Abstract
Background
Beta-carotene is the main dietary precursor of vitamin A. Potato tubers contain low levels of carotenoids, composed mainly of the xanthophylls lutein (in the beta-epsilon branch) and violaxanthin (in the beta-beta branch). None of these carotenoids have provitamin A activity. We have previously shown that tuber-specific silencing of the first step in the epsilon-beta branch, LCY-e, redirects metabolic flux towards beta-beta carotenoids, increases total carotenoids up to 2.5-fold and beta-carotene up to 14-fold.
Results
In this work, we silenced the non-heme beta-carotene hydroxylases CHY1 and CHY2 in the tuber. Real Time RT-PCR measurements confirmed the tuber-specific silencing of both genes . CHY silenced tubers showed more dramatic changes in carotenoid content than LCY-e silenced tubers, with beta-carotene increasing up to 38-fold and total carotenoids up to 4.5-fold. These changes were accompanied by a decrease in the immediate product of beta-carotene hydroxylation, zeaxanthin, but not of the downstream xanthophylls, viola- and neoxanthin. Changes in endogenous gene expression were extensive and partially overlapping with those of LCY-e silenced tubers: CrtISO, LCY-b and ZEP were induced in both cases, indicating that they may respond to the balance between individual carotenoid species.
Conclusion
Together with epsilon-cyclization of lycopene, beta-carotene hydroxylation is another regulatory step in potato tuber carotenogenesis. The data are consistent with a prevalent role of CHY2, which is highly expressed in tubers, in the control of this step. Combination of different engineering strategies holds good promise for the manipulation of tuber carotenoid content.
Background
The biofortification of potato is a viable strategy for the eradication of a series of nutritional deficiencies, since this crop stands fourth, among staple foods, in yearly per capita consumption. Several efforts are under way for the metabolic engineering of potato carotenoid content [1-3]. In a companion paper, we reported the results of the tuber-specific silencing of the first dedicated step in lutein biosynthesis, LCY-e [3]. This resulted in increases of β-carotene (up to 14-fold) and of total carotenoids (up to 2.5-fold). No changes in carotenoid content, or in endogenous carotenoid gene expression, were observed in leaves, indicating that, in agreement with previous reports [1], gene silencing remains confined in tubers.
Encouraged by this result, we decided to silence a second important regulatory step in carotenogenesis, the hydroxylation of β-carotene. This step is catalyzed by both non-heme and heme carotenoid hydroxylases. In Arabidopsis leaves, the complete complement of β-carotene hydroxylases is encoded by three genes: two encoding non-heme iron hydroxylases (CHY1, CHY2) and one encoding a cytochrome P450 (LUT5). Arabidopsis chy1chy2lut5 mutants completely lack β-xanthophylls (zeaxanthin, antheraxanthin, violaxanthin and neoxanthin), while double chy1chy2 mutants show approx. 70% reduction in the same compounds in leaves [4,5]. The tomato wf mutant, which maps to the CHY2 gene, is sufficient to severely impair flower β-xanthophyll biosynthesis, while leaf β-xanthophyll levels remain similar to those found in wild-type plants [6]. In keeping with this result, the tomato CHY2 transcript is expressed preferentially in flowers, while the CHY1 transcript is expressed preferentially in leaves [6]. These results indicate that, in different tissues, the hydroxylation of β-carotene is preferentially performed by different gene products. In order to elucidate the role of CHY genes in the hydroxylation of β-carotene in potato tubers, we took a tuber-specific gene silencing approach.
Results and discussion
In order to verify the tissue-specificity of expression of the genes controlling carotenoid biosynthesis in potato, we conducted Real Time RT-PCR experiments on leaf and tuber RNA. As can be seen (Table 1), the majority of carotenoid gene transcripts are preferentially expressed in leaves, in agreement with the higher carotenoid content of this tissue, while two housekeeping transcripts (β-TUBULIN and UBIQUITIN) are preferentially expressed in tubers. Notable exceptions to this trend are the NXS and CHY2 genes, which show higher levels of expression in tubers. The first gene [7] is the ortholog of the tomato B gene, encoding a fruit-specific lycopene β-cyclase [8] and of the pepper CCS gene, encoding a fruit-specific capsanthin-capsorubin synthase which also possesses lycopene cyclase activity [9]. The second gene is the ortholog of the tomato Wf gene, encoding a flower-specific non-heme β-carotene hydroxylase [6]. Thus, in potato the same members of the lycopene β-cyclase and β-carotene hydroxylase gene families, which in other Solanaceae are preferentially expressed in chromoplast-containing tissues, are preferentially expressed in the tuber. This is an indication that carotenogenesis in potato amyloplasts may share some regulatory mechanisms with carotenogenesis in tomato and/or pepper chromoplasts.
