between different 96-well plates, we used the L5178Y-S mouse cell line as standard, because telomere length distribution in this cell has been shown to be stable (19, 21, 22) and to reflect on the normal telomere length distribution of humans with telomeres ranging from 6–20 kb and showing an average telomere length of ≈10 kb (Fig. 1A) (17). Even though HT Q-FISH permits the analysis of thousands of telomere signals per sample (Fig. 1A), to economize data acquisition and processing, we only analyzed 1,000 nuclei per human sample, which allows both the reliable measurement of mean telomere length and the detection of nuclei with short (<6 kb) telomeres (see SI Fig. 8 for analysis of lymphocytes from a 71-year-old male healthy donor). Telomere length distributions in these donors [who were organized into 4 age groups (60–69, 70–79, 80–89, and 90–99 years of age)] show a progressive decline of mean telomere length and the appearance of cells with short telomeres in the older age groups (Fig. 3 A and B). Mean telomere length of lymphocytes from both male and female donors declined with age at a similar overall rate of 71–72 bp/year (Fig. 3C), which is in the range previously reported by others (11, 23, 24). Furthermore, HT Q-FISH showed for the first time that the abundance of nuclei with short (<6 kb) telomeres increases significantly with age (Fig. 3D). Interestingly, men and women displayed different profiles of telomere erosion. In particular, at ages ≥70 years, the mean telomere length of female lymphocytes was slightly but significantly higher than in male samples in each of the age groups studied (Fig. 3E). Concomitantly, the rates of telomere shortening were less dramatic in women of ages 60–80 years than in age-matched men (Fig. 3F). This trend was inverted, however, in women >80 years of age compared with men of the same age (Fig. 3F). This increased rate of telomere shortening in elderly women is also reflected by an increased percentage of nuclei with short (<6 kb) telomeres in female lymphocytes as compared with age-matched male controls (Fig. 3G).
Fig. 3.
Effect of age, gender, and geographic factors on telomere length in peripheral blood lymphocytes from elderly donors. (A and B) Distribution of telomere length values in lymphocytes from male (A) and female (B) donors are grouped into the indicated age groups. Mean telomere length is indicated by a straight line. The total number of nuclei analyzed (1,000 per donor) and the mean telomere length ± SD is indicated. Statistical significance was assessed by using the Wilcoxon–Mann–Whitney rank sum test. (C and D) Linear regression analysis was used to assess the correlation between age and mean telomere length (C) or percentage of nuclei with telomeres <6 kb (D) in peripheral blood lymphocytes of these donors. (E) Mean telomere length values ± SD of male and female donors in the indicated age groups. Statistical significance was calculated using the Wilcoxon–Mann–Whitney rank sum test. (F) Rate of telomere shortening, expressed as loss of bp of telomere sequences per year, in male and female lymphocytes of the indicated age groups. (G) Percentage of nuclei with telomeres <6 kb in the indicated age groups. Fisher's Exact test was used to evaluate statistical significance. (H) Distribution of telomere length values in lymphocytes from French and Italian donors grouped into the indicated age groups. Mean telomere length is indicated by a straight line. The total number of nuclei analyzed (1,000 per donor) and the mean telomere length ± SD is indicated above each telomere length distribution. Statistical significance was calculated using the Wilcoxon–Mann–Whitney rank sum test. (I) Linear regression analysis of the correlation between age and mean telomere length in lymphocytes from these donors. (J) Percentage of nuclei with telomeres <6 kb in lymphocytes from Italian and French donors of the indicated age groups. The Fisher's exact test was used to evaluate statistical significance.
Telomere length is an inherited trait (25) and can be modified by environmental factors such as stress, obesity, and smoking (11, 12). Thus, epidemiological studies on human populations with different genetic backgrounds or life styles emerge as a further potentially important application of the HT Q-FISH method. A comparison among 49 French subjects (28 men, 21 women) 60–89 years of age, and 106 age-matched Italians (40 men, 66 women) revealed significantly longer telomeres in the French subjects for all 3 age groups studied (Fig. 3H). Also, telomere shortening with age was lower in French (54 bp/year) than in Italian donors (60 bp/year) (Fig. 3I). Accordingly, the percentage of nuclei with telomeres <6 kb was significantly decreased in lymphocytes from French subjects compared with Italian donors in all age groups studied (Fig. 3J). These results indicate that telomere length is associated with geographic factors, which in turn may be the result of both environmental and genetic factors, thus supporting the notion that telomere length is inherited or may be determined by an inherited factor (25) and that telomere length may be influenced by environmental factors (12). It is tempting to discuss these data in light of the exceptionally low incidence of coronary heart disease in the French population despite high intake of cholesterol and saturated fat, the “French paradox” (26), particularly because shorter telomeres have been shown to be predictive of higher mortality by heart disease (13).
