Abstract
The definition of regions of interest for PET data analysis poses a number of complex problems. While studies have shown that regions drawn on a template can be appropriate for extracting data for normal healthy subjects, it is unclear how these results can be applied to different populations. In this study, we focused on the aging population and examined how different parameters in the template data-extraction process may affect the accuracy of the results. We first present an automated method for extracting PET counts using a region-of-interest approach within a template framework. Then, we discuss two studies in which we measure the effects of varying specific parameters in this process. In study 1 we examined three parameters that may influence this process: choice of template, region, and threshold. In study 2 we focused on the hippocampus. We considered 6 different templates, and examined how well the subject-specific hippocampal masks overlapped with each other and with the template hippocampal masks after normalization. While the data in the older cohort is more variable than the normal population, the results suggest that using an appropriate template and selecting the correct parameters for the template-based ROI method can provide template-extracted counts that are highly correlated to counts extracted using subject-specific ROIs.
Introduction
Traditionally, statistical analyses of positron emission tomography (PET) data are derived from one of two classes of analyses: region-of-interest (ROI) approaches and voxel-by-voxel approaches. ROI analyses are used to test hypotheses about specific regions. In this type of analysis, functional or anatomical regions are drawn for each individual subject based on his or her PET or structural image. This method is considered to be the gold standard for extracting data from PET images. However, it can be tedious and subjective because a neuro-anatomically trained expert must manually draw each region. In addition, defining ROIs based on PET images is subject to bias and uncertainty because the tracer uptake may be absent or decreased in some brain regions. The alternative, a voxel-by-voxel approach, requires that all images be spatially transformed to a template space. This process, known as stereotaxic normalization, assumes that each voxel corresponds to the same anatomical region across subjects. Statistical analyses are therefore performed for every voxel across all subjects. This approach to analyzing PET data is more exploratory than the ROI approach because it requires no prior hypothesis about the expected location of the effect. With current software tools, , the voxel-by-voxel process can be fast and automated (Cox et al., 1996; Friston et al., 1995; Minoshima et al., 1994), but the results are dependent on the accuracy and precision of the normalization process.
Although in the past, ROI-based methods have been labor intensive and time-consuming, requiring manual regional demarcation and segmentation of individual patient data, a number of different computerized methods have been described to partially or fully automate the process. Computerized adjustable brain atlases were developed, which could be transformed to individual PET images; this partially automated the process of identifying ROIs (Bohm et al., 1991). Later, Collins et al. (1995) presented a fully automated method for segmentation and identification of gross neuroanatomical structrures, such as caudate, white and grey. More recently, with the improvement and automation of stereotaxic normalization procedures, a number of papers have proposed using a template-based or model-based region-of interest method. In this approach, ROIs are defined on a template to which all subjects have been normalized or which has been normalized to each individual subject (Hammers et al., 2002; Yasuno et al., 2002; Hammers et al., 2003; Maldjian et al., 2003). This template-based ROI method reduces the time-intensive and subjective aspect of manually drawing subject-specific ROIs while maintaining the ability to perform hypothesis driven analyses for specific locations. Because this method is still dependent on normalization, its success remains contingent on the accuracy and precision of the normalization process. Nonetheless, it has been demonstrated that for a normal young population, there is a high correlation between the data extracted using template ROIs and the data extracted using subject-specific ROIs (Yasuno et al., 2002).
While these studies have provided a good foundation for using template-based ROI methods for the normal population, it is unclear how results can be applied to specific population groups such as the aging population. First, due to atrophy-related structural changes, the standard normal template may not be ideal for defining regions for the aging population. Second, the aging population, like other patient populations may have increased anatomical variability across subjects. Therefore, it is unclear how this increased variability would affect the accuracy of the template-extracted data. The purpose of this project was to evaluate the accuracy of template-extracted PET radioactivity counts in the context of the aging population. First, we present an automated method for extracting PET counts using a region-of-interest approach within a template framework. Then, we discuss two studies in which we measure the effects of varying specific parameters in this process.
The automated method presented here extracts PET counts using regions drawn on a template in conjunction with a gray-matter mask defined by each individual subject’s MR. In this process, the subject’s MR is first segmented to extract only the gray-matter regions. Next, the subject’s PET, MR, and gray-matter images are coregistered to each other and normalized to the template. The overlapping regions defined by the template ROI and the subject’s normalized gray image determine the final mask from which the PET counts are extracted.
In study 1 we examined three parameters that may influence this process. First, to determine if the accuracy of template-extracted counts varies across regions, several regions of interest were examined in order to sample a wide topographic range of cortex. Second, to determine the effect of the template on the accuracy of the template-extracted counts, we considered two different templates. We used a standard template derived from normal young subjects as well as a template derived from an older cohort. Third, to determine the effect of the segmentation threshold on the template-extracted counts, we considered different thresholds for the gray segmentation.
In study 2 we focused on the hippocampus. The hippocampus is particularly relevant in aging studies due to its significance in the early diagnosis of Alzheimer’s disease (AD) (de Leon et al., 2001). Moreover, the hippocampus may be especially sensitive to poor normalization because it is a small structure that is prone to high anatomical variability with aging (Mosconi et al., 2005). Therefore, our purpose in study 2 was to examine how well the counts in the hippocampal region were estimated using the template-extraction method. We considered 6 different templates. In addition, we examined how well the subject-specific hippocampal masks overlapped with each other and with the template hippocampal masks after normalization.
