Abstract
Transcriptional analysis of the tpr genes in Treponema pallidum subsp. pallidum (referred to here as simply T. pallidum) has been limited to date, and yet the expression of members of this gene family is likely relevant to the pathogenesis of syphilis. Recently, immunological studies and semiquantitative mRNA analysis led to the hypothesis of the modulation of tpr gene transcription during infection and suggested that various strains of T. pallidum might differentially express these genes. In this study we developed a real-time amplification assay to quantify the tpr mRNAs with respect to the 47-kDa lipoprotein message and to compare transcript levels among four different strains of T. pallidum. In addition, we analyzed the lymphocyte responsiveness pattern toward the Tpr antigens in late experimental syphilis to identify tpr genes that had been expressed during the course of infection. The T-cell response has been implicated in clearance of treponemes from early lesions, and some of the Tprs were identified as strong targets of the cellular immune response. We show that message for many of the tpr genes can be detected in treponemes harvested at the peak of early infection. Interestingly, tprK seems to be preferentially expressed in almost every strain, and it is uniformly the target of the strongest cellular immune response. These studies demonstrate the differential expression of certain tpr genes among strains of T. pallidum, and further studies are needed to explore the relationship between tpr gene expression and the clinical course of syphilis in infected individuals.
The Treponema pallidum repeat (tpr) genes constitute a promising branch of research on venereal syphilis and treponemal diseases in general (6, 11, 21). This paralogous gene family was identified in part by using a combination of experimental approaches (3), but all 12 members were extensively described by the genomic sequence of T. pallidum subsp. pallidum (referred to here as simply T. pallidum), Nichols strain (8). Subsequently, interest in the tpr genes has continued to rise in parallel to the corpus of data relative to these potential virulence factors (4, 5, 13, 17-19, 22). The predicted Tpr antigens can be divided into three subfamilies, based upon predicted amino acid sequence homology: subfamily I (TprC, -D, -F, and -I), subfamily II (TprE, -G, and -J), and subfamily III (TprA, -B, -H, -K, and -L). Subfamily I and II members show, within subfamilies, common amino and carboxyl termini, separated by unique central domains that differ in sequence and length. In the Nichols strain, TprC and TprD are identical, and TprF is characterized by a truncated central domain and absence of the conserved COOH-terminal region due to a ∼1-kb deletion and a frameshift mutation (3). Comparatively, less homology can be found among subfamily III Tprs (3). In the Nichols strain, tprA also contains a frameshift at nucleotide 712, which generates two open reading frames (ORFs), A1 and A2, for this gene (3). A reanalysis of the predicted cellular location of the Tpr proteins using the 2005 version of PSORT (http://www.psort.org/psortb) localizes TprA2, -C, -D, -F, -I, -J, and -K to the outer membrane of the microorganism, whereas TprA1, -B, -E, -G, -H, and -L localization cannot be hypothesized (equal probability is associated to any cellular compartment). In T. pallidum strains other than Nichols, some variants of the tpr genes have been described; for example, the Sea 81-4 strain is known to carry the tprD2 allele, as well as a hybrid tprG/J allele within the tprD and tprJ loci, respectively (6, 9). Furthermore, a frameshift and a premature termination characterize the ORF in the tprG locus of this strain.
Although the function of the Tprs is still unknown and attempts to determine the cellular location of TprI and TprK have resulted in controversial results (10, 12), several studies have highlighted the importance of these antigens during the immune response to syphilis in the rabbit model (17, 19, 22). Immunization with recombinant peptides based on TprI, TprF, and TprK sequences significantly alters lesion development after intradermal challenge (3, 10, 22); moreover, TprK possesses multiple alleles in T. pallidum isolates, conferring an impressive potential for antigenic variation and, consequently, of immune evasion (4, 5).
At present, little is known about transcriptional patterns of the tpr genes. Preliminary studies based on semiquantitative reverse transcription-PCR of the Nichols strain (3) showed a preponderance of tprK mRNA over the other tpr genes during early experimental infection (day 10), suggesting the modulation of tpr expression. This result was partially confirmed by densitometric analysis after limiting-dilution reverse transcription-PCR (12), which also showed that not all of the tpr genes are equally transcribed, although no preponderance of tprK message was found. Transcription modulation is also supported by a recent study based on microarray and real-time PCR (20), which showed the tpr genes to be differentially expressed with respect to TP0426 (V-type ATPase, A1 subunit) in the Nichols strain. No data regarding tprK expression were available in that study, however (20).
Leader et al. (14) examined the development of antibody responses toward the Tprs and demonstrated differences among strains in the time of appearance and patterns of antibody reactivity during experimental infection. A less robust and later reactivity was reported toward most of the Tprs in Nichols-infected rabbits compared to strains less adapted to rabbits (Bal 73-1 and Chicago) (14). These findings are consistent with the possibility of strain-to-strain differences in tpr expression.
