Abstract
Haemophilus ducreyi is a gram-negative bacterium that is the causative agent of chancroid. Strain 35000HP has been well characterized and is representative of the majority of H. ducreyi strains. Strain 35000HP produces a lipooligosaccharide (LOS) that contains d-glycero-d-manno-heptose in the main oligosaccharide chain extension; the lbgB gene has been shown to encode the dd-heptosyltransferase. The lbgB gene is found in a gene cluster together with the lbgA gene, which encodes for the galactosyltransferase I. These two genes are flanked by two housekeeping genes, rpmE and xthA, encoding the ribosomal protein L31 and the exonuclease III, respectively. Recently, a second group of H. ducreyi strains have been identified. Strain 33921, a representative of the class II strains, produces an LOS that lacks dd-heptose in the oligosaccharide portion of its LOS. To better understand the biosynthesis of the dd-heptose-deficient 33921 LOS, we cloned and sequenced the corresponding lbgAB genomic region from strain 33921. Similar to strain 35000HP, the 33921 genome contains xthA and rpmE. However, between these two genes we identified genes encoding two putative glycosyltransferases that were not highly homologous to the 35000HP lbgAB genes. In this study, we demonstrate that the product of one of these genes encodes a galactosyltransferase. In addition, dot blot hybridization determined that 3 of 35 strains tested had the atypical transferases present, as did 4 strains characterized as class II strains by other criterion. These data indicate that the lbgAB genes can serve as one indicator of the classification of H. ducreyi strains.
Haemophilus ducreyi is the causative agent of chancroid, a sexually transmitted genital ulcer disease. Chancroid is prevalent in certain areas of Africa, Asia, and Latin America (34); however, sporadic outbreaks occur in the United States (40). Chancroid has been linked to the transmission of the human immunodeficiency virus (HIV), especially in areas where both diseases are prevalent (21, 34, 39, 40).
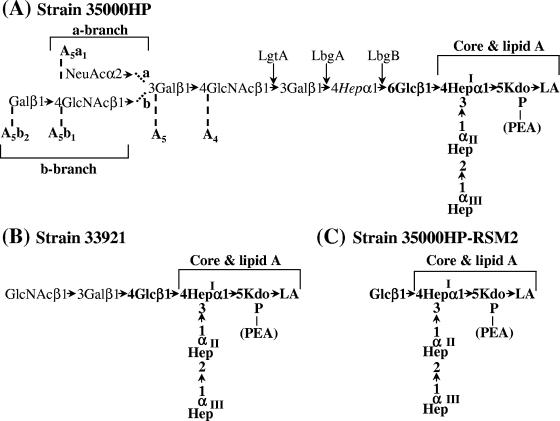
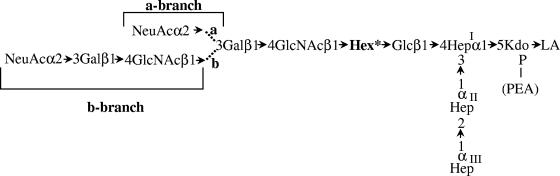
Strain 35000 and its human-passaged derivative, strain 35000HP, have been investigated extensively. Putative virulence determinants, including a hemolytic cytotoxin (2, 28, 45), cytolethal distending toxin (6-8), the serum resistance-conferring proteins DsrA and DltA (12, 24), and a hemoglobin-binding protein (11, 36), have been identified and characterized in this genetic background. The structure of the lipooligosaccharide(s) (LOS) and many of the glycosyltransferases responsible for the synthesis of the LOS have also been identified and characterized (3-5, 13, 15, 16, 25-27, 37, 38). 35000HP LOS contains three l-glycero-d-manno-heptose (ld-Hep) residues linked to a phosphorylated 3-deoxy-d-manno-octulosonic acid (Kdo). The first heptose residue is substituted with the pentasaccharide Galβ1-4GlcNAcβ1-3Galβ1-4DDHepα1-6Gluβ1-4 (Fig. 1A). The nonreducing galactose can be further substituted with sialic acid when it is available (16, 27). Most of the isolates examined produce this LOS structure (1, 32).
FIG. 1.
LOS structures of H. ducreyi strains 35000HP, 33921, and 35000HP-RSM2. The core and lipid A regions of the LOS structures are indicated. The conserved portions of the LOS are shown in boldface. The N-acetylglucosamine glycosyltransferase, LgtA, the galactosyltransferase, LbgA, and the heptosyltransferase, LbgB, are shown at their points of action. The 35000HP LOS structure is labeled with the previously developed nomenclature for the various LOS glycoforms (5). The three core heptoses (Hep) are l-glycero-d-manno-heptose; the branch heptose, in italics, is of the d-glycero-d-manno configuration.
Recently, a second class of isolates was recognized based on proteomic analysis (31) and the analysis of the genes encoding several surface proteins, including DsrA (44). One of these isolates, strain 33921, produces a LOS that lacks d-glycero-d-manno-heptose. In this LOS, the first ld-heptose residue is substituted with the trisaccharide GlcNAcβ1-3Galβ1-4Glcβ1-4 (Fig. 1B) (25). The LOS of strain 33921 does not contain sialic acid, even when sialic acid is present at high concentrations (25).
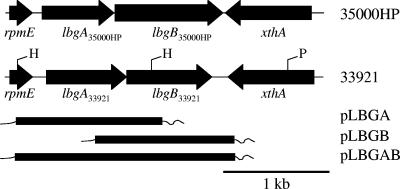
Since the LOS from strain 33921 does not contain dd-heptose, we wanted to determine whether strain 33921 had the gene encoding the dd-heptosyltransferase. The galactose I transferase and the dd-heptosyltransferase in strain 35000HP are encoded by the lbgAB genes (alternatively designated losAB genes) (15, 35, 42, 46), which are flanked by the housekeeping genes rpmE and xthA (Fig. 2). In the present study, we characterized this region of the 33921 genome. Our findings demonstrate that the gene arrangement in this region of the 33921 genome is identical to that of 35000HP; that is, two glycosyltransferases flanked by the rpmE and xthA genes (Fig. 2). However, whereas the rpmE and xthA genes are highly conserved, the genes encoding the two glycosyltransferases are not highly homologous to the lbgAB35000HP genes. We describe our findings here regarding the function of the lbgAB33921 genes.