We confirmed HT Q-FISH results by measuring telomere length in a subset of French and Italian female individuals (63–67 years old), using terminal restriction fragment analysis, which is the most commonly used technique for human population studies (23). As shown in SI Fig. 9A, the telomere length values obtained by terminal restriction fragment showed a very good correlation with those obtained by HT Q-FISH for each individual sample. Furthermore, both techniques illustrated longer telomeres in the French cohorts than in the Italian cohorts (SI Fig. 9B).
Association Between Telomere Length and Age-Related Psychosocial Parameters.
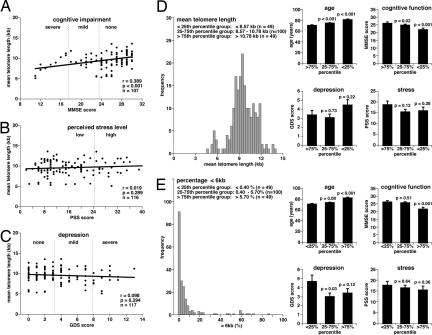
Finally, we used the HT Q-FISH method to address whether age-related psychosocial parameters, such as cognitive function [Mini Mental State Examination (MMSE)], perceived stress [perceived stress scale (PSS)], and depression [geriatric depression scale (GDS)] are associated with telomere length. These three parameters were extracted from psychological tests of elderly donors between 60 and 99 years of age (for details see Materials and Methods). Expectedly, MMSE declined with age, indicating an age-dependent loss of cognitive function, and GDS increased with age, suggesting an age-dependent increase of depression, although we did not detect any significant association of PSS with age (SI Fig. 10). Interestingly, mean telomere length in peripheral blood lymphocytes from these donors significantly correlated with their MMSE score, suggesting that telomere shortening is associated with cognitive impairment (Fig. 4A). Because cognitive impairment also correlated with age (SI Fig. 10A), we tested for an independent association between telomere length and cognitive impairment (MMSE score), performing multivariate ANOVA with adjustment for age (SI Fig. 10 D and E). Analysis of different age ranges revealed that there was a significant and independent (i.e., after adjustment for age) association of telomere length with cognitive impairment in subjects 60–69 years of age (SI Fig. 10D). Of notice, within this age range we did not observe any significant impact of age on telomere length (SI Fig. 10D). Similar results were obtained for the correlation between the percentage of nuclei with short telomeres (< 6 kb) and cognitive impairment (SI Fig. 10E). Finally, we did not detect any significant correlation between telomere length and perceived stress levels (Fig. 4B) and depression (Fig. 4C) in this patient cohort, even though depression was also increased with age in these patients (SI Fig. 10C). This finding is in contrast with a previous study showing that decreased telomerase activity and decreased telomere length significantly correlate with increased psychological stress in a group of young mothers with chronically ill children (11). A possible explanation for these differences may be the marked differences in age and in the type of stress between both patient cohorts.
Fig. 4.