In the rabbit model of syphilis infection, activated T cells and macrophages have been shown to infiltrate primary and secondary syphilitic lesions and to clear opsonized treponemes, resulting in lesion resolution (7, 15, 25). Although major T-cell antigens include T. pallidum lipoproteins and endoflagellar polypeptides, TprK, TprI, and the amino-terminal conserved subfamily I peptide were also shown to be strong targets for the cellular component of the immune response (17, 19, 22), emphasizing again the importance of these antigens in the immune response to syphilis infection.
Using a relative quantification approach based on real-time PCR, we analyzed the expression of every known tpr gene with respect to the 47-kDa lipoprotein (TP0574) mRNA in the Nichols, Sea 81-4, Chicago, and Bal 73-1 strains of T. pallidum, and we compared their expression patterns to T-cell responsiveness detectable after long-term infection of rabbits with these strains. We show that although many of the tpr genes are expressed during the early phase of infection, their mRNA levels are considerably lower compared to the 47-kDa antigen message, that transcription of the tprs is modulated among strains, and that tprK seems to be preferentially expressed in almost every strain. T-cell responsiveness patterns show which Tprs are targeted by the cellular immune response, allowing the identification of antigens for future investigation. A correlation between T-cell responsiveness and tpr transcription can be seen, although further studies are needed to examine the temporal correlation of new T-cell reactivity with gene expression.
MATERIALS AND METHODS
T. pallidum strain propagation and infection, nucleic acid extraction, and reverse transcription.
T. pallidum subsp. pallidum Nichols, Sea 81-4, Chicago, and Bal 73-1 strains were propagated in seronegative New Zealand White rabbits as previously reported (1). The Nichols strain was provided by James N. Miller (University of California, Los Angeles) in 1979. The Sea 81-4 strain was isolated in Seattle, WA, from a patient's primary lesion in 1981. The Chicago and Bal 73-1 strains were supplied by Paul Hardy and Ellen Nell, Johns Hopkins University, Baltimore, MD.
For lymphocyte proliferation assays, a total of 26 rabbits were infected with different T. pallidum strains. Seven rabbits received 5 × 107 T. pallidum (Nichols strain) per testis. Additional rabbits were infected with the same number of treponemes for Chicago (seven rabbits) and Bal 73-1 (six rabbits) isolates, whereas Sea 81-4-infected rabbits (n = 6) received 4 × 106 organisms per testis. The animals were euthanized, and splenic lymphocytes were harvested between days 180 and 240 postinfection to perform the T-cell proliferation assay as described below.
For message quantification, individual rabbits were infected with 5 × 107 T. pallidum per testis for the Nichols, Chicago, and Bal 73-1 strains and 4 × 106 organisms per testis for Sea 81-4. Treponemes were harvested from infected rabbit testes at peak orchitis (day 10 postinfection for Nichols and Chicago strains; day 20 for Bal 73-1; and day 25 for Sea 81-4) to recover the highest number of organisms before the onset of immune clearance. Some strains grow more rapidly than others. We reasoned that gene expression is more likely to be related to the clinical stage of infection rather than absolute time in days, so we chose to harvest organisms from all animals at peak orchitis. Testes were minced in 10 ml of saline for approximately 10 min, and suspensions were centrifuged for 10 min at 1,000 × g to remove host cellular debris; the resulting supernatant was split and divided into aliquots to be processed for nucleic acid extraction. For RNA isolation, 1-ml aliquots of treponemal suspensions were spun for 30 min at 12,000 rpm at 4°C, and the pellets were immediately resuspended in 400 μl of Ultraspec buffer (Biotecx Laboratories, Inc., Houston, TX). For DNA extraction, 500-μl aliquots were mixed with an equal volume of 2× lysis buffer (10 mM Tris [pH 8.0], 0.1 M EDTA, 0.5% sodium dodecyl sulfate). DNA for the analysis of the tprA ORF in the Bal 73-1, Chicago, and Sea 81-4 strains (described in more detail below) was isolated as previously described (2) using the QIAamp DNA minikit (QIAGEN, Inc., Chatsworth, CA). RNA extraction was performed according to Ultraspec manufacturer's instructions from ∼109 organisms for the Nichols strain, ∼108 organisms for the Chicago strain, and ∼107 treponemes for both Sea 81-4 and Bal 73-1. One sample for each strain harvest was extracted. Copurified DNA was digested by DNase I (Invitrogen, Carlsbad, CA) treatment. DNase I-treated RNA was checked for residual DNA contamination by qualitative amplification using T. pallidum-specific primers (TP0574 sense and antisense; Table 1) as already described (9), and negative samples were stored at −80°C until use. Reverse transcription of total RNA was performed by using the Superscript II first strand synthesis kit (Invitrogen) with random hexamers according to the provided protocol. All cDNA samples were diluted 1:5 with diethyl pyrocarbonate-treated water (Biotecx Laboratories, Inc.) to minimize the inhibitory effect on amplification due to the reagents used to synthesize cDNA. Samples were then stored in 12-μl aliquots (suitable for a single amplification reaction in quadruplicate) at −80°C until use.