Methods
Subject sampling
All data for this project were collected from subjects who participated in the Sacramento Area Latino Study on Aging (SALSA). Detailed methods for subject sampling, screening and evaluation have been described previously (Haan et al., 2003). Briefly, the SALSA project recruited 1,789 Hispanic individuals of predominantly Mexican American ethnicity who were older than 60 years of age and who came from six counties in and around Sacramento and the California central valley. A multistage approach was utilized to screen for cognitive impairment, and subjects were categorized into three groups, normal (including mild memory impairments), cognitively impaired but not demented (CIND), and demented. A total of 122 subjects underwent MRI, and 93 of these underwent PET. Of these, 5 subjects were excluded for technical reasons related to the scans, resulting in 88 subjects with a mean age of 71 (65 normal, 11 CIND, and 12 demented). All of these subjects were used for study 2, and a randomly selected subset of the subjects was used for study 1 (n = 15; mean age = 73, 8 normal, 4 CIND, and 3 demented).
Data acquisition
PET data were acquired using the Siemens-CTI ECAT EXACT (model 921) 47-section scanner (Siements, Munich/Erlangen, Germany) in a two-dimensional acquisition mode. All subjects were studied in a quiet room with eyes and ears unoccluded for 40 minutes following the injection of 5–10 mCi of 18F-fluoro-deoxy-glucose (FDG). Initially, a transmission image was obtained during 20 minutes of imaging using a rotating 68Ge source. Subsequently, emission data were acquired in a two-dimensional mode for 40 minutes. Following each PET scan, the data were reconstructed using standard two-dimensional filtered back-projection.
All MR images were collected on a GE 1.5T Signa Horizon LX NV/I System (Signal, General electric, Milwaukee, WI). A T1-weighted coronal image was acquired using a three-dimensional spoiled gradient-recalled-echo inversion-recovery prepped sequence (TR=1.9 ms; flip angle = 20°, FOV = 24 cm; matrix = 256x256; slices = 124; slice thickness = 1.6 mm).
MR Image processing
For precise identification of the regions of interest, the MR images were processed using BrainVISA (Cointepas et al., 2001) and viewed using Anatomist. First, images were reoriented using four manually specified points: the anterior commissure, posterior commissure, an interhemispheric point, and the left-most edge of the brain. After reorientation, images were skull-stripped and segmented using the utilities available in BrainVISA. The cortical surface was then triangulated and coregistered to the original MR, thus relating the three-dimensional features to the two-dimensional coronal slices on which the ROIs were subsequently drawn (Figure 1).
Figure 1. Process for defining ROIs.
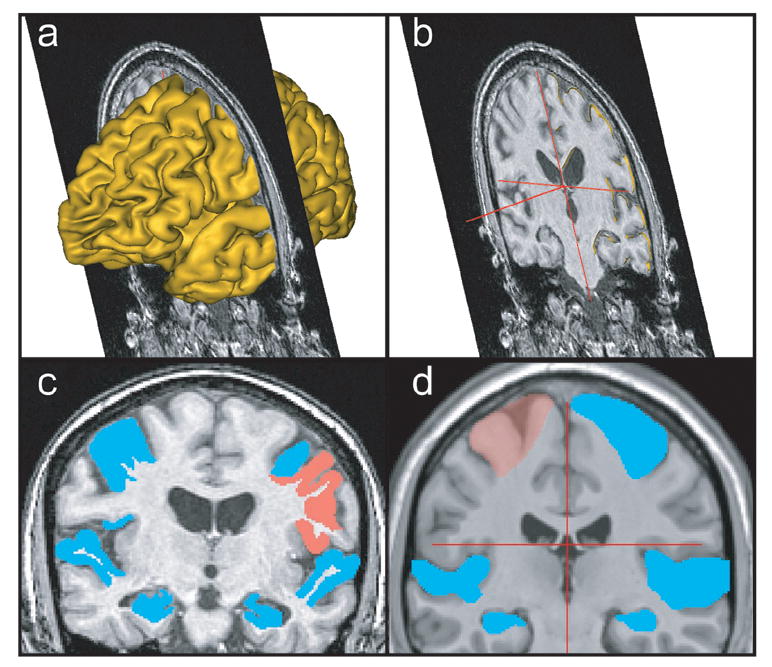
(a) The three dimensional cortical mesh of a single subject’s brain is rendered and coregistered to a single coronal slice from the subject’s high-resolution MR. Landmarks, such as sulcal boundaries, are identified using the mesh and placed on the 2-D coronal image. The mesh is then removed (b), and the final ROIs are drawn on the 2-D coronal image (c). The PcG, Hipp, and STG ROIs are shown on a single coronal slice for an individual subject. (d) The template ROIs are defined using the same method as for the single-subject ROIs except that the gray-white boundary within sulci was not delineated. The PcG, Hipp, and STG ROIs are shown for the CH2 template.
Definition of subject-specific regions of interest
For study 1, a total of eight anatomical ROIs were selected in order to examine how the accuracy of the template counts varied across regions. These regions were the left and right hippocampi, the left and right precentral gyri (PcG), the left and right middle frontal gyri (MFG), and the left and right superior temporal gyri (STG). For the cortical ROIs, we used the 3D cortical surface-rendered mesh to identify sulcal boundaries. First, markers were placed along the sulcal boundaries on the 3D representation. Then, using the markers for reference, the actual ROI labeling was performed on 2D coronal slices of the imported MR images (Figure 1a,b,c). Finally, the regions drawn on the 2D planes were combined to create 3D ROIs. For study 1, two raters independently drew the ROIs for all subjects using the anatomical landmarks described below as guidelines.
Hippocampus
Hippocampal region drawings do not rely on the surface sulci, so the 3D mesh was not used for this ROI. The anatomical boundaries for defining the hippocampus are described in detail in (Wu et al., 2002) and summarized here. Generally, the anterior boundary was identified at the level of the inferior horn of the lateral ventricle. At rostral levels, the superior boundary was formed by the amygdala (medially) and lateral ventricle (laterally). The ventral boundary was defined by the white matter deep to the hippocampus and the subiculum. Then, caudal to the amygdala, the boundaries were made by the ventricle (dorsally), the lateral adjacent white matter, and the white matter subjacent to the subiculum. The posterior boundary was defined as the first coronal slice in which the fornix was completely distinct from any gray/white matter.