Correlation between telomere shortening and age, age-related cognitive impairment, stress, and depression. (A–C) Linear regression analysis was used to assess the correlation between cognitive impairment (A), perceived stress level (B), depression (C), and mean telomere length in lymphocytes from elderly Europeans 60–99 years of age. Mean telomere length values for each subject were obtained by HT Q-FISH analysis of 1,000 interphase nuclei per subject. The MMSE was used to assess cognitive mental status. The PSS was applied to measure the perception of stress, and depressive symptoms in the elderly were quantified with the GDS. (Materials and Methods). (D) Histogram showing the mean telomere length distribution of lymphocytes from 60- to 99-year-old donors. The 25th and 75th percentile values of this distribution were used to group donors into subjects with long (>10.78 kb), intermediate length (8.57–10.78 kb), and short (<8.57 kb) telomeres. The three groups (the graphs shown to the right) were analyzed for age, cognitive function (MMSE score), depression (GDS score), and perceived stress (PSS score). Data are mean values ± SEM, and the statistical significance of differences was assessed using the Wilcoxon–Mann–Whitney rank sum test. (E) Histogram showing the distribution of percentage of nuclei with telomeres <6kb in lymphocytes of the 60- to 99-year-old donors. The 25th and 75th percentile values of the distribution were used to group donors into subjects with a high (>5.7%), intermediate (0.4–5.7%), and low (<0.4%) abundance of nuclei with short telomeres, corresponding to the >75th percentile, 25–75th percentile, and <25th percentile groups, respectively. The three groups (graphs shown to the right) were analyzed for age, cognitive function (MMSE score), symptoms of depression (GDS score), and perceived stress (PSS score). Data are mean values ± SEM, and statistical significance was assessed using the Wilcoxon–Mann–Whitney rank sum test.
Together, these data suggest that telomere length can be used as a predictor of age and age-related symptoms such as cognitive impairment. To further corroborate this association, HT Q-FISH data for 198 elderly subjects 60–99 years of age range were used to determine 25th and 75th percentiles for telomere length (Fig. 4D) and the percentage of nuclei with short (<6 kb) telomeres (Fig. 4E). The 25th and 75th percentile limits of these distributions were used to divide subjects into three groups, namely the <25th (49 subjects), 25–75th (100 subjects), and >75th (49 subjects) percentile groups and to analyze whether these groups differed from each other in age and in age-related psychosocial parameters (MMSE, GDS, and PSS). Fig. 4D shows that mean telomere length values in the 25–75th (8.57–10.78 kb) and <25th (<8.57 kb) percentile groups associated significantly with increased age as compared with the >75th percentile group with the highest telomere length values (>10.78 kb). Assignment of subjects to the 25–75th and <25th percentile groups further correlated with decreased cognitive function as compared with the >75th percentile group. We did not find, however, any significant associations of mean telomere length with depression and stress scores. Similarly, an increased percentage of nuclei with telomeres <6 kb was found to associate with increased age (Fig. 4E). In particular, in 49 out of 198 subjects (<25th percentile) we detected <0.4% of interphase nuclei with a mean telomere length value <6 kb, in 49 out of 198 subjects (>75th percentile) the abundance of <6 kb nuclei was >5.7%, and in 100 subjects (25–75th percentile) the abundance of <6 kb nuclei ranged from 0.4% to 5.7%. Individuals with intermediate (25–75th percentile group) and elevated (>75th percentile group) levels of nuclei had a significantly higher average age than those in the <25th percentile group, i.e., subjects with lymphocyte populations containing low levels (<0.4%) of nuclei with short telomeres. A highly significant correlation was also found between the abundance of short telomeres and impaired cognitive function (Fig. 4E). Together, these data indicate that telomere erosion, measured either as decreased mean telomere length or increased abundance of cells with short telomeres, correlates significantly with age and age-related cognitive impairment. These results are in agreement with recent reports also showing that telomere length is predictive of cognitive impairment (27, 28).
Discussion
Here we report on the development and validation of a HT quantitative telomere FISH assay of interphase nuclei, HT Q-FISH, which provides the same high quality average telomere length data as conventional Q-FISH analysis of metaphase spreads on slides. Similarly to metaphase Q-FISH, HT Q-FISH is performed with very high stringency hybridization conditions, which only allow detection of perfect TTAGGG repeats, therefore reflecting on the length of distal telomeric repeats and not of subtelomeric repeats. Furthermore, HT Q-FISH allows the determination of the frequency of cells with very short telomeres within a given population, which in turn are indicative of telomere dysfunction. Because HT Q-FISH is performed on tissue culture plates, it can be used for any cell type that normally grows in culture in or that can be attached to the plate (i.e., polylysine coated plates in the case of human lymphoid cells). HT Q-FISH generates large data sets for each sample, conferring high accuracy and statistical power to this method. Importantly, HT Q-FISH allows the simultaneous determination of telomere length in 96 samples, with at least 1,000 nuclei per sample being captured in 2 h, therefore minimizing error due to intersession variability of signal intensity and lack of homogeneity of the light field over time, which are common drawbacks of conventional Q-FISH. HT Q-FISH, thus, represents a crucial improvement to the application of telomere FISH analysis to large-scale studies. This advance is illustrated by our analysis of human peripheral blood lymphocytes, revealing that age, gender, and geographic factors impinge on telomere length. We also show that telomere shortening correlates significantly with cognitive impairment in elderly subjects. We, therefore, anticipate that the HT Q-FISH method will be a powerful tool to expand our knowledge on the role of telomere length in human disease and a powerful prognostic tool in the context of human population and epidemiological studies. We also envision that the HT Q-FISH technique will provide a useful screening platform for the development of telomere-based therapeutic strategies.