TABLE 1.
Primers used for plasmid standard construction and message quantificationa
| Gene | Primer sequence (5′-3′)
|
Amplicon size (bp) | |
|---|---|---|---|
| Sense | Antisense | ||
| TP0574 (47-kDa lipoprotein) | CGTGTGGTATCAACTATGG | TCAACCGTGTACTCAGTGC | 313 |
| Subfamily I | |||
| tprC/D | CAAGAGAGAGCTATCCTCAAAG | GTTTAGCAGTGACAACTCTTGG | 289 |
| tprD2 | CACTAGTCTTGGGGACACGC | TACGTGAATTGCAACCAGGA | 278 |
| tprF | GACCCTGCCGATGCAGGTAAT | TCAGCAAGCACCCCCTGTTC | 266 |
| tprI | GACCCTGCCGATGCAGGTAAT | TAAGCACGATGTCCGACTGACT | 336 |
| Subfamily II | |||
| tprE | CGGCAAAGTCCTGTTCGGCAA | GCTCAACACGCTGTCGTATAGTA | 358 |
| tprG | GAAGGTGTTCATTACCGACCCT | TTGTAGCCTCAGCCGTAAGCTT | 359 |
| tprJ | TCTTCACACCCCGCAGGGAA | CGTTATTTCCGTTCGCATCATC | 364 |
| tprG/J | CCTACAATGTCGCCTTCGAT | CAATGTGCCCTGAGGAACC | 277 |
| Subfamily III | |||
| tprA | |||
| Cloning | TACCTACCGGGATACGAACAGT | TGCAAGGCATGGGTGTAATCAT | 315 |
| Quantification | ATACGAACAGTGCGAGAGCA | TCATCTCCCGAACGAGTTTC | 286 |
| tprB | AGTCACCACCAGGTGTGTGG | GACACAAGCTTAGAAAGAGAATCGT | 370 |
| tprH | GCAGAAGCTCGATAGTGTCAAG | GTGTGCTCCCATACGTAGGAAA | 280 |
| tprK | AGTTTGCGTCTAACACCGACTG | TCGCATGGCCATGTTGAGAAAT | 410 |
| tprL | GGTGGTTTCCCATTTGGAAGG | CAAGTAGTCTGTAAGCTGCCTG | 295 |
The primers used to construct the plasmid standards contain suitable restriction tags at their 5′ ends to allow cloning into the pCRII-TOPO vector (Invitrogen) unless direct cloning was performed. The following tags were used: EcoRI (5′-CCGGAATTC) for tprD2, tprA, and tprG; XhoI (5′-CCGCTCGAG) for TP0574; XbaI (5′-CTAGTCTAGA) for tprG/J, tprI, and tprE; NotI (5′-ATAAGAATGCGGCCGC) for tprC/D and tprL; KpnI (5′-CGGGGTACC) for tprK; and SpeI (5′-CTAGACTAGT) for tprB. Palindromes are underlined. tprF, tprJ, and tprH were inserted by direct cloning.
Real-time quantification of tpr messages.