FIG. 2.
Map of the lbgA, lbgB region in strains 35000HP and 33921. The positions and direction of transcription of the genes are designated. Restriction enzyme sites relevant to the 33921 clones used for sequence determination are labeled as H for HincII and P for PvuII. Another HincII site is located approximately 400 bp 5′ to the xthA gene in strain 33921. The plasmids containing portions of the 33921 lbgAB region used in the complementation studies are also illustrated. The thick line represents the portion of the 33921 lbgAB region in each clone, and the thin line represents adjacent vector sequences. The plasmid pLBGBR contains the same insert as pLBGB in the reverse orientation relative to the plasmid backbone.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in the present study are listed in Table 1. H. ducreyi strains were grown on chocolate agar or in brain heart infusion broth as described previously (28). Streptomycin was used at 20 μg/ml as necessary. Chloramphenicol was used at 1 μg/ml on transformation plates or at 0.5 μg/ml when clones were isolated. All Escherichia coli strains were grown on Luria-Bertani plates or in Luria-Bertani broth. When necessary, this medium included X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 40 μg/ml and/or the appropriate antibiotics. Ampicillin was used at 50 μg/ml, kanamycin was used at 20 μg/ml, and streptomycin was used at 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or site of isolation | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Host strain used for cloning | Invitrogen |
| DH5αpcnB | pcnB, zad::Tn10 | G. Barcak (30) |
| H. ducreyi | ||
| 35000HP | Human-passaged virulent isolate | 33 |
| 33921 | Kenya | ATCC |
| CIP542 ATCCa | Hanoi, Vietnam | ATCC |
| DMC64 | Bangladesh | 44 |
| DMC111 | Bangladesh | 44 |
| HMC37 | England | 10 |
| HMC65 | Raleigh, NC | 10 |
| HMC112 | Kenya | P. Totten |
| SSMC71 | Bangladesh | 44 |
| 35000HP-RSM2 | lbgB35000HP mutant | 46 |
| 35000HP-RSM223 | lbgB35000HP, lgtA mutant | This study |
| 35000HP-RSM2(pLBGA) | lbgB35000HP mutant + lbgA33921 | This study |
| 35000HP-RSM2(pLBGB) | lbgB35000HP mutant + lbgB33921 | This study |
| 35000HP-RSM2(pLBGBR) | lbgB35000HP mutant + lbgB33921 (insert in the reverse orientation) | This study |
| 35000HP-RSM223(pLBGA) | lbgB, lgtA mutant + lbgA33921 | This study |
| 35000HP-RSM223(pLBGAB) | lbgB, lgtA mutant + lbgAB33921 | This study |
| Plasmids | ||
| pWKS30 | Low-copy-number plasmid vector | 43 |
| pLS88 | H. ducreyi shuttle vector | 9 |
| pCR2.1 | TA cloning vector | Invitrogen |
| pLSSK | H. ducreyi shuttle vector | 45 |
| pUC4DEcat | Source of cat cassette | 17 |
| pRSM2207 | pWKS30 + HincII fragment of 33921 | This study |
| pRSM2287 | pWKS30 + PvuII fragment of 33921 | This study |
| pRSM2279 | lbgB33921 + pCR2.1 | This study |
| pLBGB | lbgB33921 + pLS88 | This study |
| pRSM2306 | lbgA33921 + pCR2.1 | This study |
| pLBGA | lbgA33921 + pLS88 | This study |
| pLBGBR | lbgB33921 + pLS88, insert in the reverse orientation compared to pLBGB | This study |
| pRSM2307 | lbgAB33921 + pCR2.1 | This study |
| pLBGAB | lbgAB33921 + pLSSK | This study |
| pRSM2379 | lgtA + pRSM2072 | 37 |
| pRSM2727 | pRSM2379 with cat gene cloned into the lgtA gene | This study |
Recombinant DNA methodology.
Plasmids were isolated by utilizing purification kits (QIAGEN, Chatsworth, CA). Restriction enzymes were purchased from New England Biolabs (Beverly, MA), and T4 DNA ligase was purchased from Invitrogen (Grand Island, NY). Standard recombinant DNA methods were used.
DNA sequence was determined through cycle sequencing using ABI Prism dye terminator cycle sequencing ready reaction kits (Applied Biosystems, Foster City, CA). Excess dye terminators were removed by passage through Centriflex gel filtration cartridges (Edge BioSystems, Gaithersburg, MD), and the sequence was determined on an ABI 377 or an ABI 3130 DNA sequencer. Sequence analysis and comparisons were performed with Lasergene software (DNAStar, Madison, WI).
Determination of the sequence of the rpmE-xthA genomic region in strain 33921.
To determine whether strain 33921 had the lbgB gene, which encodes for the dd-heptosyltransferase, we probed HincII-digested 33921 chromosomal DNA with a probe containing portions of the lbgB and xthA genes from 35000HP. A band approximately 2.2 kb in size was detected (data not shown). To isolate the DNA that reacted with the probe, a partial genomic library was generated by digesting 33921 chromosomal DNA with HincII and collecting 2- to 2.4-kb fragments. This HincII digested chromosomal DNA was ligated into the low-copy vector pWKS30 (43) and transformed into the pcn strain of E. coli. The library was then rescreened by colony blotting. A positive clone was identified from this screen and designated pRSM2207. This clone was sequenced, and a homologue of the xthA gene was identified, as well as a portion of a glycosyltransferase gene. In order to complete the sequence of this region, PvuII-digested DNA from strain 33921 was screened by Southern analysis using a probe containing the newly identified partial 33921 glycosyltransferase. A band of approximately 8 kb hybridized to the probe (data not shown). PvuII-digested genomic DNA was separated on an agarose gel; 6- to 9-kb fragments were isolated and then cloned into EcoRV-digested pWKS30. These clones were transformed into the E. coli pcn strain. Colony blots were performed on the transformants using the partial 33921 glycosyltransferase probe. A positive clone was identified and designated pRSM2287. Additional sequencing was performed on this clone to complete the sequence of the glycosyltransferase and the adjacent genes. Two glycosyltransferases were identified between the rpmE and xthA genes. Since the location is the same genomic context as the lbgAB genes from strain 35000HP, they were designated lbgA33921 and lbgB33921, respectively. The sequence from this region of the strain 33921 genome has been assigned the GenBank accession number DQ646555.