Materials and Methods
Cells.
For set-up and validation of HT Q-FISH, we used the following panel of human and mouse cell lines (ATCC numbers in parenthesis): two HeLa cell lines, HeLa and HeLa2 (CCL-2); MCF-7 (HTB-22); 293T (CRL-11268); HeLa 1211 (HeLaI.211); mouse L5178Y-S (CRL-9518); mouse L5178Y-R (CRL-1722); primary MEF from wild-type and third generation G3 Terc−/− telomerase-deficient mice in a C57BL6 background; and primary MEF from wild-type mice on a mixed C57Bl6/SV129 background. Human lymphocyte samples were obtained from the Italian National Research Center on Aging (Ancona, Italy) in the context of the European research consortium ZincAge.
Telomere Restriction Fragment Analysis.
Telomere restriction fragment analysis was performed as described by using MboI digested DNA (5).
Flow-FISH Analysis.
Telomere fluorescence of at least 5,000 cells, gated at G1/G0, was measured by using a FACScalibur cytometer with CellQuest software (BD Biosciences, Franklin Lakes, NJ) as described in refs. 20 and 29. Telomere fluorescence values were converted into kb by external calibration with the L5178Y-S and L5178Y-R lymphocyte cell lines with known telomere lengths of 10.2 kb and 79.7 kb, respectively (30).
Q-FISH on Metaphases (Metaphase Q-FISH).
Q-FISH on metaphases was performed as described in refs. 17, 18, and 30). Cy3 and DAPI images were captured at 100x magnification using a COHU CCD camera on a Leica DMRA (Leica, Heidelberg, Germany) microscope. Telomere fluorescence signals were integrated from 10 metaphases and quantified by using the TFL-TELO program (gift from Peter Lansdorp, Vancouver, Canada) (17). Telomere fluorescence values were converted into kb by external calibration with the L5178Y-S and L5178Y-R lymphocyte cell lines with known telomere lengths of 10.2 and 79.7 kb, respectively (30).
Sample Preparation for HT Q-FISH.
Cells were cultured in clear-bottom black-walled 96-well plates (Greiner, Longwood, FL). In the case of lymphocytes, plates were precoated with a 0.001% (wt/vol) (poly)l-lysine solution (Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C. PolyL-lysine was removed before cell addition (30,000–90,000 lymphocytes/well). When cultures reached subconfluency in the case of adherent cells, or in the case of lymphocytes when cells attached to the wells, cells were washed three times with PBS and then fixed during 1 h at room temperature (RT) by slowly filling up the wells with methanol/acetic acid (3/1, vol/vol) three times. After fixation, cells were processed as described for metaphase Q-FISH. Wells were then sealed (Alumaseal; Sigma–Aldrich) and stored at 4°C in the dark. Samples were processed by HT microscopy as described in HT Microscopy within 48 h after sample preparation.
Where indicated (Fig. 2), HT Q-FISH telomere length values were normalized by the fluorescence intensity of centromere repeats in each nuclei. In particular, interphase nuclei were processed as described Sample Preparation for HT Q-FISH, except that samples were simultaneously stained for telomeres with an FITC-labeled PNA probe [(C3TA2)3] and for centromeres with a Cy3-labeled PNA probe directed against mouse major satellite repeats (5′-TCG CCA TAT TCC AGG TC-3′).
HT Microscopy.