A relative quantification protocol using external standards was chosen to analyze the tpr message levels at the time of bacterial harvest. This approach normalizes the amount of message from one or more target genes to the mRNA of a chosen reference gene (in this case, TP0574, the 47-kDa lipoprotein) present in the same sample and allows comparison of the levels of different messages to each other. TP0574 (the 47-kDa lipoprotein gene) was selected among a panel of T. pallidum highly transcribed genes (20), including flaA (TP0249), the tpp17 gene, (TP0435), groEL (TP0030), and the A subunit of V-type ATPase (TP0426) after analysis of their transcription patterns among the strains studied here. Our results (not shown) suggested that the expression of both the tpN47 gene and the V-type ATPase is characterized by the least degree of variability among these strains at the peak of infection, allowing the use of either one as reference genes to normalize tpr gene expression. On the contrary, the groEL, flaA, and tpp17 genes appeared to vary in greater extent among these strains. To obtain the standards, specific sequences of each tpr gene were amplified from Nichols DNA and cloned into three separate plasmids (pCRII-TOPO vector; Invitrogen), one for each tpr subfamily. Although only a limited set of tpr genes have been sequenced in T. pallidum strains other than Nichols (9, 22), data obtained in our laboratory (not shown) are consistent with the absence of heterogeneity in the tpr primer binding sites among the strains examined in the present study. Furthermore, identical melting temperatures for the amplicons obtained during real-time amplification did not suggest any differences in the sequences of the amplified tpr gene fragments among the Nichols, Sea 81-4, Bal 73-1, and Chicago strains. The sequences of tpr genes not present in the Nichols genome (tprD2 and tprG/J hybrid) were amplified from a suitable strain and cloned into a fourth plasmid. Both direct cloning (according to the provided protocol) and a general cloning procedure (16) with restriction endonucleases were used to make the plasmid constructs (primers shown in Table 1). The partial sequence of the 47-kDa antigen was also inserted into each plasmid for normalization. To assess the sequence identity and lack of amplification errors, final constructs were sequenced in both directions with the Applied Biosystems dye terminator sequencing kit (Perkin-Elmer, Foster City, CA) using the primer-walking approach. Plasmids were then purified using the QIAGEN plasmid minikit and linearized using an appropriate restriction site (EcoRV) within the polylinker region to locate the cloned fragments at the extremities of the open construct. Digestion was performed according to the enzyme manufacturer (Promega, Madison, WI) for 12 h. Digested plasmids were subsequently purified with the PCR purification kit (QIAGEN), and their concentrations were measured spectrophotometrically using a ND-1000 instrument (NanoDrop Technologies, Wilmington, DE). The plasmid copy number was calculated taking into account the size of the original plasmid plus the cloned inserts and the concentration of the construct. To generate standard curves, the plasmids were serially diluted (tenfold) over the appropriate concentration range (106 to 100 copies/μl) in cDNA synthesis mixture to reproduce the same chemical environment present in the actual samples. To achieve a reliable standard curve for each target message, the plasmids were amplified in four replicates for each standard dilution point over the complete range, except for the 101 and 100 dilution points, which were amplified in five replicates. The threshold value for the maximum acceptable error associated with a standard curve was set to 0.0500. Amplification reactions and data collection were carried out by using the LightCycler (Roche, Basel, Switzerland) system. All reactions were performed according to the manufacturer's instructions with the Roche FastStart DNA master plus SYBR green kit (Roche), which does not require MgCl2 optimization. The same tpr-specific primers (without restriction tags) used for construction of the plasmid standard were also used for message (cDNA) quantification (Table 1), except for the tprA gene, which required primers internal to the cloned sequence to improve amplification efficiency (Table 1). For each reaction, optimal primer concentration and reaction conditions are reported in Table 2. Quadruplicate amplifications were performed for each gene using the cDNA preparation from each strain; a known concentration of the relevant plasmid DNA was amplified concurrently in each run as an internal control. These amplifications used 3 μl of the relevant cDNA preparation or 3 μl of plasmid standard as templates. The results were analyzed by using the LightCycler 3.5 software (Roche) according to the manufacturer's instructions. Differences between levels of tpr expression within strains were compared by using the Student t test, with significance set at a P value of <0.05.
TABLE 2.
Real-time amplification conditionsa
| Gene | Primer concn (μM) | Annealing conditions (temp [°C], time [s]) | Extension time (s)b | Acquisition temp (°C) |
|---|---|---|---|---|
| TP0574 (47-kDa lipoprotein) | 1.0 | 60, 8 | 13 | 88 |
| tprD | 2.00 | 60, 8 | 11 | 88 |
| tprD2 | 0.50 | 64, 8 | 12 | 87 |
| tprF | 0.50 | 64, 8 | 11 | 88 |
| tprI | 0.50 | 62, 8 | 13 | 87 |
| tprE | 1.00 | 65, 5 | 15 | 89 |
| tprG | 0.50 | 64, 8 | 14 | 88 |
| tprJ | 1.00 | 64, 8 | 12 | 88 |
| tprG/J | 1.00 | 64, 8 | 11 | 89 |
| tprA | 0.50 | 63, 8 | 12 | 89 |
| tprB | 0.75 | 62, 5 | 15 | 87 |
| tprH | 1.00 | 62, 8 | 11 | 90 |
| tprK | 0.75 | 60, 5 | 16 | 89 |
| tprL | 3.00 | 62, 6 | 12 | 90 |
Every amplification reaction was carried out for 45 cycles.
The extension temperature was 72°C.
Cloning, expression, and purification of recombinant proteins.