Cloning of the lbgAB genes.
The lbgA33921 gene was amplified from chromosomal DNA by PCR using primers 1 (5′-AATTAAAACACGCTCAACAGTAGA-3′) and 2 (5′-ATCCCATAAATCAATAAGACTACC-3′). Similarly, the lbgB33921 gene was amplified with primers 3 (5′-TGGGGAAATACAACAGGT-3′) and 4 (5′-AGAAATCAGAGCTATGGAAAAACC-3′), and the lbgAB33921 genes were amplified by using primers 1 and 4. The amplicons were TA cloned into vector pCR2.1 (Invitrogen) and then sequenced to verify that there were no PCR-induced errors. An EcoRI fragment containing the lbgA gene was isolated and cloned into the shuttle vector pLS88 (9) to create plasmid pLBGA (Fig. 2). Similarly, the EcoRI fragment containing the lbgB gene was cloned into pLS88 (Fig. 2). Plasmids containing the insert in both orientations relative to the plasmid backbone were saved as pLBGB and pLBGBR. The lbgAB33921 gene cluster was cloned as a NotI-to-HindIII fragment into pLSSK, a pLS88 derivative containing the lacZα region of pBluescript SK(−) (45) that had been digested with the same enzymes to create pLBGAB (Fig. 2).
Expression of strain 33921 glycosyltransferases in strains 35000HP-RSM2 and 35000HP-RSM223.
The pLBGA, pLBGB, and pLBGBR plasmids were transformed into strain 35000HP-RSM2, a previously described derivative of strain 35000HP containing an insertionally inactivated lbgB, the dd-heptosyltransferase gene, as described by Young et al. (46). The resulting strains were designated 35000HP-RSM2(pLBGA), 35000HP-RSM2(pLBGB), and 35000HP-RSM2(pLBGBR), respectively.
A derivative of strain 35000HP containing mutations in both the lbgB35000HP gene and the lgtA gene, encoding the N-acetylglucosamine glycosyltransferase, was constructed. We insertionally inactivated the lgtA gene in pRSM2379 (37) with a chloramphenicol resistance cassette (17). The insertionally inactivated lgtA gene was then introduced into strain 35000HP-RSM2 as described by Sun et al. (37), using chloramphenicol as the selectable marker. The strain containing both mutations was designated 35000HP-RSM223. The correct genotype of this mutant was confirmed by Southern blot analysis (data not shown). Plasmids pLBGA and pLBGAB were transformed into this mutant: the transformants were designated 35000HP-RSM223(pLBGA) and 35000HP-RSM223(pLBGAB), respectively.
Preparation of LOS and SDS-PAGE analysis.
LOS was extracted from H. ducreyi that was grown on chocolate agar plates. Cells were suspended, washed with phosphate-buffered saline (PBS) containing 0.15 mM CaCl2 and 0.5 mM MgCl2, and extracted by using the hot phenol micromethod (18). Neuraminidase-treated LOS samples were reconstituted in 5 μl of 2× neuraminidase buffer (100 mM sodium acetate, 8 mM calcium chloride [pH 5.5]) and incubated overnight at 37°C with 5 mU of neuraminidase isolated from Vibrio cholerae (5 μl of a 1-U/ml solution; Roche, Indianapolis, IN). Samples of the extracted LOS were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% gel as previously described (23) and were visualized by silver staining (41).
Mass spectrometry analysis.
Water soluble O-deacylated LOS (O-LOS) samples were prepared by treating the LOS with 50 μl of anhydrous hydrazine, followed by acetone precipitation (29). Lyophilized O-LOS samples were desalted by reconstitution in double-distilled H2O (ddH2O) and drop dialysis using 0.025-μm-pore-size nitrocellulose membranes (Millipore, Bedford, MA). Samples were then lyophilized and reconstituted in 5 to 10 μl of ddH2O. Before the samples were loaded onto the target, they were mixed 1:1 (vol/vol) with matrix (50 mg of 2,5-dihydroxybenzoic acid [Laser Biolabs, Sophia-Antipolis Cedex, France]/ml in 70% acetonitrile). For the analysis of O-LOS, matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) using a ThermoElectron (San Jose, CA) vMALDI-LTQ linear-ion trap mass spectrometer was performed. The vMALDI source uses an SI N2 laser (337.3 nm) with a 20-Hz firing rate. The instrument was run in negative-ion mode with a laser intensity of 40, and spectra were recorded using the automated gain control to control the number of laser shots and the automatic spectrum filter tool.
β-Galactosidase treatment of O-LOS.
O-LOS from one to two plates of bacteria was treated with β(1-4)galactosidase (Prozyme, San Leandro, CA) isolated from Streptococcus pneumoniae. O-LOS was incubated with 10 or 20 mU of enzyme, 10 μl of the provided 5× buffer (250 mM sodium phosphate [pH 6)], and the volume was brought to a total of 50 μl with ddH2O. Incubations were done at 37°C for 20 to 24 h. Samples were boiled for 5 min to stop the reaction. The samples were concentrated by using YM-3 Microcon filters (Millipore) as previously described (22). Samples were reconstituted in 5 to 10 μl of ddH2O, and 1 μl of each sample was spotted onto the MALDI target plate. Once samples had dried they were overlaid with 0.5 to 1 μl of 2,5-dihydroxybenzoic acid matrix (see above). Samples were analyzed by MALDI-MS using an Applied Biosystems (Framingham, MA) Voyager DE time-of-flight mass spectrometer. Mass spectra were determined in linear negative-ion mode with a nitrogen laser (337 nm) under delayed extraction conditions: a 165-ns delay time, with a grid voltage of 94% of full acceleration voltage (20 kV). Mass spectra were acquired, averaged (typically 100 laser shots), and internally calibrated.