Table 1.
Tissue-specific expression of carotenoid biosynthesis genes in potato
| Leaves | Tubers | |
| Tubulin | 5.07 ± 2.17 | 46.18 ± 19.54 |
| Ubiquitin | 271.42 ± 83.97 | 417.30 ± 108.97 |
| Psy1 | 161.33 ± 27.44 | 55.85 ± 22.64 |
| Psy2 | 33.82 ± 5.31 | 10.28 ± 1.62 |
| Pds | 46.9 ± 16.41 | 3.71 ± 0.13 |
| Zds | 3.18 ± 1.20 | 0.19 ± 0.04 |
| CrtISO | 29.04 ± 12.09 | 7.07 ± 1.54 |
| Lcy-e | 1115.65 ± 482.19 | 3.15 ± 0.89 |
| Lcy-b | 20.52 ± 6.38 | 4.73 ± 0.93 |
| Lut1 | 7.93 ± 2.32 | 4.73 ± 1.36 |
| Chy1 | 70.65 ± 27.59 | 14.13 ± 1.06 |
| Chy2 | 49.39 ± 21.24 | 61.48 ± 19.07 |
| Lut5 | 121.34 ± 32.15 | 25.26 ± 12.89 |
| Zep | 841.85 ± 114.78 | 15.93 ± 1.22 |
| Nxs | 0.85 ± 0.05 | 14.24 ± 6.59 |
Transcript levels were studied via Real Time RT-PCR, using gene-specific oligonucleotides on RNAs isolated from a minimum of 4 different tubers or leaves from 2 different wild-type plants. Numbers indicate attograms gene-specific cDNA/20 ng total RNA. For details, see Methods.
Given that both CHY genes show strong levels of expression in the tuber, we decided to choose, as a silencing fragment, a region showing high (>80%) sequence identity between the two genes (data not shown). This region was amplified from tuber cDNA using specific oligonucleotides (see Methods), inserted, in antisense orientation, under the control of the tuber-specific patatin B33 promoter [3,10] and introduced in potato (cv. Desirée) using Agrobacterium-mediated transformation [11]. Transgenic plants were selected on kanamycin, the presence of the transgene was confirmed via PCR (see Methods), and six independent transgenic lines (AS-h, two plants per line) were acclimated in the greenhouse for tuberization. As controls, we used the original Desirée line used for transformation and one line transformed with a PB33:GUS construct [3]. At the end of the life cycle, tubers were harvested and tuber production was evaluated. None of the transgenic lines showed major alterations in tuber weight or number (data not shown).
The carotenoid composition of tubers was analyzed through HPLC (Table 2). The GUS line and two AS-h lines (lines 5 and 6) did not show relevant changes in carotenoid content, with respect to wild-type Desirée. Four AS-h lines (lines 1 to 4) showed significant (p ≤ 0.001) increases in total tuber carotenoids, as well as changes in carotenoid composition. In these lines, consistent with the hypothesized silencing of CHY genes, the levels of β-carotene showed significant increases (up to 38-fold). Levels of downstream β-β-xanthophylls showed more variable trends: the immediate product of β-hydroxylation, zeaxanthin, decreased 2- to 8-fold, while violaxanthin increased significantly in two out of four lines and neoxanthin increased significantly in one line. The final product of the competing ε-β-branch, lutein, increased 4- to 7-fold in all four "expressor" lines. The colorless biosynthetic intermediate, phytofluene, increased in two of the lines, while the other early intermediates (phytoene, ζ-carotene) were below detection in all lines. Consistently with the tuber-specific nature of the promoter used for the silencing construct, no significant variations in carotenoid or chlorophyll content, with respect to the Wt, were observed in leaves of GUS or AS-h lines (data not shown).