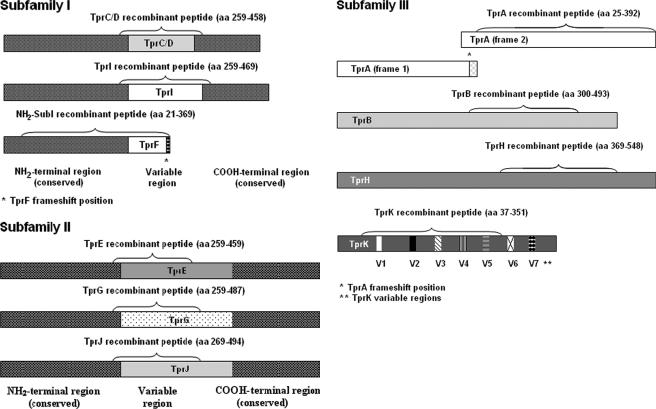
Recombinant peptides representing unique portions of individual Tprs were expressed as previously described (3) with two exceptions: TprL could not be expressed, and the amino-terminal-subfamily I (NH2-SubI) recombinant peptide comprises the conserved amino-terminal region of all subfamily I molecules, as well as a piece of the TprF central region. A schematic representation of the Tpr regions expressed as recombinant peptides is shown in Fig. 1. Note that there is no TprF-specific peptide, since the TprF central region is shared with TprI; the TprI peptide, however, contains regions unique to TprI. In addition, the amino-terminal portion of TprA (before the frameshift) was not able to be expressed successfully, so the lymphocyte studies were performed with a recombinant peptide representing the portion of TprA downstream of the frameshift. Briefly, PCR products obtained with tpr-specific primers (Table 3) were cloned into the TOPO-TA cloning vector (Invitrogen), subsequently transferred into the pRSET/C vector (Invitrogen), and expressed in BL21(DE3)/pLysS cells (Invitrogen). Cultures were induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the optical density at 600 nm reached 0.4 to 0.6 absorbance units (Sigma, St. Louis, MO) and harvested after 4 h of incubation at 37°C. Purification was performed by nickel affinity chromatography under denaturing conditions using the ProBond purification system (Invitrogen). Inclusion bodies containing insoluble recombinant proteins were isolated by successive rounds of sonication and centrifugation and then resuspended in 1× binding buffer (0.5 M NaCl, 20 mM Tris-HCl, 5 mM imidazole [pH 7.9]) containing 6 M urea. After 1 h of incubation in ice, suspensions were centrifuged, and the supernatants were passed through a 0.45-μm-pore-size filter. Purification was followed by dialysis against phosphate-buffered saline (PBS) containing decreasing concentrations of urea and eventually against PBS alone. The products were tested for size and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and quantified by using a bicinchoninic acid assay kit (Pierce, Rockford, IL).
FIG. 1.
Schematic representation of the Tpr antigens. The fragments expressed as recombinant peptides for the T-cell proliferation assay are indicated. Identical shading within the same Tpr subfamily indicates regions of homology among the antigens.
TABLE 3.
Primers used to clone and express recombinant Tprs
| Gene | Primer sequence (5′-3′)
|
Expressed aaa | Length (aa) | |
|---|---|---|---|---|
| Sense | Antisense | |||
| TprA | ATGGGTTTGGTAGTGACCTCTCT | TCACCAAGTGATCTTGCACCC | 25-392 | 368 |
| TprB | CGCGTTTGACGCTTTCCCCG | ACACAAGCTTAGAAAGAGAATCG | 300-493 | 194 |
| Tp/D | CGACTCACCCTCGAACCA | GGTGAGCAGGTGGGTGTAG | 259-458 | 200 |
| TprE | CGACTCACCCTCGAACCA | GGTGAGCAGGTGGGTGTAG | 269-459 | 191 |
| NH2-SubIb | TATGCAGGCGTACTCACTCCG | TCAGCAAGCACCCCCTGTTCC | 21-369 | 349 |
| TprG | CGACTCACCCTCGAACCA | GGTGAGCAGGTGGGTGTAG | 269-487 | 219 |
| TprH | CGCGCATAACGCTCACTCC | GTCTATAAGGTGTGTATACGCG | 369-548 | 180 |
| TprI | CGACTCACCCTCGAACCA | GGTGAGCAGGTGGGTGTAG | 259-469 | 211 |
| TprJ | CGACTCACCCTCGAACCA | CGTTATTTCCGTTCGCATCATC | 269-494 | 226 |
| TprK | ATATTGAAGGCTATGCGGAGCTG | CCTCAAGGAAAAGAAGTATCAGG | 37-351 | 315 |
The amino acid (aa) designations are based upon the start site identified in the T. pallidum Nichols strain genome (8), with two exceptions. tprA consists of two overlapping reading frames. Our numbers refer to the amino acid designation of the second ORF. The translational start site for tprK has been determined to be different from that listed in the T. pallidum genome (12). The expressed amino acids correspond to the experimentally determined start site.
The NH2-subfamily I recombinant (NH2-SubI) peptide comprises the conserved amino-terminal region of subfamily I molecules, as well as the TprF central region. aa, amino acids.