RESULTS AND DISCUSSION
Previously, our laboratory noted that the two-dimensional SDS-PAGE profiles of proteins from strains 35000HP and 33921 were significantly different (31). In order to independently access the relatedness of the two isolates, we sequenced the 16S gene from strain 33921 (GenBank accession no. AY513483) and determined that it is 99% identical to the gene from strain 35000HP (GenBank accession no. NC_002940). In a different study, White et al. observed that a murine monoclonal antibody directed against the DsrA protein from strain 35000HP failed to bind to strains CIP542 ATCC and HMC112 (44). Sequence analysis of the DsrA proteins from strains CIP542 ATCC and 35000HP determined that they are 48% identical. That study also showed that strains that did not bind the DsrA antibody also seemed to have a truncated LOS structure when visualized on a silver-stained SDS-PAGE (44). These studies, among others, led to the hypothesis that there were at least two classes of H. ducreyi strains.
The structure of the LOS from the atypical strain 33921 has been reported (25). This LOS lacks the d-glycero-d-manno-heptose found in the LOS of strain 35000HP. Since lbgB encodes for the dd-heptosyltransferase, we were interested in determining whether the lbgB gene was absent in strain 33921 or whether the lack of dd-heptose in the LOS was the result of phase-variable gene expression. The lbgAB genes in strain 35000HP are localized between the rpmE and xthA genes (Fig. 2). We therefore determined the sequence of this region of the chromosome of strain 33921. The gene arrangement in this region is the same as that observed in strain 35000HP (15, 35). That is, two glycosyltransferase genes are present between the rpmE and xthA genes in the chromosome of both strains. The two newly identified glycosyltransferase genes in strain 33921 were designated lbgA33921 and lbgB33921 (Fig. 2). The derived amino acid sequences of the rpmE genes from strains 35000HP and 33921 are 97% identical. Similarly, the derived amino acid sequences of the two xthA genes are also 97% identical. In contrast to the high level of homology observed between the flanking genes, the glycosyltransferases are not highly homologous. The derived amino acid sequence of the strain 33921 glycosyltransferase gene, designated lbgA33921, is 37% identical to the derived amino acid sequence of the lbgA35000HP gene. Similarly, the derived amino acid sequence of the strain 33921 glycosyltransferase gene, designated lbgB33921, is 23% identical to the derived amino acid sequence of the lbgB35000HP gene.
The predicted gene product of the lbgA33921 gene is most similar to the Lob1 protein of Histophilus somni (19) and the Lex2B protein of Haemophilus influenzae (20). The Lob1 gene contains a number of tandem repeat nucleotide sequences that are thought to mediate phase variation of the LOS glycoforms in H. somni (19). Since no tandem repeat nucleotide sequences were observed in the lbgA33921 gene, there is no indication that this gene is phase variable. The predicted gene product of the lbgB33921 gene is most similar to a glycosyltransferase group 8 member, Hi0258 (accession no. P43974), from H. influenzae strain KW20 (14).
Since the two housekeeping genes, xthA and rpmE, are 5′ and 3′ of two glycosyltransferase genes in the chromosomes of both strain 35000HP and strain 33921, the lack of dd-heptose in the LOS of 33921 appears to be due to the absence of the lbgB35000HP gene. No repeat regions or remnants of insertion sequences were identified in this region.
The identification of two new glycosyltransferases in strain 33921 prompted us to examine a series of isolates to determine whether the lbgAB33921 genes could be identified in additional isolates. Thirty-five isolates were subjected to dot blot hybridization using probes containing the lbgAB sequences from strain 35000HP or from 33921 (data not shown). Three of the 35 strains tested from a series of random isolates in the Totten collection reacted with the lbgAB33921 probe, while the remainder reacted with the lbgAB35000HP probe (data not shown). No isolate reacted with both probes. The strains that reacted with the lbgAB33921 probe were HMC37, HMC65, and HMC112. To determine the sequence similarity of the strains that reacted with the lbgAB33921 probe, we amplified the lbgAB region from HMC37, HMC65, and HMC112 using primers 5 (5′-AAAGAAATTACGGCAACA-3′) and 6 (5′-AGCTATGGAAAAACCCTCTG-3′). Since White et al. (44) reported that strain HMC112 was a member of a second class of H. ducreyi organisms, we also amplified this region from other proposed class II strains CIP542 ATCC, DMC64, DMC111, and SSMC71 using the same primers. The sequence of each of these amplicons was 100% identical to the sequence determined for the lbgAB33921 genes. These data suggest that strain 33921 and the two newly characterized isolates from the Totten collection are all class II organisms.
To determine the role that the lbgAB33921 genes play in LOS biosynthesis, we made several attempts to insertionally inactivate the lbgAB33921 genes in strain 33921 but were unable to genetically manipulate this strain. White et al. were also unable to genetically manipulate their class II isolates (44). To overcome the inability to genetically modify strain 33921, we complemented defined glycosyltransferase mutants constructed in the strain 35000HP background. The lbgA33921 gene and the lbgB33921 genes were amplified by PCR, cloned into pCR2.1, and sequenced. These genes were then cloned into the shuttle vector pLS88. The resulting plasmids were designated pLBGA, pLBGB, pLBGBR, and pLBGAB (see Table 1 and Fig. 2 for details).