Table 2.
HPLC analysis of tuber carotenoids (ng/g dry weight)
| Phytofluene | Lutein | β-carotene | Zeaxanthin | Antheraxanthin | Violaxanthin | Neoxanthin | Other Xanth. | Esters | Total | |
| Wild-type | 61.69 ± 5.60 | 426.00 ± 124.44 | 2.25 ± 1.17 | 320.87 ± 111.63 | 1415.67 ± 335.02 | 1598.73 ± 468.72 | 468.76 ± 161.43 | 76.91 ± 22.60 | 517.06 ± 178.15 | 4887.95 ± 1421.16 |
| Gus 2 | 54.30 ± 5.28 | 501.81 ± 126.33 | 3.11 ± 2.15 | 311.42 ± 98.85 | 1317.06 ± 149.48 | 1282.35 ± 403.34 | 488.24 ± 121.55 | 91.14 ± 21,55 | 615.01 ± 98.73 | 4664.44 ± 1022.00 |
| Fold Variation | 0.88 | 1.18 | 1.38 | 0.97 | 0.93 | 0.80 | 1.04 | 1.18 | 1.19 | 0.95 |
| As-h1 | 162.57 ± 2.88 | 1590.36 ± 257.09 | 85.30 ± 2.33 | 168.37 ± 308.48 | 2198.74 ± 983.80 | 3452.99 ± 749.84 | 1509.40 ± 269.05 | 177.44 ± 54.47 | 4918.76 ± 920.25 | 14263.95 ± 2992.78 |
| Fold Variation | 2.63*** | 3.73 | 37.91*** | 0.52* | 1.55 | 2.16* | 3.22*** | 2.31* | 9.51*** | 2.92*** |
| As-h2 | 55.64 ± 0.58 | 2637.17 ± 570.03 | 34.08 ± 11.71 | 147.90 ± 39.91 | 945.83 ± 156.85 | 802.98 ± 87.20 | 832.59 ± 240.51 | 367.85 ± 19.18 | 4587.20 ± 1069.79 | 10411.26 ± 2195.20 |
| Fold Variation | 0.90 | 6.19*** | 15.15** | 0.46* | 0.67 | 0.50 | 1.78 | 4.78*** | 8.87*** | 2.13** |
| As-h3 | 74.70 ± 8.85 | 2982.04 ± 881.18 | 56.34 ± 16.90 | 37.09 ± 5.84 | 1034.68 ± 402.16 | 11422.26 ± 2470.47 | 754.32 ± 228.82 | 329.06 ± 87.63 | 5068.07 ± 1181.94 | 21758.57 ± 5274.94 |
| Fold Variation | 1.21* | 7.00*** | 25.04*** | 0.12*** | 0.73 | 7.14*** | 1.61 | 4.28*** | 9.80*** | 4.45*** |
| As-h4 | 67.64 ± 24.22 | 1808.90 ± 239.24 | 32.65 ± 6.50 | 153.71 ± 39.99 | 1102.94 ± 287.98 | 3377.31 ± 371.15 | 764.34 ± 171.39 | 255.47 ± 17.75 | 1789.33 ± 51.88 | 9352.31 ± 1186.71 |
| Fold Variation | 1.10 | 4.25*** | 14.51** | 0.48** | 0.78 | 2.11 | 1.63 | 3.32*** | 3.46 | 1.91** |
| As-h5 | 63.30 ± 5.84 | 764.54 ± 244.69 | 2.71 ± 7.75 | 218.53 ± 140.68 | 1382.88 ± 203.52 | 2018.71 ± 282.39 | 456.06 ± 55.76 | 126.36 ± 3.23 | 1293.42 ± 564.92 | 6326.51 ± 1502.94 |
| Fold Variation | 1.03 | 1.79 | 1.20 | 0.68 | 0.98 | 1.26 | 0.97 | 1.64 | 2.50 | 1.29 |
| As-h6 | 73.12 ± 19.18 | 575.11 ± 71.52 | 4.31 ± 0.43 | 331.03 ± 28.14 | 1551.15 ± 108.39 | 1780.21 ± 509.03 | 617.62 ± 103.11 | 120.02 ± 35.45 | 924.89 ± 132.96 | 5977.47 ± 989.06 |
| Fold Variation | 1.18 | 1.35 | 1.92 | 1.03 | 1.10 | 1.11 | 1.32 | 1.56 | 1.79 | 1.22 |
Carotenoids were measured via diode array HPLC (see Methods) on a minimum of 4 different tubers from 2 different plants. Fold variation with respect to the wild-type is reported for each carotenoid compound and for each line. Asterisks indicate significance of the fold variation in an ANOVA test (*: p ≤ 0.05;**: p ≤ 0.01; ***: p ≤ 0.001). For details, see Methods.