Quantitative image acquisition and analysis were performed on a BD Pathway Bioimager (BD Biosciences), using the Attovision software, Version 1.14 (BD Biosciences). Images were captured for each well, using a UAPO/340C ×20/0.75 objective, in the DAPI channel (excitation, 380/10 nm; epifluorescence dichroic, Fura/FITC; and emission, 435LP) for nucleus staining, and in the Cy3 channel (excitation, 548/20 nm; epifluorescence dichroic, Fura/TRITC; emission, Fura/TRITC) for telomere staining. Settings for exposure and gain remained constant between captures. To eliminate eventual local variations in signal intensity, for each channel, nine independent images (3 × 3 mounting) were captured at three different positions within each well. After image acquisition, the DAPI image was used to define a nuclear area or ROI for each cell to measure telomere fluorescence intensity in the Cy3 image within each predefined ROI. In particular, the automated image acquisition procedure requires the previous setting of parameters that define the characteristics of a set of image pixels that are interpreted by the software as a single DAPI-stained cell nucleus; i.e., to identify the ROI within an image. The definition of a ROI is based on the assumption that a cell nucleus is constituted of a common, clearly delimited, ellipsoid area of adjacent pixels with signal intensities above the background and within the size and shape limits typical of the analyzed cell type (31). Additional algorithms are applied to decompose cell aggregates into single cell nuclei (watershed segmentation). The reliability of ROI identification was validated for each cell type by testing that >95% of automatically identified nuclei were in concordance with visual inspection by a human observer. Results were exported from the Attovision software (BD Biosciences) as text files, containing for each well the list of all ROIs and their corresponding Cy3 fluorescence intensities, expressed as average value per pixel within a given ROI. A data analysis macro was written in-house using Visual Basic for Microsoft Excel 2000 (Microsoft, Redmond, WA) to manage and export data and build frequency histograms (available upon request from the authors). Telomere fluorescence intensity values were normalized between plates, using the mean for intensity values of L5178Y-S cells that had been included as calibration standard in each plate. Telomere fluorescence values were converted into kb by using L5178Y-S and L5178Y-R cells with stable and known telomere lengths of 10.2 and 79.7 kb, respectively (30).
When fluorescence intensities of major satellite repeats (centromeres) were used as internal control to normalize telomere length values (Fig. 2), images were captured in the DAPI channel (excitation, 380/10 nm; epifluorescence dichroic, Fura/FITC; and emission, 435LP) for nucleus staining, in the Cy3 channel (excitation, 548/20 nm; epifluorescence dichroic, Fura/TRITC; emission, Fura/TRITC) for centromere staining, and in the FITC channel (excitation, 480/10 nm; epifluorescence dichroic, Fura/FITC; emission, Fura/FITC) for telomere detection. Images of samples that had been hybridized with either Cy3- or FITC-labeled PNA probes were captured in each channel to confirm negligible spectral bleeding between Cy3 and FITC. The DAPI image was used to define a nuclear area or ROI for each cell to measure centromere and telomere fluorescence intensities in the Cy3 and FITC image, respectively, within each predefined ROI. Telomere fluorescence values were normalized by dividing individual telomere fluorescence intensities of each nucleus (FITCi) by the corresponding centromere fluorescence intensity in the same nucleus (Cy3i), and multiplying these values by the mean centromere fluorescence intensity obtained for the total number of nuclei analyzed in the same well (ΣCy3i). Fluorescence intensities were converted into kb by using L5178Y-S and L5178Y-R cells as calibration standards, with stable and known telomere lengths of 10.2 and 79.7 kb (30), respectively.
Psychosocial Data.
Psychosocial data on cognitive function, stress, and depression of elderly individuals was obtained from lymphocyte donors in the context of the ZincAge project (see Cells), using the tests and questionnaires described in ref. 32. In particular, the MMSE (33), was used to assess cognitive mental status, including orientation, attention, concentration, immediate and short-term recall, language, and the ability to follow simple verbal and written commands. Score of 24–30 indicate normal cognitive function, 18–23 indicate mild cognitive impairment, and 0–17 indicate severe cognitive impairment. The GDS (34) assesses depressive symptoms in the elderly (positive/sad mood, boredom, memory problems, energy level, and staying at home). A score of 0–4 indicates a normal state, a score of 5–9 indicates mild depression, and a score of 10–15 indicates moderate–severe depression. The PSS (35) was used to measure the perception of stress. The score range is 0–56, and a score above mean (25 in the case of the population studied) is an indication of a high stress level.