35000HP-RSM2 was the first strain that was complemented with the lbgAB33921 genes. This strain contains a mutation in lbgB35000HP (46), the gene that encodes the dd-heptosyltransferase; therefore, the most complex LOS glycoform produced by strain 35000HP-RSM2 is Glcβ1-4(Hep)3-Kdo-lipid A (Fig. 1) (15, 42). Since the structure of the LOS of strain 33921 is GlcNAcβ1-3Galβ1-4Glcβ1-4(Hep)3Kdo-lipid A (Fig. 1), the terminal glucose of the LOS glycoform of strain 35000HP-RSM2 should serve as a substrate for the addition of galactose when this strain is complemented with the strain 33921 galactosyltransferase. The plasmids pLBGA, pLBGB, and pLBGBR were electroporated into 35000HP-RSM2. Transformants were selected on chocolate agar plates containing streptomycin and designated 35000HP-RSM2(pLBGA), 35000HP-RSM2(pLBGB), and 35000HP-RSM2(pLBGBR), respectively.
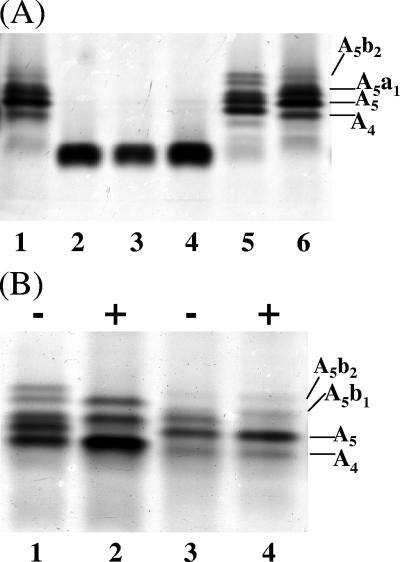
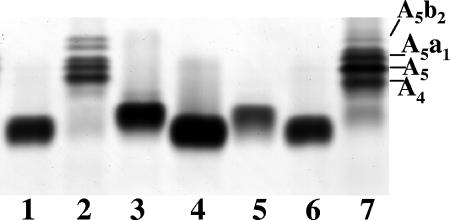
Purified LOS was extracted from the transformants and strains 35000HP and 35000HP-RSM2 and characterized by silver-stained SDS-PAGE (Fig. 3). These analyses demonstrated that LOS from strain 35000HP-RSM2(pLBGA) contained a complex series of glycoforms (Fig. 3A, lane 5). This series of glycoforms is similar to those produced by 35000HP LOS (Fig. 3A, lanes 1 and 6). The slight differences seen in the LOS from these two strains are most likely due to the absence of the dd-Hep in the LOS from 35000HP-RSM2(pLBGA). Since 35000HP LOS is known to be extended by the addition of sialic acid, the LOS glycoforms present in the 35000HP-RSM2(pLBGA) were examined for evidence of sialylation. 35000HP-RSM2(pLBGA) LOS and 35000HP LOS were neuraminidase treated and examined by silver-stained SDS-PAGE. Figure 3B shows these LOS samples both with (lanes 2 and 4) and without (lanes 1 and 3) neuraminidase treatment. In both strains neuraminidase treatment alters the glycoforms visualized on the SDS-PAGE. Neuraminidase treatment alters the presence of one band in the 35000HP LOS (removal of the terminal NeuAc from the a-branch structure [see Fig. 1]). Neuraminidase-treated 35000HP-RSM2(pLBGA) LOS has two bands missing instead of one as in the 35000HP LOS sample. These two bands in the 35000HP-RSM2(pLBGA) LOS likely correspond to the addition of sialic acid onto both the a- and b-branches of the 35000HP-RSM2(pLBGA) LOS. A sialylated b-branch was not detected in 35000HP LOS.
FIG. 3.
(A) Silver-stained SDS-PAGE of LOS isolated from 35000HP (lanes 1 and 6), 35000HP-RSM2 (lane 2), 35000HP-RSM2(pLBGB) (lane 3), 35000HP-RSM2(pLBGBR) (lane 4), and 35000HP-RSM2(pLBGA) (lane 5). This SDS-PAGE gel shows that only the addition of the lbgA gene changes the LOS glycoforms produced by strain 35000HP-RSM2. (B) Silver-stained SDS-PAGE of LOS isolated from 35000HP-RSM2(pLBGA) (lanes 1 and 2) and 35000HP (lanes 3 and 4). Lanes designated with a “+” are LOS samples that were treated with neuraminidase; lanes designated with a “−” are LOS samples that were not treated with neuraminidase. Neuraminidase treatment of the samples indicates that both the 35000HP LOS and 35000HP-RSM2(pLBGA) LOS have sialylated glycoforms. Various 35000HP LOS glycoforms are labeled in the figures (A5b2, A5b1, A5a1, A5, and A4); see Fig. 1 for details.
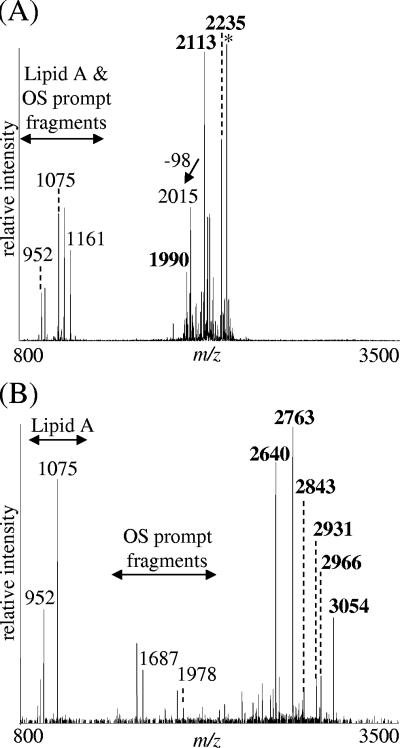
MS was used to further investigate the LOS structure of strain 35000HP-RSM2(pLBGA). Figure 4B shows a typical spectrum of 35000HP-RSM2(pLBGA) O-LOS. The major deprotonated molecular ions, [M-H]−, present at m/z 2,640 and 2,763 in this spectrum correspond to an LOS structure consisting of Hex3, HexNAc, Hep3, Kdo(P), and di-N-acyl lipid A (lipid A′) with one or two phosphoethanolamine (PEA) substitution(s), respectively. A predicted structure of the LOS for 35000HP-RSM2(pLBGA) is shown in Fig. 5. The major 35000HP-RSM2(pLBGA) glycoform can be further extended by the addition of sialic acid (at m/z 2,931 and 3,054, with one or two PEA substitutions, respectively) and is referred to as the a-branch (Fig. 5). Alternatively, the major glycoform can be further extended by the addition of a HexNAc (at m/z 2,843 and 2,966, with one or two PEA substitutions, respectively). This addition of the HexNAc is most likely the start of the b-branch LOS structure (Fig. 5).