The AS-h silencing transcript was detected at high levels (0.4- to 27-fold tubulin) in the tubers of the four AS-h lines showing significant changes in carotenoid content (Table 3). The highest levels of expression are observed in line AS-h 3, which has the highest total carotenoid levels. In tubers of the two AS-h lines with unchanged carotenoid content ("non-expressor" lines), as well as in leaves of all lines, the silencing transcript was below the levels of detection of the Real Time RT-PCR assay. Variability in the expression of introduced genes among independent transformants is a common phenomenon in plant transformation [12] and has been found, at comparable frequencies, in the case of the B33:AS-e and B33:GUS constructs [3].
Table 3.
Trangene expression.
| Line | Organ | AS-chy expression |
| (Fold Tubulin) | ||
| Wild-type | Leaf | nd |
| Tuber | nd | |
| Gus 2 | Leaf | nd |
| Tuber | nd | |
| AS-h1 | Leaf | nd |
| Tuber | 7.428 ± 1.228 | |
| AS-h2 | Leaf | nd |
| Tuber | 10.721 ± 3.152 | |
| AS-h3 | Leaf | nd |
| Tuber | 27.630 ± 8.457 | |
| AS-h4 | Leaf | nd |
| Tuber | 0.412 ± 0.046 | |
| AS-h5 | Leaf | nd |
| Tuber | nd | |
| AS-h6 | Leaf | nd |
| Tuber | nd |
AS-h transgene expression was measured via Real Time RT-PCR and normalized for the β-TUBULIN transcript. For details see Methods. nd = not detectable.
Expression of endogenous carotenoid genes is often altered as a consequence of manipulations modifying the levels of biosynthetic intermediates in the pathway. This phenomenon has been observed both in tomato leaves [13,14] and in potato tubers [2,3]. We measured the expression of carotenoid gene transcripts (PSY1, PSY2, PDS, ZDS, CrtISO, LCY-b, LCY-e, CHY1, CHY2, LUT1, LUT5, ZEP, NXS) in tubers, using Real Time RT-PCR. The tuber transcript levels, normalized first for the β-tubulin transcript and then for the Wt transcript levels, are shown in Figure 1. The biosynthetic steps catalyzed by these genes are shown in Figure 2. The GUS line, as well as "non expressor" lines AS-h 5 and 6, showed only minor variations in gene expression with respect to the Wt line. This indicates that culture conditions, somaclonal effects due to regeneration procedures, or the presence of the silencing transgene by itself, do not cause any major variability in endogenous carotenoid gene expression. The endogenous CHY2 gene is silenced in the four lines showing alterations in carotenoid content (AS-h 1 to 4). Line AS-h 3, which is the one showing the highest increase in total carotenoids (Table 2), also shows the most efficient silencing of endogenous CHY2. Alongside CHY2, also the CHY1 transcript is silenced, albeit to different extent, in transgenic tubers of lines AS-h1 to 4. This result indicates that the homology between the two transcripts in the region chosen for silencing is sufficiently high to warrant cross-silencing.
Figure 1.