Lymphocyte proliferation assay.
Spleens were harvested from infected rabbits between day 180 and day 240 postinfection; spleens from uninfected rabbits (n = 9) were used as negative controls. All splenic lymphocytes were cultured as previously described (1, 15). Briefly, cells were cultured in RPMI supplemented with glutamine (2 mM final concentration), penicillin (100 U/ml [final concentration]), streptomycin (100 μg/ml [final concentration]), and heat-inactivated normal rabbit serum (1% final concentration). Approximately 5 × 105 cells in 200 μl of culture medium were plated in quadruplicate for each animal and each antigen in flat-bottom 96-well plates (Costar, Cambridge, MA), followed by incubation at 37°C in a 5% CO2 atmosphere. Recombinant proteins were tested at final concentrations of 2 and 10 μg/ml (cells from rabbits at day 180 were tested only with 2 μg of peptide/ml); 10 μl of a sonicated suspension of T. pallidum/well (equivalent to 107 T. pallidum per well) was used as a treponeme-specific positive control, and 0.5 μg (as determined by titration) of the mitogen concanavalin A (ICN, Irvine, CA)/well was used as a positive T-cell control. PBS was used to measure background reactivity. Three days after exposure to recombinant peptide or control antigens, cells were pulsed with 0.5 μCi of [3H]thymidine per well and harvested after 24 h to measure the radioactive thymidine incorporation. The geometric mean of quadruplicate wells with no antigens (determined as the background value) was subtracted from the geometric means of the quadruplicate experimental wells. The reported data represent the geometric means (± the standard error [SE]) of values for each antigen for each group of rabbits. Proliferation values greater than 5,000 cpm were considered to be significantly reactive. Lymphocytes from all rabbits responded appropriately to concanavalin A stimulation with at least 36,000 cpm; spleen cells from infected rabbits responded to T. pallidum sonicate with at least 14,000 cpm (data not shown).
PCR amplification, cloning, sequencing, and sequence analysis.
The T. pallidum genome sequence (8) was used to design primers in the 5′ and 3′ flanking regions of the tprA gene to amplify the corresponding DNA regions from the Chicago, Sea 81-4, and Bal 73-1 strains (sense prime, 5′-CGTATGCTTTTACCCGCTGT; antisense primer, 5′- TGCAACCATCTTCGATTACG), and amplification was performed as previously described (11). The products were cloned into the pCRII-TOPO cloning vector (Invitrogen) according to the manufacturer's instructions. Plasmid DNA was extracted by using the QIAGEN plasmid minikit (QIAGEN), and at least two clones for each strain were sequenced with the Applied Biosystems dye terminator sequencing kit (Perkin-Elmer, Foster City, CA) using an internal primer (5′-TAAGGAACCTAGAGTGTGCG). A single tprA ORF clone from the Sea 81-4 strain was completely sequenced by using the primer-walking approach as already reported (11). Sequences were aligned by using the Multiple Alignment Program (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html), and shading of identical bases was done with the Boxshade 3.21 program (http://www.ch.embnet.org/software/BOX_form.html).
RESULTS
Real-time quantification of tpr messages.
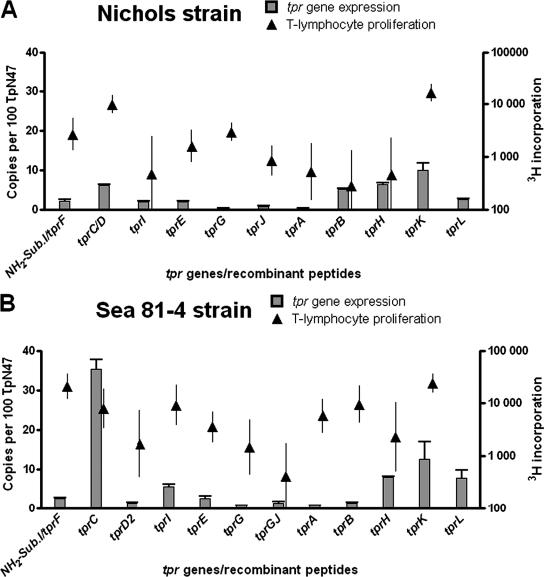
Our relative quantification approach expresses mRNA levels as the ratio between the target message (tpr) and a reference message (TP0574, the 47-kDa lipoprotein). The 47-kDa gene is expressed at a higher level compared to the tpr genes in every strain that we analyzed, and it was shown to be one of the most highly expressed genes in T. pallidum Nichols strain (20). In our study, the number of copies of every tpr message is expressed per 100 copies of 47-kDa antigen message (Fig. 2, scale on left y axes). The tpr expression pattern found in the Nichols strain at peak orchitis (day 10 postinfection; shown as bars in Fig. 2A) reveals that among subfamily I members, tprC/D messages are predominant and are higher than tprI and tprF (P = 0.013), although it should be remembered that the tprC and tprD loci are identical in the Nichols strain. Our analysis cannot determine whether one or both loci were being transcribed. tprE appears to be slightly but still significantly more highly transcribed than the other subfamily II genes. In Nichols, tprK is the most highly transcribed of all tpr genes, albeit at 10% of the level of TpN47. Compared to other subfamily III members, tprK transcription is significantly higher than tprA and tprL (P < 0.05) but not higher than tprB and tprH (P > 0.05) (Fig. 2A).
