Abstract
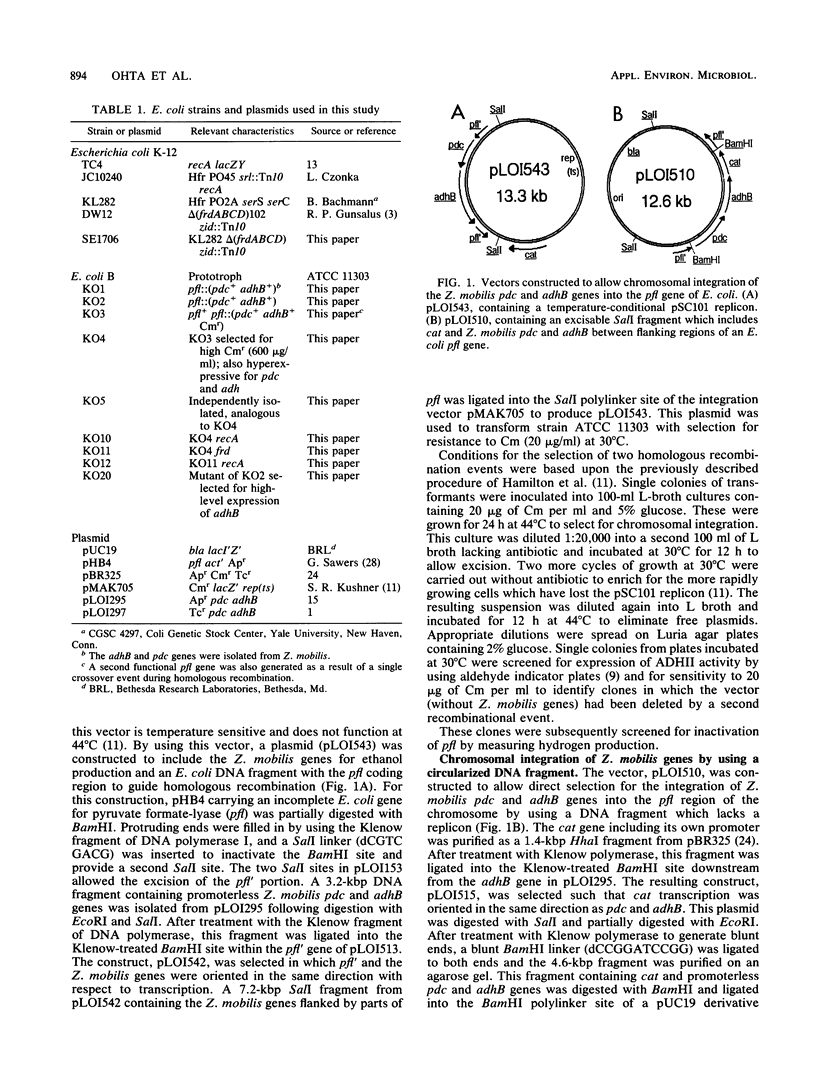
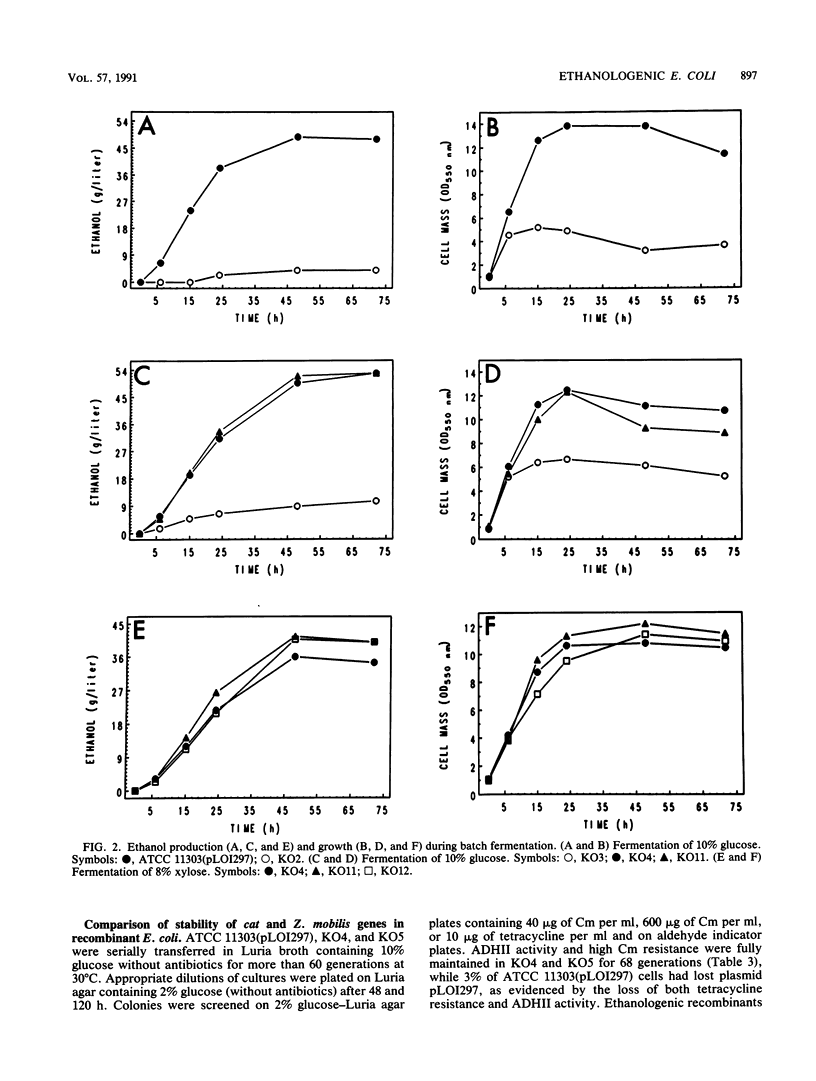
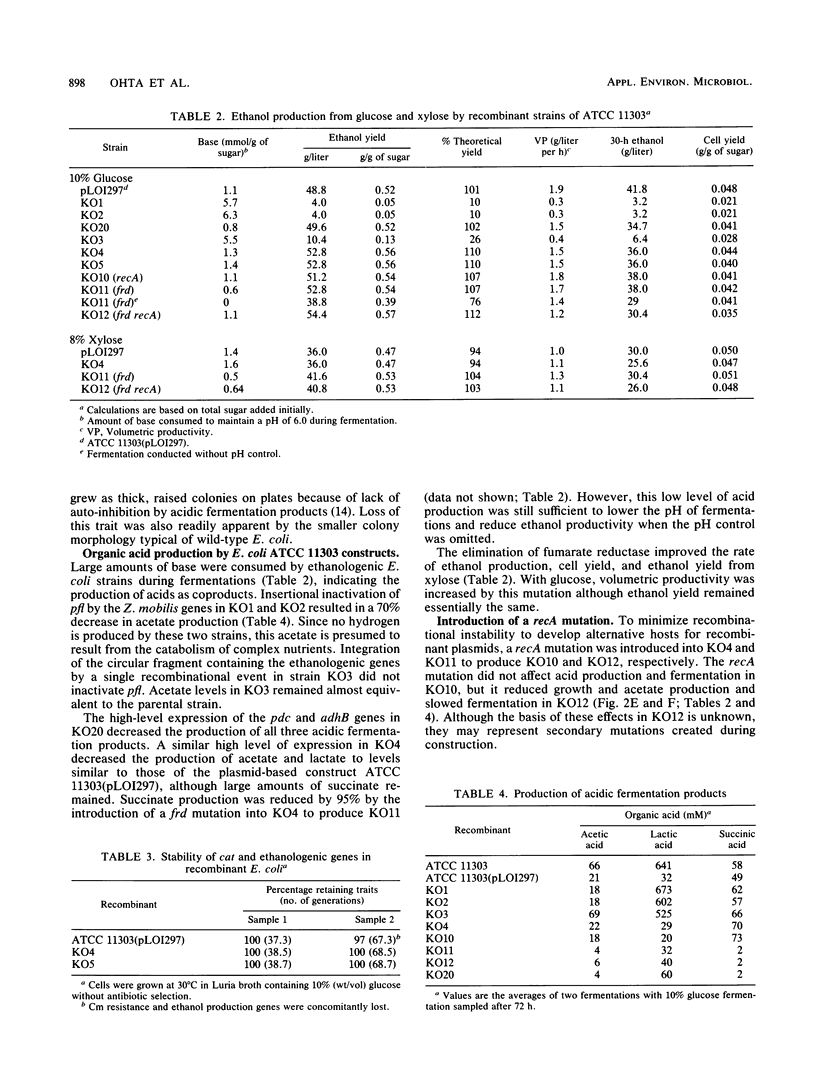
Zymomonas mobilis genes for pyruvate decarboxylase (pdc) and alcohol dehydrogenase II (adhB) were integrated into the Escherichia coli chromosome within or near the pyruvate formate-lyase gene (pfl). Integration improved the stability of the Z. mobilis genes in E. coli, but further selection was required to increase expression. Spontaneous mutants were selected for resistance to high level of chloramphenicol that also expressed high levels of the Z. mobilis genes. Analogous mutants were selected for increased expression of alcohol dehydrogenase on aldehyde indicator plates. These mutants were functionally equivalent to the previous plasmid-based strains for the fermentation of xylose and glucose to ethanol. Ethanol concentrations of 54.4 and 41.6 g/liter were obtained from 10% glucose and 8% xylose, respectively. The efficiency of conversion exceeded theoretical limits (0.51 g of ethanol/g of sugar) on the basis of added sugars because of the additional production of ethanol from the catabolism of complex nutrients. Further mutations were introduced to inactivate succinate production (frd) and to block homologous recombination (recA).