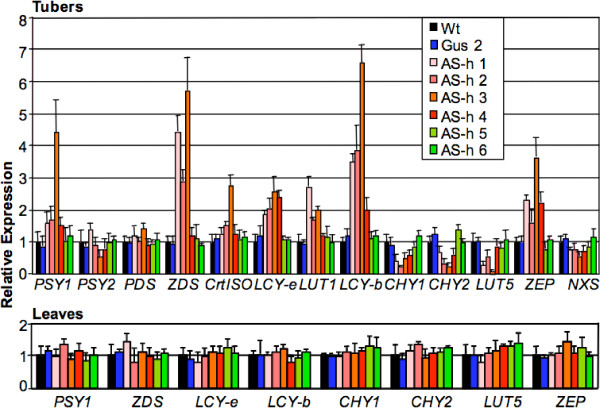
Endogenous carotenoid gene expression. Transcript levels were measured through Real Time RT-PCR and were first normalized for expression of the housekeeping β-tubulin gene, and then for the expression levels in the Wt. Data show the average and SE (error bars) of determinations from at least 4 different tubers (or leaves) from 2 different plants. For details see Methods.
Figure 2.
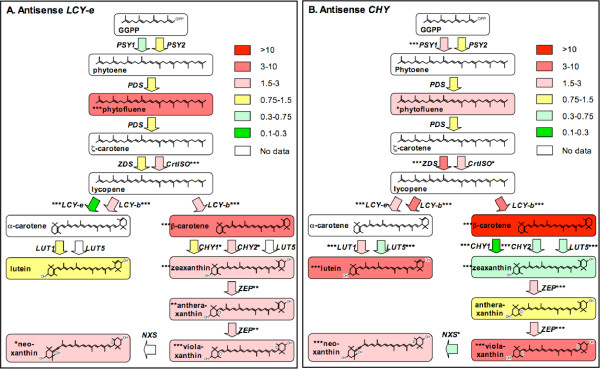
Schematic representation of metabolite and gene expression changes in engineered tubers. Boxes represent the metabolic intermediates, arrows represent the genes catalyzing the various reactions. Fold induction or repression with respect to the wild-type – averaged over three transgenic lines- is represented by different hues of red or green, respectively (see legend). White means that no data are available. Asterisks indicate significance of the fold variation with respect to the Wt in an ANOVA test (*: p ≤ 0.05; **: p ≤ 0.01; ***: p ≤ 0.001). (A) AS-e lines 1,2,3 [3]. (B) AS-h lines 1,2,3 (this paper).
The silencing of CHY transcripts causes, directly or indirectly, an extensive remodeling of the expression of the endogenous carotenoid genes. Alongside CHY1 and CHY2, also LUT5 and, to a lesser extent, NXS are repressed in lines AS-h1 to 4. The repression of LUT5 is likely to have a cooperative effect with the silencing of CHY1 and CHY2 in mediating β-carotene accumulation, since this gene, like CHY1 and CHY2, encodes a β-ring hydroxylase [5] (Figure 2). Also, the induction of lycopene beta-cyclase (LCY-b) is likely to be enhancing β-carotene content. LUT5 and LCY-b are, respectively, maximally repressed and induced in line AS-h3, which has the highest total carotenoid content. ZDS, LCY-e, LCY-b, LUT1 and ZEP are induced in lines showing changes in carotenoid content (AS-h 1 to 4). CrtISO is induced only in line AS-h 3, which is one showing the highest total carotenoid levels.
We conducted a comparative assessment of gene expression and of metabolite composition in the carotenoid pathway in the tubers of AS-e lines [3] and of AS-h lines (this paper). The results are shown in Figure 2 and can be summarized as follows:
• The two lycopene cyclases, LCY-b and LCY-e, are induced as a result of either manipulation. A notable exception are of course AS-e plants, in which the LCY-e transcript is silenced as a result of the introduced transgene. We cannot distinguish, at the present moment, whether this induction in LCY-b and LCY-e transcripts is a consequence of the increase in total carotenoid levels or of the increase of a specific intermediate. Pharmacological experiments with inhibitors of various steps in carotenoid biosynthesis [13,14] could, to a certain extent, discriminate between the different possibilities.