Precentral gyrus (PcG)
The anterior boundary of the PcG was defined by the precentral sulcus. The interhemispheric fissure defined the superior boundary; regions within the fissure were excluded. The posterior border was defined by the central sulcus, and the inferior border was defined by the lateral sulcus, excluding the insula.
Middle frontal gyrus (MFG)
The middle frontal gyrus was bound superiorly by the superior frontal sulcus and inferiorly by the inferior frontal sulcus. The posterior boundary was the precentral sulcus. The anterior boundary was defined to be the coronal slice at the 40% mark from the genu of the corpus callosum to the anterior-most slice with cortex. Because our subjects were from an older cohort, there was considerable atrophy in their frontal lobes. As a result, the topology of the anterior portion of the middle frontal gyrus was highly variable. In ambiguous cases in which neither the inferior nor superior sulci were clearly identifiable, the boundaries were defined by the most prominent sulci in the anterior portion of frontal lobe that were the most likely extension of their respective sulci. This was one of the most difficult ROIs to label due to the high variability across subjects.
Superior temporal gyrus (STG)
The boundaries for the STG were defined superiorly by the lateral sulcus, medially by the limiting fissure of the insula, and inferiorly by the superior temporal sulcus. The posterior boundary was the same as the last slice of the posterior boundary of the hippocampus, and the anterior boundary was defined as the most anterior coronal slice to still show the temporal pole.
Definition of the “gold standard”
In both study 1 and study 2, the “gold standard” was defined as the counts extracted from the PET image by using the regions that were drawn on the subject’s MR then coregistered to the PET using the MR. Because the gray-white boundaries were difficult to determine while drawing on the ROIs, all of the subject-specific regions were masked by the subject’s segmented gray image with a threshold of 50%. The counts extracted from this composite mask were used as the “gold standard”. In study 1, there were two independent raters. In this case, rater 1 was arbitrarily defined as the “gold standard,” and results from rater 2 were compared with those of rater 1.
Definition of template regions of interest
The template regions of interest were defined using the same boundary guidelines that were used for the individual subject regions, however the gray-white boundary within sulci was not delineated in the template regions of interest (Figure 1d). Masks were made in this way so that template regions could accommodate localized differences between the subject anatomy and the template anatomy, and the conjunction of the gray-matter segmented subject specific MRI with the template ROI would define the precise cortical ROI for each individual. The template ROIs were drawn on both the CH2-masked and MDT2-masked templates. The creation of these templates is described in the Templates section.
Templates
For study 1, we considered two template images: CH2-masked and MDT2-masked. We used templates based upon single-subjects for this study because drawing ROIs on single subject templates was more reliable than drawing ROIs on averaged templates, in which contrast and anatomical detail are lost. For study 2, we considered 6 different templates. These include the two templates used in study 1 as well as the standard MNI template, the CH2-gray, a SALSA template created from subjects in the SALSA cohort, and a SALSA-gray template. These 6 templates are described below. See Figure 2 for a summary of the templates.
Figure 2. Summary of templates.
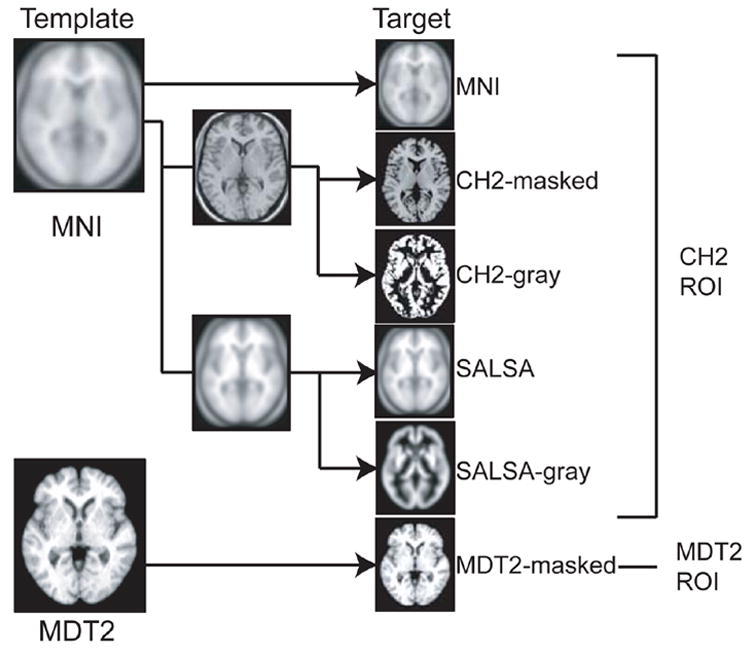
All of the target images used in studies 1 and 2 are derived from either the MNI template (shown in the upper left) or the MDT2 template (shown in the lower left). The first target image (right column, from top to bottom) is the standard MNI template. The second and third target images are derived from the CH2 template, which was generated by coregistering 27 scans from a single subject to the MNI template. The second target image, CH2-masked, is the CH2 template masked so that the skull and CSF are excluded. The third target image, CH2-gray, is the gray component of the segmented CH2 template. SALSA, the fourth target image, was generated by coregistering and normalizing 151 older subjects to the MNI template. The fifth target image, SALSA-gray, was generated by coregistering and normalizing the segmented gray images for each individual subject to the MNI gray-matter prior. Finally, the sixth target image, MDT2, was created by using a C-spline warping method to warp a single older subject’s image to the average warp of 24 older subjects’ image. See Methods for a more detailed description of each target image.