FIG.2.
tpr gene expression (▪) in T. pallidum subsp. pallidum Nichols (A), Sea 81-4 (B), Bal 73-1 (C), and Chicago (D) strains compared to T-cell proliferation (▴) in rabbits infected with these strains. For tpr expression, the reported values represent the mean ± the SE of quadruplicate assays for each cDNA sample; the number of copies of tpr transcript are expressed per 100 copies of the 47-kDa lipoprotein message. For T-lymphocyte proliferation, the 3H incorporation after stimulation with the specific Tpr recombinant peptide is reported, representing the geometric means ± the SE of quadruplicate values for each antigen for each rabbit; at least six rabbits were examined for each strain. Proliferation values above 5,000 cpm were considered to be significantly reactive. The NH2-subfamily I recombinant peptide (NH2-SubI) encompasses the conserved amino-terminal region of subfamily I molecules, as well as the TprF central region. The gene expression value was obtained using tprF-specific primers (shown in Table 1).
In the Sea 81-4 strain, the tprD locus carries an alternative allele (tprD2), instead of a copy of the tprC gene, so it is possible to differentiate transcription from the tprC and tprD loci. At peak orchitis in Sea 81-4 infections, the expression of the tprC allele is significantly higher than any other tpr in this strain (bars in Fig. 2B), and this is the only strain examined that demonstrated such a high level of tprC transcription. In contrast, the tprD2 allele expression level is the lowest of all subfamily I members (Fig. 2B). No significant differences in transcription levels are seen among subfamily II members, which were all very low. As in Nichols, tprK expression in Sea 81-4 is higher than the other subfamily III tpr alleles, although the tprH and tprL levels are not significantly different from that of tprK (P > 0.05), whereas tprA and tprB are less expressed (Fig. 2B).
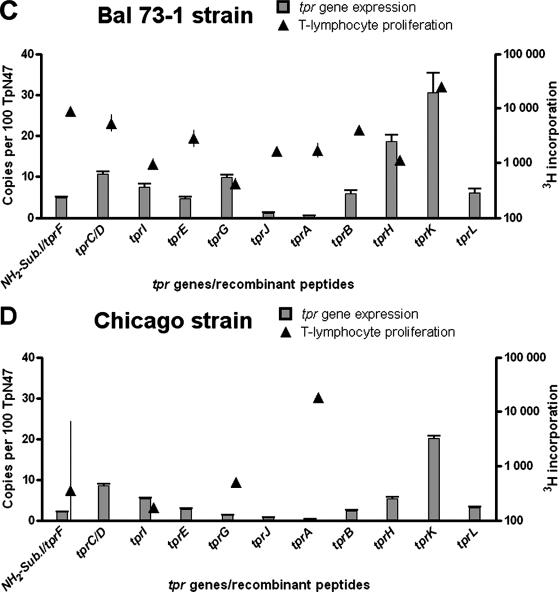
The Bal 73-1 strain expression pattern was also analyzed at peak orchitis (day 20 postinfection; Fig. 2C). We observed in this strain the highest expression level for tprK, a level statistically significant compared to each of the remaining tpr genes except for tprH (P = 0.09). This relatively high level of tprH transcription was seen only in Bal73-1; significant levels of tprG expression were also seen only in this strain (Fig. 2C). In subfamily I, again tprC/D are expressed more highly than other subfamily I genes.
The expression pattern for the Chicago strain (Fig. 2D) at peak orchitis (day 10 postinfection) again shows that tprK is expressed at a significantly higher level with respect to the other tpr genes. Among subfamily I members, tprC/D and tprI appear to be preferentially expressed with respect to tprF. No significant differences were seen among subfamily II members, which were poorly transcribed.
Development of responsiveness to Tpr antigens during the course of infection.
Lymphocyte proliferative responses to all recombinant Tpr peptides are shown as solid triangles in Fig. 2 (scale on right y axes), superimposed on the tpr expression data. Significant levels of responsiveness (>5,000 cpm) to the unique portion of TprC/D was seen for rabbits infected with all of the strains except for Chicago (Fig. 2D). Reactivity to the subfamily I peptide (representing the common amino-terminal region of all subfamily I molecules, along with the TprF central fragment) was also seen in rabbits infected with the Sea 81-4 and Bal 73-1 strains and, more weakly (2,620 cpm), in Nichols-infected rabbits but not in rabbits infected with the Chicago strain. Responsiveness to the unique central domain of TprI was seen only in lymphocytes from rabbits infected with Sea 81-4.