• Another gene showing induction in AS-e and AS-h tubers is ZEP. Zeaxanthin is a rare carotenoid in cultivated potato [15], and the fact that the immediately downstream gene is induced as a result of perturbations in carotenoid content may partially explain this fact. This gene has been silenced in a tuber-specific fashion, resulting in accumulation of zeaxanthin [1].
• A third gene showing induction in both cases is CrtISO. Its gene product is involved in the isomerization of cis double bonds during the synthesis of lycopene [16-18].
• The overall pool of xanthophylls derived from α- and β-carotene increases in both cases; in AS-e lines, lutein levels remain relatively stable, while those of β-β-xanthophylls show a moderate increase; in AS-h lines, the product of hydroxylation, zeaxanthin, shows a moderate decrease, antheraxanthin levels are unaltered, while all other compounds show moderate (neoxanthin) to strong (violaxanthin and lutein) increases.
• Early compounds in the pathway are below detection in the Wt and in transgenic lines, with the exception of phytofluene, which shows from moderate to strong increases in both AS-e and AS-h lines.
• By far, the highest increase obtained in both cases is that of β-carotene, one of the main goals of our engineering effort; these results, together with those of Ducreux et al. [2] and Lu et al. [19], clearly show that β-carotene, although it is a very rare compound in cultivated potato [15], can accumulate to remarkable levels after metabolic engineering.
What is the relative contribution of different carotenoid hydroxylases to beta-carotene hydroxylation in tubers? Table 1 shows that the transcripts for all three genes controlling β-carotene hydroxylation in Arabidopsis [5] are expressed albeit at different levels in potato tubers, in the order CHY2>LUT5>CHY1. Comparison of carotenoid content (Table 2) with gene expression (Figure 1) in the different lines suggests that maximal accumulation of total carotenoids, and repression of zeaxanthin content, is observed in line AS-h 3, in which CHY2 and LUT5 are maximally repressed. Thus, both gene expression data in wild-type tubers, and correlations between gene silencing and metabolite levels in transgenic ones, suggest that CHY2 is the most important contributor to tuber β-carotene hydroxylation. However, this remains a hypothesis, until accurate measurements are obtained of the levels and activities of the corresponding enzymes.
Recently, Lu et al. [19] showed that the cauliflower Or gene, encoding a DnaJ-related protein, is able to dramatically increase total carotenoid and β-carotene levels in transgenic potato tubers. This approach is complementary to more "classical" ones, like the one reported here, that rely on the alteration of expression, in tubers, of structural genes in the carotenoid pathway, [1][2][3][20][21]. A combination of different approaches holds good promise for the further increase of the provitamin A levels of potato.
Conclusion
Using an antisense construct under the control of the tuber-specific patatin promoter, we obtained the simultaneous, tuber-specific silencing of the potato CHY1 and CHY2 transcripts. β-carotene increased and zeaxanthin decreased accordingly in a tuber-specific fashion, while phytofluene, violaxanthin, neoxanthin, lutein and total carotenoids also showed increases. This modification in carotenoid content is paralleled by modifications in endogenous carotenoid gene expression. Modeling of carotenoid gene expression and intermediate metabolite levels in the two cases showed several overlaps with the changes already observed in LCY-e-silenced tubers [3]. CrtISO, LCY-b and ZEP were induced in both cases, suggesting that the levels of these transcripts may be sensing the similar changes in metabolite abundance induced by the two types of manipulations (Figure 2). By far the metabolite showing the highest increase in both cases was β-carotene, confirming that this compound is relatively stable in potato tubers.
Methods
Unless otherwise indicated, molecular biology methods are as described [22]. A 0,56 Kb CHY2 cDNA fragment was amplified from potato (cv Desirée) tuber cDNA using the primers As-chy Up1 and As-chy Dw2 (Table 4). These primers inserted, respectively, Sac I and Bam HI sites upstream and downstream of the cDNA fragment. After intermediate cloning in the pBSK+ vector and re-sequencing, the fragment was inserted, in antisense orientation, to replace the GUS gene in the pBI33:GUS vector [3].
Table 4.