MNI
The Montreal Neurological Institute (MNI) template is the international standard as defined by the International Consortium of Brain Mapping (ICBM) and is the default T1 template provided by commonly used functional imaging packages such as SPM2. This template was created in a multi-stage process. First, 241 normal T1-weighted MR scans were manually scaled to match to the Talairach atlas using manually defined reference points. These scaled images were averaged to create the template for the first stage. Next 305 normal MRI scans were coregistered to the first template using an automated 9-parameter linear transformation and then averaged to create the MNI305. Finally, 152 normal scans were coregistered to the MNI305 template and averaged to create the current MNI template. For review, see (Brett et al., 2002).
CH2-masked
The CH2 template was created by coregistering and averaging 27 scans from a single individual to the MNI305 template, resulting in a highly detailed single-subject T1 template (Holmes et al., 1998). A brain mask for this template was generated by segmenting the template and combining the gray- and white- segmented images. The CH2 template was then masked with the brain mask to create the CH2-masked template.
CH2-gray
Segmenting the CH2 template using the standard segmentation parameters defined in SPM2 generated the CH2-gray template.
SALSA
T1-weighted MR scans of 151 older subjects from the SALSA cohort were normalized to the MNI template using a multistage method described in (Senjem et al., 2005). Briefly, each scan was coregistered to the MNI template using a 12-parameter affine registration. The coregistered image was then segmented and the gray-matter image was normalized to the MNI gray-matter prior. The non-linear warping parameters generated in the normalization process were applied to the coregistered image. These warped images were averaged across subjects and smoothed to create the SALSA template. Note that all of the SALSA subjects who had an MR were used to create the template, including CIND and demented subjects.
SALSA-gray
The SALSA-gray template was created in the same way as the SALSA template. However, after the original MRs were warped by the nonlinear parameters, each MR was segmented again to produce a new gray-matter image. These new gray-matter images were averaged across subjects and smoothed to create the SALSA-gray template (Senjem et al., 2005). The same subjects as for the SALSA template were used here.
MDT2
The MDT2 template was created from T1-weighted MR scans of 25 normal older subjects. First, a single subject’s MR was selected as a proto-template. The proto-template was normalized to the 24 other scans using a C-spline warping method. The deformation field resulting from the warp indicated how each voxel in the proto-template should deform to fit the subject. These deformation fields were averaged across all subjects and smoothed. The final deformation field, which was an average of 24 deformation fields, was applied to the proto-template. This resulted in a detailed single-subject scan warped by the averaged-deformation field of 24 subjects. This template was created by the Imaging of Dementia and Aging laboratory (IdeALab) at the University of California, Davis.
Brain pipeline
We developed an in house software program to automate the process of extracting PET counts from individual subject data using regions of interest drawn on a template and gray-matter segmented images derived from the subject’s MR. The procedure is outlined below:
Coregister the subject’s PET to the subject’s MR
Segment and mask the subject’s MR
Normalize subject’s masked MR to the template
Overlay the template masks with the subject’s segmented image
Extract the PET counts from the intersection of the Template mask and the segmented image
First, using a 12-parameter affine transformation, the subject’s PET image is coregistered to the subject’s MR. The transformation parameters are determined using the coregistration tool in SPM2. Next, the subject’s MR is segmented (Ashburner and Friston, 1997; Ashburner and Friston, 2005). The extracted gray- and white- segmented images are summed to form a brain mask. This brain mask is applied to the subject’s MR to create a brain-masked MR, stripped of the skull and surrounding CSF. Third, the subject’s MR is normalized to the template using the default non-linear spatial normalization settings in SPM2. If the template is already brain-masked, the brain-masked MR is used as the source. Otherwise the original MR image is used as the source. The normalization parameters derived in this process are then applied to the coregistered PET and the segmented images while preserving the image concentrations. Finally, after normalization, a new mask is generated from the intersection of the normalized segmented gray image (derived from the subject’s MR) and the template mask. This conjunction mask represents subject-specific gray matter within the region of interest, as defined by the template mask. The PET data, representing the averaged counts per voxel within the subject specific region of interest are extracted from the normalized PET image using the conjunction mask (Figure 3).
Figure 3. Flow chart demonstrating template-based ROI analyses.
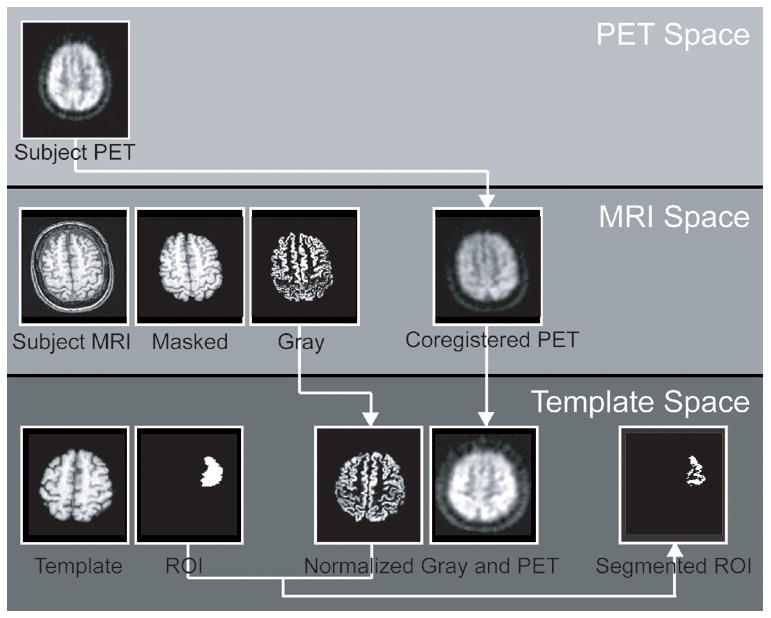
The raw images originate in three separate spaces; subject PET space, subject MRI space, and Template space. First, the subject’s PET image is transformed to MRI space by coregistering it to the subject’s MR image. Within MRI space, the subject’s MR image is masked to exclude skull and CSF and segmented to produce a gray image. The subject’s PET image, and segmented gray image are transformed to Template space by normalizing the masked MRI to the masked template image and applying the transformation parameters to the segmented gray image and the PET image. In Template space, PET counts are extracted using the intersection of the template ROI and the subject’s segmented gray image.