FIG. 4.
vMALDI-LTQ-MS spectra of O-deacylated LOS isolated from 35000HP-RSM2 (A) and 35000HP-RSM2(pLBGA) (B). Samples were run in the negative ion mode. A loss of 98 indicates the neutral loss of phosphoric acid, H3PO4, presumably by β-elimination. All of the deprotonated molecular ions, [M-H]−, corresponding to the major O-LOS glycoforms are shown in boldface in each spectrum. Peaks labeled with an asterisk indicate glycoforms that contain salt adducts (potassium) resulting in an increase of 38 Da.
FIG. 5.
Predicted LOS structure for 35000HP-RSM2(pLBGA). Similar to the LOS from 35000HP, 35000HP-RSM2(pLBGA) seems to be able to generate both the a-branch and b-branch structures. “Hex*” indicates a hexose residue that was subsequently determined to be a galactose.
In contrast to the LOS from 35000HP-RSM2(pLBGA), both SDS-PAGE analyses (Fig. 3A, lanes 3 and 4) and MS analyses (data not shown) demonstrated that the LOS glycoforms produced by strains 35000HP-RSM2(pLBGB) and 35000HP-RSM2(pLBGBR) containing the lbgB33921 gene, cloned in both orientations relative to the vector backbone, were identical in size to the glycoform produced by strain 35000HP-RSM2.
The MS data for strain 35000HP-RSM2(pLBGA) were consistent with the hypothesis that the lbgA33921 gene encoded a galactosyltransferase with specificity for the nonreducing terminal glucose found in the LOS produced by 35000HP-RSM2. The more complex glycoforms seen in the 35000HP-RSM2(pLBGA) LOS were likely synthesized by 35000HP glycosyltransferases. This would mean that the 35000HP N-acetylglucosamine glycosyltransferase (LgtA) had a relaxed specificity, recognizing either Galβ1-4Glc [for 35000HP-RSM2(pLBGA)] or Galβ1-4ddHepα1-6Glc (its normal substrate in 35000HP). To determine more conclusively whether lbgA33921 encodes a galactosyltransferase, a strain with mutations in both lgtA and lbgB35000HP was generated. This strain was designated 35000HP-RSM223. Since the glycosyltransferase, LgtA, was probably responsible for the addition of the HexNAc to the Galβ1-4Glc in 35000HP-RSM2(pLBGA), we reasoned that if lbgA33921 was expressed in the lgtA lbgB35000HP double-mutant strain 35000HP-RSM223, the LOS glycoform would terminate in Galβ1-4Glc. To test this hypothesis, strain 35000HP-RSM223 was transformed with pLBGA and designated 35000HP-RSM223(pLBGA).
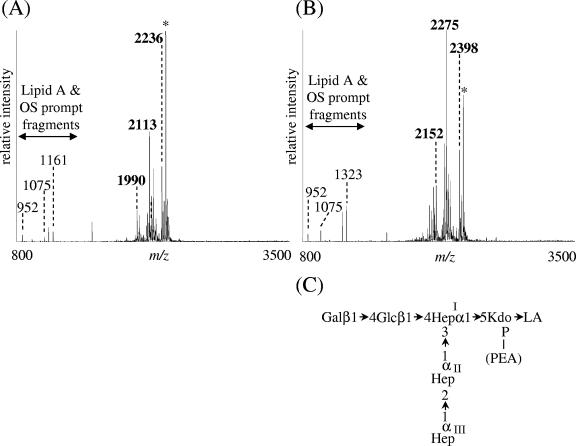
LOS was extracted from strain 35000HP-RSM223(pLBGA) and compared to LOS isolated from 35000HP, 35000HP-RSM2, 35000HP-RSM2(pLBGA), and 35000HP-RSM223. Silver-stained SDS-PAGE analyses (Fig. 6) demonstrated that the LOS glycoform produced by 35000HP-RSM223(pLBGA) (lane 3) is slightly larger than the glycoform produced by 35000HP-RSM2 (lanes 1 and 6) but less complex than the glycoforms produced by 35000HP-RSM2(pLBGA) (lane 2). To investigate the 35000HP-RSM223(pLBGA) LOS structure in more detail, MS analyses of O-LOS generated from 35000HP-RSM223 and 35000HP-RSM223(pLBGA) were acquired (see Fig. 7A and B, respectively). The major deprotonated molecular ions, [M-H]−, in the spectrum from 35000HP-RSM223 O-LOS were at m/z 1,990, 2,113, and 2,236, corresponding to a structure containing Hex, Hep3, Kdo(P), lipid A′ with zero, one, or two PEA substitution(s), respectively. The major molecular ions detected in the spectrum from 35000HP-RSM223(pLBGA) O-LOS were at m/z 2,152, 2,275, and 2,398, corresponding to a structure containing Hex2, Hep3, Kdo(P), lipid A′, with zero, one, or two PEA substitution(s), respectively. These data demonstrate that the difference between the major glycoforms expressed by 35000HP-RSM223 and the major glycoforms expressed by 35000HP-RSM223(pLBGA) is the addition of one hexose (+162 Da). We predicted that this hexose is a galactose, based on the known LOS structure from 33921 (25). This was confirmed by treatment of the O-LOS from 35000HP-RSM223(pLBGA) with β-galactosidase. MS analyses of the O-LOS demonstrated that after β-galactosidase treatment the mass of the O-LOS was decreased by 162 Da, a finding consistent with the loss of a single hexose moiety (data not shown). The predicted LOS structure for 35000HP-RSM223(pLBGA) is shown in Fig. 7 panel C and demonstrates that lbgA33921 encodes a galactosyltransferase.