Primers used
| Primer name | Sequence | Use |
| AS-chy Up1 | TAGAGCTCGGGATTACTTC | AS-chy cloning |
| AS-chy Dw2 | ATGGATCCTCCTTTTCCAA | AS-chy cloning |
| AS-h Up | GTTAAGGGAACTTCTCCAC | PCR screening |
| Nos-test 2 | CGCGTATTAAATGTATAATTG | PCR screening |
| AS-h RT Up | ACCCTCCATTTGCCACGAA | Real-time assay |
| AS-h RT Dw | TTATATGATAATCATCGCAAGACCG | Real-time assay |
| Chy1 Up | CTTGGCCCAAAACCCACTT | Real-time assay |
| Chy1 Dw | CCTCAAATTGAGGTTTCAGCTTCT | Real-time assay |
| Chy2 Up | TTTTGCTGTCTCGAAGAAAGCC | Real-time assay |
| Chy2 Dw | AGCCAACAGGCAGCTAAACTCT | Real-time assay |
| Lut5 Up | GTCTCAAGCAAGCAACTTCGTG | Real-time assay |
| Lut5 Dw | GATAAAAGGTCCATGTGAGCACTG | Real-time assay |
| Nxs Up | CTTGGAGGAGACTTCTTTGGTGA | Real-time assay |
| Nxs Dw | CGGAAGTGGTCCTCCCATAG | Real-time assay |
Sequences of the primers used for cloning of the gene fragment, for PCR screening of the putative transgenic plants, and for Real Time RT-PCR quantitation of transcript levels. For further details, see Methods.
Potato (cv Desirée) was transformed as previously described [11]. Plantlets growing on kanamycin were tested by PCR, using primers AS-h Up and Nos-test 2 (Table 4). PCR-positive, rooted plantlets were adapted in greenhouse in pots (diameter: 25 cm) in a soil mixture composed of 1/3 sand and 2/3 of sterile soil (Terraplant 2, BASF). Photoperiod was set at 14 hours of light and 10 hours of dark, with temperature set at 24°C during the light period and at 16°C during the dark period. In the advanced phase of growth, the day temperature was kept around 20°C in order to promote tuberization. During tuberization, irrigation was reduced in order to prevent tuber decay.
Tubers from the lower 2/3 of the pot ("deep" tubers) were collected separately from superficial ones, washed in water, briefly dried at room temp, cut in pieces and frozen at -80°C. Tuber productivity for each line was estimated as the total number of tubers produced and as their total weight. All carotenoid and RT-PCR measurements were conducted on at least 4 different "deep" tubers per each line, to prevent possible alterations in carotenoid composition/gene expression resulting from light accidentally illuminating the superficial tubers.
Total RNA was isolated from frozen tissue and analyzed through Real Time RT-PCR using previously published methods [13,23]. Two independent RNA extractions and four cDNAs (two from each RNA) were used for the analyses; RT-PCR conditions and gene-specific primers were as in Diretto et al (2006) with the addition of the following genes: Lut5 (ESTID18235) and NXS (AJ272136). Real Time assay oligos for these genes are described in Table 4. In order to discriminate the introduced AS-h mRNA from the endogenous CHY mRNA, the former was amplified using primers AS-h RT Up and AS-h RT Dw, while the latter was amplified using primers Chy1 Up and Dw, and Chy2 Up and Dw (Table 4).
HPLC analysis was performed exactly as described previously [3].
Statistical analysis (one-way ANOVA) was performed using the PAST software [24].
Authors' contributions
GG, RT and PB planned and supervised the work. FM prepared the constructs for transformation, RT performed the transformations and maintained the lines in vitro, DP and GD grew and sampled the plants, GD performed the Real Time RT-PCR assays and the statistical analysis, RW performed the HPLC's. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
Work supported by EU projects ProVitA, EU-SOL and Develonutri, by the HarvestPlus program, and by the Italian Ministry of Research (FIRB project). GD acknowledges the Univ. of L'Aquila for a doctoral fellowship and Prof. Laura Spanò for supervision. We thank Velia Papacchioli for maintenance of the plant in vitro material and Carlo Rosati for comments on the manuscript and help with statistical analysis.
Contributor Information
Gianfranco Diretto, Email: gianfranco.diretto@casaccia.enea.it.
Ralf Welsch, Email: welschra@web.de.