Results
Study 1
In study 1 we examined 3 parameters that may affect the accuracy of the template-extracted counts. These parameters are the anatomical location, the choice of template, and the threshold of the segmented gray image. All extracted counts using the template regions were compared against the “gold standard”, which were counts extracted using regions carefully drawn on each individual subject by rater 1 (n=15). In addition, the extracted counts using subject-specific regions drawn by rater 2 (n=10) were compared to the first rater in order to measure variability of inter-rater differences.
Effect of Template
We normalized PET and MR images to two different target images, the standard CH2 template derived from a single young healthy subject and the MDT2 template derived from an older subject. Both templates were masked using a brain mask. The percentage error was calculated by dividing the absolute difference between the template-extracted counts and the “gold standard” by the “gold standard”. The average percent error of the counts across all regions and all subjects was 3.4% for the CH2-masked template, 1.5% for the MDT2-masked template, and 1.1% for the second rater. Paired comparisons were used to test whether the percent error for each template was significantly different from each other and from the percent error of the second rater. The p-values are shown in table 1, and the percent error metrics are shown with standard error bars in figure 4 (top). The percent error of the counts extracted using the CH2-masked template was significantly greater than that found when using the MDT2-masked template for all regions except for the left Hipp. In addition, the percent error of the MDT2-masked template was comparable to the percent error of the second rater, with the exception of the right Hipp. The percent error of the CH2-masked template was significantly different from the second rater in the left and right Hipp, the left PcG, and the right STG. Figure 5 shows a scatter plot of the counts extracted using the MDT2-masked template and the gold standard (r = .992), and a scatter plot of the counts extracted using the CH2-masked template and the gold standard (r = .967).
Table 1. Paired comparisons of the percent error (template).
Paired comparisons were used to test whether the difference between each method and the “gold standard” was significantly greater than the difference between rater 2 and “gold standard”. The percent error between the MDT2-masked and CH2-masked templates were also compared.
| L Hipp | r Hipp | l PcG | r PcG | l MFG | r MFG | l STG | r STG | |
|---|---|---|---|---|---|---|---|---|
| MDT2 vs R2 | 0.115 | 0.001 | 0.383 | 0.763 | 0.374 | 0.455797 | 0.202 | 0.362 |
| CH2 vs R2 | 0.013 | 0.008 | 0.007 | 0.024 | 0.063 | 0.085934 | 0.330 | 0.003 |
| MDT2 vs CH2 | 0.106 | 0.007 | 0.003 | 0.015 | 0.019 | 0.032761 | 0.035 | 0.021 |
Figure 4. Percent error between template-based ROIs and the subject-specific ROIs.
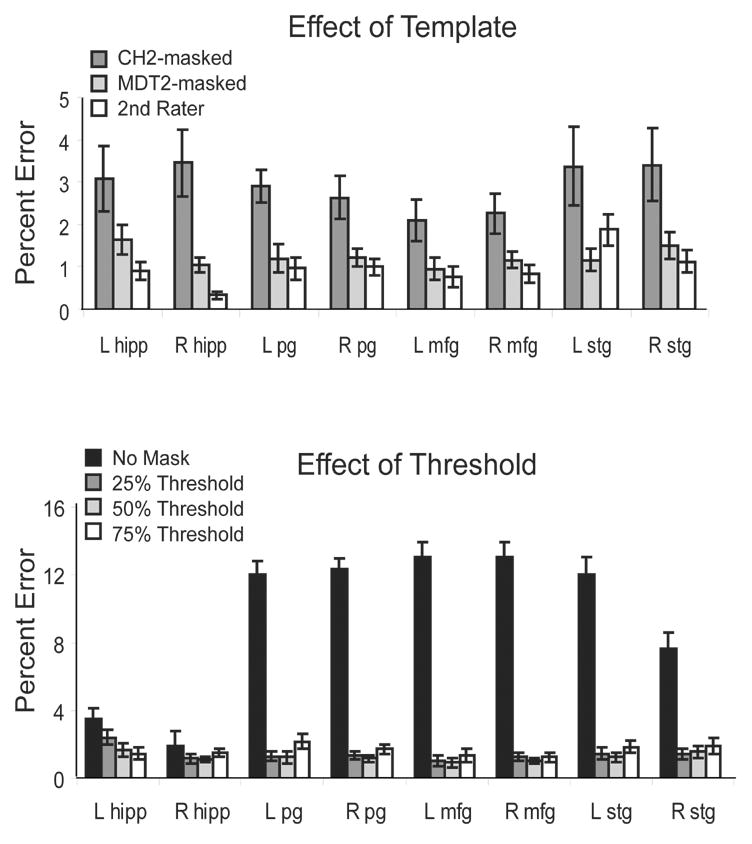
The percent error reflects the difference between each method and the “gold standard” normalized by the “gold standard”. Here, the “gold standard” is defined as the counts extracted using subject-specific ROIs drawn by the 1st rater. (top) The MDT2-masked template and the 2nd rater were comparable for all regions except for the right Hipp. The percent error of the CH2-masked template was significantly greater than that of the 2nd rater and the CH2-masked template for all regions except the left Hipp, where the percent error of the CH2-masked template was comparable to the MDT2-masked template. (bottom) The percent error for the MDT2-masked template is shown for different levels of thresholding on the segmented gray image. Using zero thresholding resulted in the largest percent error across all regions.
Figure 5. Scatter plots of template-based ROIs versus subject specific ROIs.