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterthum F., Ingram L. O. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol. 1989 Aug;55(8):1943–1948. doi: 10.1128/aem.55.8.1943-1948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaut M., Whittaker K., Valdovinos A., Ackrell B. A., Gunsalus R. P., Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989 Aug 15;264(23):13599–13604. [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Christiansen L., Pedersen S. Cloning, restriction endonuclease mapping and post-transcriptional regulation of rpsA, the structural gene for ribosomal protein S1. Mol Gen Genet. 1981;181(4):548–551. doi: 10.1007/BF00428751. [DOI] [PubMed] [Google Scholar]
- Clark D. P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989 Sep;5(3):223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek K. M., Ingram L. O. Determination of the intracellular concentration of ethanol in Saccharomyces cerevisiae during fermentation. Appl Environ Microbiol. 1986 Jan;51(1):197–200. doi: 10.1128/aem.51.1.197-200.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. O., Conway T., Clark D. P., Sewell G. W., Preston J. F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987 Oct;53(10):2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. O., Conway T. Expression of Different Levels of Ethanologenic Enzymes from Zymomonas mobilis in Recombinant Strains of Escherichia coli. Appl Environ Microbiol. 1988 Feb;54(2):397–404. doi: 10.1128/aem.54.2.397-404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Alterthum F., Ingram L. O. Effects of environmental conditions on xylose fermentation by recombinant Escherichia coli. Appl Environ Microbiol. 1990 Feb;56(2):463–465. doi: 10.1128/aem.56.2.463-465.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Rödel W., Plaga W., Frank R., Knappe J. Primary structures of Escherichia coli pyruvate formate-lyase and pyruvate-formate-lyase-activating enzyme deduced from the DNA nucleotide sequences. Eur J Biochem. 1988 Oct 15;177(1):153–158. doi: 10.1111/j.1432-1033.1988.tb14356.x. [DOI] [PubMed] [Google Scholar]
- Sankar P., Shanmugam K. T. Biochemical and genetic analysis of hydrogen metabolism in Escherichia coli: the hydB gene. J Bacteriol. 1988 Dec;170(12):5433–5439. doi: 10.1128/jb.170.12.5433-5439.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G., Böck A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988 Nov;170(11):5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G., Böck A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989 May;171(5):2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]