FIG. 6.
Silver-stained SDS-PAGE of LOS isolated from 35000HP-RSM2 (lane 1), 35000HP-RSM2(pLBGA) (lane 2), 35000HP-RSM223(pLBGA) (lane 3), 35000HP-RSM223 (lane 4), 35000HP-RSM223(pLBGAB) (lane 5), 35000HP-RSM2 (lane 6), and 35000HP (lane 7). This SDS-PAGE gel indicates that the addition of the lbgA gene to the double mutant, 35000HP-RSM223, allows a larger glycoform to be made. Various 35000HP LOS glycoforms are labeled in the figure (A5b2, A5a1, A5, and A4); see Fig. 1 for details.
FIG. 7.
vMALDI-LTQ-MS spectra of O-deacylated LOS isolated from 35000HP-RSM223 (A) and 35000HP-RSM223(pLBGA) (B). (C) Predicted LOS structure of 35000HP-RSM223(pLBGA). All of the deprotonated molecular ions, [M-H]−, corresponding to the major O-LOS glycoforms are shown in boldface in each spectrum. Peaks labeled with an asterisk indicate glycoforms that contain salt adducts (potassium) resulting in an increase of 38 Da.
Strain 35000HP-RSM223 was also transformed with a plasmid containing both lbgA33921 and lbgB33921 and was designated 35000HP-RSM223(pLBGAB). Both silver-stained SDS-PAGE (Fig. 6, lane 5) and MS analyses (data not shown) demonstrated that the LOS from this strain was unchanged compared to the LOS from 35000HP-RSM2(pLBGA). Therefore, it is currently unclear what role, if any, lbgB33921 plays in the biosynthesis of 33921 LOS.
In the present study, we demonstrated that 33921 had a genome gene arrangement similar to that of 35000HP, with the rpmE and xthA genes flanking two glycosyltransferases, lbgAB. However, the glycosyltransferases present in these two strains are not highly homologous. Unlike strain 35000HP, this region of DNA from strain 33921 does not contain a dd-heptosyltransferase. Both lbgA35000HP and lbgA33921 are galactosyltransferases (42); however, they recognize different acceptor sugars. LbgA35000HP recognizes a terminal dd-heptose (42), whereas the LbgA33921 transferase recognizes a terminal glucose as its acceptor. Various H. ducreyi strains probed with the lbgAB genes from either 35000HP or 33921 demonstrated that strains reacted with one, but never both probes. These data demonstrate that the lbgAB genes can serve as good indicators of whether a strain is a class I or class II strain. Therefore, this information may be used as a tool for future characterization of H. ducreyi strains.
Acknowledgments
This study was supported by Public Health Service grants AI34967 and AI38444 (R.S.M.) and AI31254 (B.W.G.). National Institutes of Health grant HD34615 supported, in part, the Core DNA Sequencing Facility at Children's Research Institute. J.A.B. was the recipient of a National Research Service Award (F32-AI09813).
We thank Rachna Mungur, Shuhua Sun, Laurie Tarantino, and Jing Wang for technical assistance; Pat Totten for H. ducreyi strains and helpful comments; Chris Elkins for H. ducreyi strains; and Gerard Barcak for the E. coli DH5αpcnB strain.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Ahmed, H. J., A. Frisk, J. E. Mansson, E. K. Schweda, and T. Lagergard. 1997. Structurally defined epitopes of Haemophilus ducreyi lipooligosaccharides recognized by monoclonal antibodies. Infect. Immun. 65:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa, M. J., P. DeGagne, and P. A. Totten. 1996. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect. Immun. 64:2349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, B. A., S. R. Lumbley, and E. J. Hansen. 1999. Characterization of a WaaF (RfaF) homolog expressed by Haemophilus ducreyi. Infect. Immun. 67:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, B. A., M. K. Stevens, and E. J. Hansen. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect. Immun. 66:4290-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozue, J. A., M. V. Tullius, J. Wang, B. W. Gibson, and R. S. Munson, Jr. 1999. Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J. Biol. Chem. 274:4106-4114. [DOI] [PubMed] [Google Scholar]
- 6.Cope, L. D., S. Lumbley, J. L. Latimer, J. Klesney-Tait, M. K. Stevens, L. S. Johnson, M. Purven, R. S. Munson, Jr., T. Lagergard, J. D. Radolf, and E. J. Hansen. 1997. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 94:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes-Bratti, X., C. Karlsson, T. Lagergard, M. Thelestam, and T. Frisan. 2001. The Haemophilus ducreyi cytolethal distending toxin induces cell cycle arrest and apoptosis via the DNA damage checkpoint pathways. J. Biol. Chem. 276:5296-5302. [DOI] [PubMed] [Google Scholar]
- 8.Deng, K., and E. J. Hansen. 2003. A CdtA-CdtC complex can block killing of HeLa cells by Haemophilus ducreyi cytolethal distending toxin. Infect. Immun. 71:6633-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon, L. G., W. L. Albritton, and P. J. Willson. 1994. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid 32:228-232. [DOI] [PubMed] [Google Scholar]
- 10.Dutro, S. M., G. E. Wood, and P. A. Totten. 1999. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect. Immun. 67:3317-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 63:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filiatrault, M. J., B. W. Gibson, B. Schilling, S. Sun, R. S. Munson, Jr., and A. A. Campagnari. 2000. Construction and characterization of Haemophilus ducreyi lipooligosaccharide (LOS) mutants defective in expression of heptosyltransferase III and β1,4-glucosyltransferase: identification of LOS glycoforms containing lactosamine repeats. Infect. Immun. 68:3352-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, B. W., A. A. Campagnari, W. Melaugh, N. J. Phillips, M. A. Apicella, S. Grass, J. Wang, K. L. Palmer, and R. S. Munson, Jr. 1997. Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179:5062-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, B. W., W. Melaugh, N. J. Phillips, M. A. Apicella, A. A. Campagnari, and J. M. Griffiss. 1993. Investigation of the structural heterogeneity of lipooligosaccharides from pathogenic Haemophilus and Neisseria species and of R-type lipopolysaccharides from Salmonella typhimurium by electrospray mass spectrometry. J. Bacteriol. 175:2702-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, E. J., J. L. Latimer, S. E. Thomas, M. Helminen, W. L. Albritton, and J. D. Radolf. 1992. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J. Bacteriol. 174:5442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inzana, T. J. 1983. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J. Infect. Dis. 148:492-499. [DOI] [PubMed] [Google Scholar]