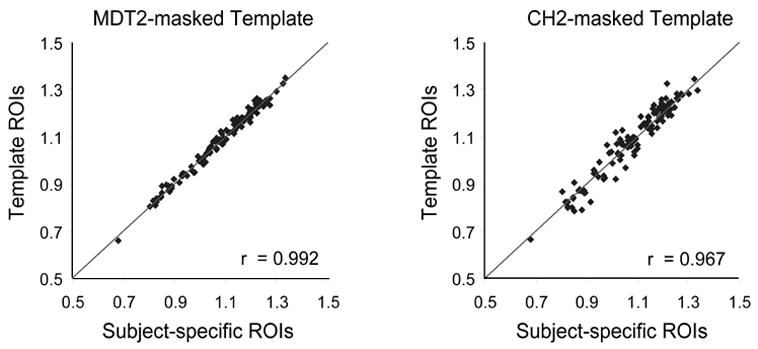
These plots illustrate the correlation between the MDT2-masked template and the “gold standard” (left) and between the CH2-masked template and the “gold standard” (right).
Effect of Location
For both templates, an ANOVA revealed no significant differences in the percent discrepancy across regions. However, for the 2nd rater, the ANOVA revealed a significant difference across regions (p = 0.002). Post-hoc analysis indicates that the left STG had significantly greater error than the right Hipp.
Effect of Threshold
In step 4 of the brain pipeline, the template ROI was masked with the subject’s segmented gray image. The intensity values of the segmented gray image ranged from zero to one, indicative of the likelihood of the voxel representing a gray-matter region. We considered different threshold levels of the segmented gray image to determine if the threshold affected the accuracy of the extracted counts. We used thresholds at the 25%, 50%, and 75% intensity levels, as well as using no threshold such that all of the counts within the template ROI were extracted. The percent error for the MDT2-masked template is shown for all regions in figure 4 (bottom). Using a threshold of zero resulted in the greatest errors and was significantly greater for all regions except for the right Hipp. The differences between using the 25%, 50%, and 75% thresholds were small, however; in all cases where there was a significant difference between the thresholds, the error for the 50% threshold was less. P-values are shown for the paired comparisons of the percent error between all thresholds and the 50% threshold in Table 2 (significant values are highlighted in red).
Table 2. Paired comparisons of percent error (threshold).
Paired comparisons were used to test whether there was a significant difference between different thresholds for the segmented gray image. All values are shown for the MDT2-masked template.
| l Hipp | r Hipp | l PcG | r PcG | L MFG | r MFG | l STG | r STG | |
|---|---|---|---|---|---|---|---|---|
| 25% vs 50% | 0.046 | 0.776 | 0.811 | 0.361 | 0.696 | 0.252 | 0.108 | 0.465 |
| 75% vs 50% | 0.533 | 0.016 | 0.003 | 0.073 | 0.099 | 0.373 | 0.033 | 0.349 |
| None vs 50% | 0.024 | 0.411 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Study 2
In study 2, we focused specifically on the hippocampus. In our cohort of 88 subjects, the left and right hippocampi were drawn for each individual subject using their T1-weighted MRI. We tested several different templates to use as targets in the normalization process. These templates were the MNI, CH2-masked, CH2-gray, SALSA, SALSA-gray, and MDT2-masked. In this study we examined two aspects of normalization: 1) accuracy, as in how well the extracted counts matched the “gold standard”; and 2) correspondence, as in how well the subject’s hippocampal masks overlapped across the group. In all cases, we used a segmented-gray threshold of 50%.
Accuracy
The graph in figure 6a illustrates the effect of using different templates to extract the PET counts within the hippocampus. Templates that resulted in the greatest percent error as compared to the “gold standard” are the standard MNI template, the SALSA template, and the SALSA-gray template. Templates that resulted in the least percent error are the CH2-masked template, the CH2-gray template and the MDT2-masked template. Of all the templates, we found that the MDT2-masked template resulted in the least percent error, which was significantly lower than all of the other templates. Moreover, the PET values extracted using the MDT2 template mask were highly correlated with the counts extracted using individual subject masks (r = 0.968). The scatter-plot of this relationship is shown in Figure 6b.
Figure 6. Percent error between template-based ROIs and subject-specific ROIs for the hippocampus.
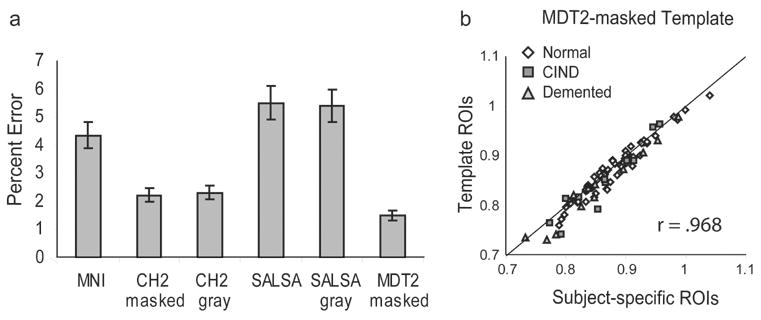
(a) This bar-graph illustrates the percent error between each of the 6 templates and the “gold standard”. The SALSA, SALSA-gray, and MNI templates have the highest percent error, whereas the CH2-masked, CH2-gray and MDT2-masked templates have the lowest percent error, with the error of the MDT2-masked template significantly lower than all other templates. (b) The scatter plot illustrates the correlation between the MDT2-masked template and the subject-specific ROIs (r = 0.968). All 88 subjects are included; white circles = normal; dark gray squares = CIND; light gray triangles = AD.