Raffaela Tavazza, Email: raffaela.tavazza@casaccia.enea.it.
Fabienne Mourgues, Email: fabienne.mourgues@trisaia.enea.it.
Daniele Pizzichini, Email: daniele.pizzichini@casaccia.enea.it.
Peter Beyer, Email: peter.beyer@biologie.uni-freiburg.de.
Giovanni Giuliano, Email: giuliano@casaccia.enea.it.
References
- Romer S, Lubeck J, Kauder F, Steiger S, Adomat C, Sandmann G. Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng. 2002;4:263–272. doi: 10.1006/mben.2002.0234. [DOI] [PubMed] [Google Scholar]
- Ducreux LJ, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J Exp Bot. 2005;56:81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]
- Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, Papacchioli V, Beyer P, Giuliano G. Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 2006;6:13. doi: 10.1186/1471-2229-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D. Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell. 2003;15:1320–1332. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A, Dall'osto L, Fraser PD, Bassi R, Giuliano G. Elucidation of the beta-carotene hydroxylation pathway in Arabidopsis thaliana. FEBS Lett. 2006;580:4718–4722. doi: 10.1016/j.febslet.2006.07.055. [DOI] [PubMed] [Google Scholar]
- Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18:1947–1960. doi: 10.1105/tpc.105.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Hugueney P, Schledz M, Welsch R, Frohnmeyer H, Laule O, Beyer P. Identification of a novel gene coding for neoxanthin synthase from Solanum tuberosum. FEBS Lett. 2000;485:168–172. doi: 10.1016/S0014-5793(00)02193-1. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta -carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Hugueney P, d'Harlingue A, Kuntz M, Camara B. Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J. 1994;6:45–54. doi: 10.1046/j.1365-313X.1994.6010045.x. [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin gene. Embo J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazza R, Tavazza M, Ordas RJ, Ancora G, Benvenuto E. Genetic transformation of potato (Solanum tuberosum); an efficient method to obtain transgenic plants. Plant Science. 1988;59:175–181. doi: 10.1016/0168-9452(89)90135-0. [DOI] [Google Scholar]
- Hobbs SL, Kpodar P, DeLong CM. The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol. 1990;15:851–864. doi: 10.1007/BF00039425. [DOI] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona V, Aracri B, Kosturkova G, Bartley GE, Pitto L, Giorgetti L, Scolnik PA, Giuliano G. Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 1996;9:505–512. doi: 10.1046/j.1365-313X.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- Nesterenko S, Sink KC. Carotenoid profiles of potato breeding lines and selected cultivars. HortScience. 2003;38:1173–1177. [Google Scholar]
- Isaacson T, Ohad I, Beyer P, Hirschberg J. Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol. 2004;136:4246–4255. doi: 10.1104/pp.104.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14:321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Giliberto L, Rosati C. Carotenoid isomerase: a tale of light and isomers. Trends Plant Sci. 2002;7:427–429. doi: 10.1016/S1360-1385(02)02329-4. [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O'Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, Kochian LV, Kupper H, Earle ED, Cao J, Li L. The Cauliflower Or Gene Encodes a DnaJ Cysteine-Rich Domain-Containing Protein That Mediates High-Levels of {beta}-Carotene Accumulation. Plant Cell. 2006. [DOI] [PMC free article] [PubMed]
- Morris WL, Ducreux LJ, Fraser PD, Millam S, Taylor MA. Engineering ketocarotenoid biosynthesis in potato tubers. Metab Eng. 2006;8:253–263. doi: 10.1016/j.ymben.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Morris WL, Ducreux LJ, Hedden P, Millam S, Taylor MA. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: implications for the control of the tuber life cycle. J Exp Bot. 2006 doi: 10.1093/jxb/erl061. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual (Second Edition) Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Carbone F, Pizzichini D, Giuliano G, Rosati C, Perrotta G. Comparative profiling of tomato fruits and leaves evidences a complex modulation of global transcript profiles. Plant Sci. 2005;169:165–175. doi: 10.1016/j.plantsci.2005.03.011. [DOI] [Google Scholar]
- http://folk.uio.no/ohammer/past/