Supplementary Material
Acknowledgments
We thank Drs. F. Marcellini, E. Mocchegiani, G. Herbein, G. Dedoussis, D. Monti, L. Rink, and J. Jate for patient recruitment. We thank J. García (CNIO, Madrid) for the development of Visual Basic Excel Macros and data management, and R. L. Milne (Spanish National Cancer Centre, Madrid) for statistical analysis. This work was supported by the European Union (A.C.). The M.A.B. laboratory is supported by the Ministerio de Ciencia Y Tecnologia (SAF2005-00277, GEN2001-4856-C13-08), the Regional Government of Madrid (GR/SAL/0597/2004), the European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842), and the Josef Steiner Cancer Research Award 2003.
Abbreviations
- GDS
geriatric depression scale
- HT
high-throughput
- MEF
mouse embryonic fibroblasts
- MMSE
mini mental state examination
- Q-FISH
quantitative fluorescence in situ hybridization
- PNA
peptide nucleic acid
- PSS
perceived stress scale
- ROI
region of interest.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609367104/DC1.
References
- 1.Chan SW, Blackburn EH. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 3.Flores I, Benetti R, Blasco MA. Curr Opin Cell Biol. 2006;18:254–260. doi: 10.1016/j.ceb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 6.Blasco MA. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell JR, Wood E, Collins K. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 8.Mason PJ, Wilson DB, Bessler M. Curr Mol Med. 2005;5:159–170. doi: 10.2174/1566524053586581. [DOI] [PubMed] [Google Scholar]
- 9.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 10.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 11.Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. Proc Natl Acad Sci USA. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, Aviv A, Spector TD. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 13.Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 14.Shay JW, Wright WE. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 15.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 16.Poon SS, Martens UM, Ward RK, Lansdorp PM. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemann MT, Strong MA, Hao LY, Greider CW. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 19.Samper E, Flores JM, Blasco MA. EMBO Rep. 2001;2:800–807. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Nat Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 23.Iwama H, Ohyashiki K, Ohyashiki JH, Hayashi S, Yahata N, Ando K, Toyama K, Hoshika A, Takasaki M, Mori M, et al. Hum Genet. 1998;102:397–402. doi: 10.1007/s004390050711. [DOI] [PubMed] [Google Scholar]
- 24.Rufer N, Brummendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L, Schulzer M, Lansdorp PM. J Exp Med. 1999;190:157–167. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordfjall K, Larefalk A, Lindgren P, Holmberg D, Roos G. Proc Natl Acad Sci USA. 2005;102:16374–16378. doi: 10.1073/pnas.0501724102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrieres J. Heart. 2004;90:107–111. doi: 10.1136/heart.90.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Ruiz C, Dickinson HO, Keys B, Rowan E, Kenny RA, Von Zglinicki T. Ann Neurol. 2006;60:174–180. doi: 10.1002/ana.20869. [DOI] [PubMed] [Google Scholar]
- 28.Harris SE, Deary IJ, MacIntyre A, Lamb KJ, Radhakrishnan K, Starr JM, Whalley LJ, Shiels PG. Neurosci Lett. 2006;406:260–264. doi: 10.1016/j.neulet.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 29.Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIlrath J, Bouffler SD, Samper E, Cuthbert A, Wojcik A, Szumiel I, Bryant PE, Riches AC, Thompson A, Blasco MA, et al. Cancer Res. 2001;61:912–915. [PubMed] [Google Scholar]
- 31.Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. Cytometry A. 2003;56:23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- 32.Marcellini F, Giuli C, Papa R, Malavolta M, Mocchegiani E. Rejuvenation Res. 2006;9:333–337. doi: 10.1089/rej.2006.9.333. [DOI] [PubMed] [Google Scholar]
- 33.Folstein MF, Folstein SE, McHugh PR. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 34.Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, Leirer VO. J Psychiatr Res. 1982;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]
- 35.Cohen S, Kamarck T, Mermelstein R. J Health Soc Behav. 1983;24:385–396. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.