Correspondence
To determine how well the hippocampi overlap across subjects after normalization, all of the normalized hippocampal masks were overlapped with each other. The overlapped masks are shown in Figure 7. We measured the percentage of voxels for which at least 90% of the subjects overlapped. These values are displayed in Table 3. The template that resulted in the greatest overlap after normalization was the SALSA-gray. The CH2-gray and MDT2-masked also resulted in a considerable 90% overlap across subjects. To quantify how well the subject masks overlapped with the template masks, we used a fit measure that summed the total number of overlapping voxels (weighted by the number of overlaps) within the template mask minus the weighted sum of the voxels outside of the template mask, divided by the total weighted sum of overlapping voxels. This fit measure is normalized from −1 to 1, such that a value of −1 indicates that all hippocampal voxels of all subjects lie outside of the template hippocampal mask, and a value of 1 indicates that all hippocampal voxels of all subjects lie inside the template hippocampal mask. These values are displayed in Table 3. Figure 7 illustrates the outline of the template hippocampal masks along with the overlapping subject masks. The template with the greatest fit is the MDT2-masked.
Figure 7. Overlap of hippocampal masks across subjects.
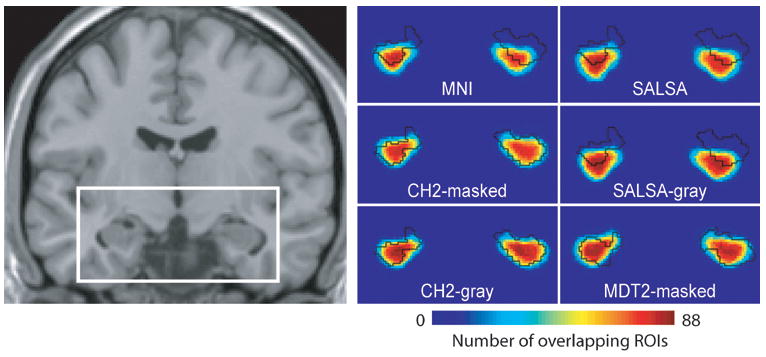
Each individual subject’s hippocampal mask is transformed to the template space and overlapped with each other. This figure illustrates how well the hippocampal masks overlap with each other, and with the template hippocampal mask (outlined in black). The white rectangle on the coronal slice indicates the anatomical location of the color-scale maps. Red indicates a high percentage overlap across subjects, blue indicates a low percentage overlap across subjects.
Table 3.
Fit and percent overlap of hippocampal masks of all subjects.
| MNI | CH2 masked | CH2 gray | SALSA | SALSA gray | MDT2 masked | |
|---|---|---|---|---|---|---|
| Fit | 0.404 | 0.668 | 0.671 | 0.298 | 0.312 | 0.679 |
| % Overlap | 5.603 | 2.569 | 12.16 | 8.588 | 17.62 | 11.28 |
Discussion
In this paper, we present an automated method for extracting PET data from specific regions of interest. While studies have shown that regions drawn on a template can be appropriate for extracting data for normal healthy subjects, it is unclear how these results can be applied to different populations. In this study, we focused on the aging population and examined how different parameters in the template data-extraction process may affect the accuracy of the results.
We found that the target image to which all subjects were normalized had a large effect on the accuracy of the counts extracted from the regions. In the first study, we compared a template based on a single individual in the healthy young population, which was coregistered to an averaged template of 305 healthy young subjects, to a template based on a single older subject, which was warped to the average space of 24 older subjects. We found that using an older template resulted in a significantly reduced percent error of extracted counts compared to using a young template for all regions except the left hippocampus. Moreover the older template performed comparably to the second rater for all regions except for the right hippocampus. This suggests that the error from using a template region to extract PET data maybe within the range of inter-rater variability.
In addition, we examined the effect of the segmented gray threshold on the extracted counts. Similar to the method presented by Yusano et al., (2002) the automatic template extraction method that we presented uses the subject-specific segmented gray image to refine the generalized template ROIs. We found that using no gray threshold resulted in a percentage error of (8–12%), which was far greater than the percentage error measured when using a threshold for the segmented gray image. This reiterates the importance of incorporating the individual’s anatomical information even within an automated template framework.
To further investigate the effect of the template, we considered 6 templates in study 2. The three single-subject templates (MDT2-masked, MDT2-gray, and CH2-masked) had significantly reduced errors compared to the group-averaged templates, which includes the standard MNI template. Within the single-subject templates, the MDT2 template performed significantly better than the young template. This may be because the MDT2 template, which was derived from older subjects, was more representative of our older cohort. In comparison to the CH2 template, the MDT2 template is atrophied and the ventricles are larger. Surprisingly, among the group-averaged templates, the SALSA templates resulted in significantly greater error than the MNI template, even though the SALSA template was composed of the same subjects used in the study. This may be due to the high variability in the older cohort. Averaging images from a highly variable group would have resulted in a template image with greater smoothing and reduced spatial information, which may have affected the fidelity of the normalization.
We also examined whether there would be any difference between gray-to-gray, brain-to-brain, or head-to-head normalization. There was no difference in the mean percentage error between CH2-masked and CH2-gray as well as between SALSA and SALSA-gray, suggesting no benefit in using either a gray-only, or brain-only template for extracting data. However, the two gray-only templates resulted in the greatest overlap of masked voxels, revealing that using a gray-only template improves the overall normalization process. The discrepancy between these results may be resolved by examining the overlap of all hippocampal masks after normalization. The SALSA-gray template resulted in the greatest number of overlapping voxels across all subjects, possibly because the template was partially defined by the subjects in the study. However, the region of overlap for the hippocampal masks after normalizing to the SALSA-gray template was actually shifted relative to the template ROI. Therefore the data extracted using the template ROI were not completely representative of the data within the subject’s ROI. Because the hippocampus is a small region, it is particularly sensitive to millimeter shifts between the template ROI and the subject ROI.