- 19.Inzana, T. J., J. Hensley, J. McQuiston, A. J. Lesse, A. A. Campagnari, S. M. Boyle, and M. A. Apicella. 1997. Phase variation and conservation of lipooligosaccharide epitopes in Haemophilus somnus. Infect. Immun. 65:4675-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarosik, G. P., and E. J. Hansen. 1994. Identification of a new locus involved in expression of Haemophilus influenzae type b lipooligosaccharide. Infect. Immun. 62:4861-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jessamine, P. G., and A. R. Ronald. 1990. Chancroid and the role of genital ulcer disease in the spread of human retroviruses. Med. Clin. N. Am. 74:1417-1431. [DOI] [PubMed] [Google Scholar]
- 22.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melaugh, W., A. A. Campagnari, and B. W. Gibson. 1996. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J. Bacteriol. 178:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melaugh, W., N. J. Phillips, A. A. Campagnari, R. Karalus, and B. W. Gibson. 1992. Partial characterization of the major lipooligosaccharide from a strain of Haemophilus ducreyi, the causative agent of chancroid, a genital ulcer disease. J. Biol. Chem. 267:13434-13439. [PubMed] [Google Scholar]
- 27.Melaugh, W., N. J. Phillips, A. A. Campagnari, M. V. Tullius, and B. W. Gibson. 1994. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry 33:13070-13078. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, K. L., W. E. Goldman, and R. S. Munson, Jr. 1996. An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol. Microbiol. 21:13-19. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, N. J., C. M. John, L. G. Reinders, B. W. Gibson, M. A. Apicella, and J. M. Griffiss. 1990. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 19:731-745. [DOI] [PubMed] [Google Scholar]
- 30.Pierson, V. L., and G. J. Barcak. 1999. Development of Escherichia coli host strains tolerating unstable DNA sequences on ColE1 vectors. Focus 21:18-19. [Google Scholar]
- 31.Scheffler, N. K., A. M. Falick, S. C. Hall, W. C. Ray, D. M. Post, R. S. Munson, Jr., and B. W. Gibson. 2003. Proteome of Haemophilus ducreyi by 2-D SDS-PAGE and mass spectrometry: strain variation, virulence, and carbohydrate expression. J. Proteome Res. 2:523-533. [DOI] [PubMed] [Google Scholar]
- 32.Schweda, E. K., J. A. Jonasson, and P. E. Jansson. 1995. Structural studies of lipooligosaccharides from Haemophilus ducreyi ITM 5535, ITM 3147, and a fresh clinical isolate, ACY1: evidence for intrastrain heterogeneity with the production of mutually exclusive sialylated or elongated glycoforms. J. Bacteriol. 177:5316-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spinola, S. M., A. Orazi, J. N. Arno, K. Fortney, P. Kotylo, C. Y. Chen, A. A. Campagnari, and A. F. Hood. 1996. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J. Infect. Dis. 173:394-402. [DOI] [PubMed] [Google Scholar]
- 34.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79:818-826. [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens, M. K., J. Klesney-Tait, S. Lumbley, K. A. Walters, A. M. Joffe, J. D. Radolf, and E. J. Hansen. 1997. Identification of tandem genes involved in lipooligosaccharide expression by Haemophilus ducreyi. Infect. Immun. 65:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, S., N. K. Scheffler, B. W. Gibson, J. Wang, and R. S. Munson, Jr. 2002. Identification and characterization of the N-acetylglucosamine glycosyltransferase gene of Haemophilus ducreyi. Infect. Immun. 70:5887-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, S., B. Schilling, L. Tarantino, M. V. Tullius, B. W. Gibson, and R. S. Munson, Jr. 2000. Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J. Bacteriol. 182:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telzak, E. E., M. A. Chiasson, P. J. Bevier, R. L. Stoneburner, K. G. Castro, and H. W. Jaffe. 1993. HIV-1 seroconversion in patients with or without genital ulcer disease: a prospective study. Ann. Intern. Med. 119:1181-1186. [DOI] [PubMed] [Google Scholar]
- 40.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 42.Tullius, M. V., N. J. Phillips, N. K. Scheffler, N. M. Samuels, R. S. Munson, Jr., E. J. Hansen, M. Stevens-Riley, A. A. Campagnari, and B. W. Gibson. 2002. The lbgAB gene cluster of Haemophilus ducreyi encodes a β-1,4-galactosyltransferase and an α-1,6-dd-heptosyltransferase involved in lipooligosaccharide biosynthesis. Infect. Immun. 70:2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 44.White, C. D., I. Leduc, B. Olsen, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi Outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 73:2387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 67:3740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, R. S., K. Fortney, J. C. Haley, A. F. Hood, A. A. Campagnari, J. Wang, J. A. Bozue, R. S. Munson, Jr., and S. M. Spinola. 1999. Expression of sialylated or paragloboside-like lipooligosaccharides are not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect. Immun. 67:6335-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]