In the best-case scenario, which was using the MDT2-masked template in conjunction with the segmented gray thresholded at 50%, we found that the average percent error for all regions was only 1.5% with respect to the gold standard. For the hippocampus in a cohort of 88 older subjects, the maximum percent error was 7.1% and the correlation coefficient between the template data and gold standard was 0.968. To compare these results to the normal population, Yasuno found in a cohort of 22 subjects that the maximum percent error between the template and the gold standard was 4.8% for the hippocampi and that the correlation coefficient was greater than 0.99. These results reflect that the data in the older cohort is more variable than the normal population. However the high correlation coefficient suggests that using a template-based ROI method could be useful provided that the appropriate template and parameters are selected. These results provide future direction for template-based hypothesis-testing research using PET data in studies of aging and dementia.
The goal of this paper was to identify the parameters within the normalization and segmentation process that may affect the accuracy of an automated ROI-based analysis, and not to compare the accuracy across different software packages. While we use the SPM2 software package, a number of other methods are available that provide similar tools. These have been compared in recent publications (Gold et al., 1997; Boesen et al, 2004). We have noted that some parameters could increase the variability of the results, particularly the choice of template for a patient population. We expect that this would be true regardless of the software analysis package chosen since the issue of anatomical distortion as a function of aging and atrophy is a universal problem. However, software may perform differently based on the specific algorithms used, so that a complete analysis of normalization method, templates, and segmentation methodology should be performed with any new patient population and software used.
Acknowledgments
This work was supported by NIH grants U01AG024904, AG12975, and DK60753. The authors are grateful to Dr. Mary Haan, Principal Investigator of SALSA for support and advice, and to Drs. Evan Fletcher and Charles DeCarli for the MDT2 template.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J, Friston K. Multimodal image coregistration and partitioning--a unified framework. Neuroimage. 1997;6:209–217. doi: 10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Boesen K, Rehm K, Schaper K, Stoltzner S, Woods R, Lüders E, Rottenberg D. Quantitative comparison of four brain extraction algorithms. NeuroImage. 2004;22:1255–1261. doi: 10.1016/j.neuroimage.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Bohm C, Greitz T, Seitz R, Eriksson L. Specification and selection of regions of interest (ROIs) in a computerized brain atlas. J Cereb Blood Flow Metab. 1991;11:A64–68. doi: 10.1038/jcbfm.1991.39. [DOI] [PubMed] [Google Scholar]
- Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nature Reviews Neuroscience. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Cointepas Y, Mangin JF, Garnero L, Poline JB, Benali H. BrainVISA: Software platform for visualization and analysis of multi-modality brain data. Neuroimage. 2001;13:S98–S98. [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1996;3:190. [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- de Leon MJ, Convit A, Wolf OT, Tarshish CY, DeSanti S, Rusinek H, Tsui W, Kandil E, Scherer AJ, Roche A, Imossi A, Thorn E, Bobinski M, Caraos C, Lesbre P, Schlyer D, Poirier J, Reisberg B, Fowler J. Prediction of cognitive decline in normal elderly subjects with 2-[(18)F]fluoro-2-deoxy-D-glucose/poitron-emission tomography (FDG/PET) Proc Natl Acad Sci U S A. 2001;98:10966–10971. doi: 10.1073/pnas.191044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold S, Christian B, Amdt S, Zeien G, Cizadio T, Johnson DL, Flaum M, Andreasen NC. Functional MRI statistical software packages: A comparative analysis. Human Brain Mapping. 1997;6(2):73–84. doi: 10.1002/(SICI)1097-0193(1998)6:2<73::AID-HBM1>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline JB, Frith CD, Frackowiak RSJ. Statistical Parametric Maps in Functional Imaging: A General Linear Approach. Human Brain Mapping. 1995;1:189–210. [Google Scholar]
- Haan MN, Mungas DM, Gonzales HM, Ortiz TA, Acharya A, Jagust WJ. Prevalence of dementia in older Latinos: the influence of type 2 diabetes, stroke, and genetic factors. J Am Geriatr Soc. 2003;51:169–177. doi: 10.1046/j.1532-5415.2003.51054.x. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Koepp MJ, Duncan JS. Three-dimensional probabilistic atlas of the human brain. Neuroimage. 2002;16:S86–S86. [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Human Brain Mapping. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. Journal of Computer Assisted Tomography. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Minoshima S, Koeppe RA, Frey KA, Kuhl DE. Anatomic Standardization: Linear Scaling and Non-linear warping of Functional Brain Images. J Nucl Med. 1994;35:1528–1537. [PubMed] [Google Scholar]
- Mosconi L, Tsui WH, De Santi S, Li J, Rusinek H, Convit A, Li Y, Boppana M, de Leon MJ. Reduced hippocampal metabolism in MCI and AD - Automated FDG-PET image analysis. Neurology. 2005;64:1860–1867. doi: 10.1212/01.WNL.0000163856.13524.08. [DOI] [PubMed] [Google Scholar]
- Senjem ML, Gunter JL, Shiung MM, Petersen RC, Jack CR. Comparison of different methodological implementations of voxel-based morphometry in neurodegenerative disease. Neuroimage. 2005;26:600–608. doi: 10.1016/j.neuroimage.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Mungas D, Petkov CI, Eberling JL, Zrelak PA, Buonocore MH, Brunberg JA, Haan MN, Jagust WJ. Brain structure and cognition in a community sample of elderly Latinos. Neurology. 2002;59:383–391. doi: 10.1212/wnl.59.3.383. [DOI] [PubMed] [Google Scholar]
- Yasuno F, Hasnine AH, Suhara T, Ichimiya T, Sudo Y, Inoue M, Takano A, Ou T, Ando T, Toyama H. Template-based method for multiple volumes of interest of human brain PET images. Neuroimage. 2002;16:577–586. doi: 10.1006/nimg.2002.1120. [DOI] [PubMed] [Google Scholar]