In contrast to subfamily I peptides, all of the subfamily II recombinant antigens (TprE, -G, and -J) failed to elicit robust proliferation of splenocytes from infected rabbits, a finding consistent with the low levels of transcription of those genes. Modest lymphocyte responses (∼3,500 cpm) to TprE were seen only in rabbits infected with Sea 81-4 and Bal 73-1 (Fig. 2).
Only TprK, among subfamily III peptides, elicited a consistent and robust T-cell response, independent of the infecting strain. In addition, a significant level of lymphocyte reactivity was elicited by TprA and TprB peptides in Sea 81-4-infected rabbits.
tprA sequence analysis in T. pallidum strains.
In the Nichols strain, the tprA ORF contains an authentic frameshift within a region characterized by three dinucleotide repeats (CT) at positions 706 to 711 (8); thus, it was expected that there would be no T-cell response to the TprA recombinant peptide representing the portion of the molecule downstream from the frameshift. We were surprised to see a significant T-cell response to this TprA peptide in animals infected with the Sea 81-4 strain. Subsequent sequence analysis of the Sea 81-4 strain showed that the tprA ORF in Sea 81-4 contains four CT repeats (Fig. 3), which reverse the frameshift and predict the expression of a full-length TprA protein. As in the Nichols strain, only three repeats are present in the Bal73-1 and Chicago strains (Fig. 3), resulting in premature termination. Sequence analysis of the whole tprA gene in Sea 81-4 showed the absence of other changes potentially responsible for a similar frameshift mutation and/or premature stop elsewhere in the ORF.
FIG. 3.
Alignment of the tprA region where the frameshift is located (nucleotides 703 to 760, according to the Nichols tprA ORF sequence). Four CT dinucleotide repeats are found in the Sea 81-4 strain in comparison to the other strains. This different sequence architecture in the Sea 81-4 strain reverts the frameshift and generates a single ORF within the tprA locus. *, Frameshift position in Nichols, Chicago and Bal 73-1 tprA sequence (nucleotide 712).
DISCUSSION
The investigation of the tpr gene family in T. pallidum has yielded some intriguing new information for our evolving understanding of syphilis pathogenesis; however, many of these studies have focused on the Nichols strain. Examination of other strains has resulted in the identification of new alleles at the tprC, tprD, tprG, and tprJ loci (6, 10) and has led to the recognition of sequence and antigenic variation in tprK (4). Tantalo et al. (23) demonstrated that different strains of T. pallidum have different propensities for invading the central nervous system after intravenous infection in rabbits, and it has been recognized that different strains cause lesions of differing severity in the rabbit model (24). The biological bases for these differences in pathogenesis have not yet been carefully explored. In the present study, we explore the relative transcription levels of the tpr family of putative virulence factors in four strains of T. pallidum, with comparison to the cellular immune responses that they induce during infection.
To date, semiquantitative and quantitative approaches have shown that all of the tprs are transcribed to some degree during early experimental infection with the Nichols strain (3, 12), although discrepancies were reported for the relative level of the tprK message. Using both microarray and real-time PCR, Smajs et al. (20) confirmed that the tpr genes are not equally transcribed, although tprK data were not included in that report and strains other than Nichols were not examined. In the present study we compared the expression of the tpr genes in four strains of T. pallidum subsp. pallidum (Nichols, Sea 81-4, Bal 73-1 and Chicago), demonstrating that tpr transcription patterns and levels vary among strains.
Levels of tprK message were higher than for other tpr alleles in three of the four strains examined; although tprK was highly transcribed in the remaining Sea 81-4 strain, the message for tprC was higher. The high levels of tprK are consistent with our previous findings that TprK induces an early and very high level of humoral and cellular immune response during experimental infection (18, 19). The differential levels of tprK transcription among the four strains correspond to the data reported by Leader et al. (14), which showed earlier and higher TprK antibody reactivity in rabbits infected with Chicago and Bal 73-1 compared to Nichols infection. No data are available on the TprK humoral response in rabbits infected with Sea 81-4. The evidence that the expression of this gene can be modulated in different strains argues for the existence of a mechanism of transcriptional regulation of tprK. Although phase variation or modulation can be hypothesized for tprG, tprF, tprJ, and tprI based on the architecture of the sequences upstream of their transcription start site (9), no similar organization was found for tprK transcription start site in any of the strains analyzed. All of the hypotheses that could provide a better understanding for these differences in tprK mRNA levels, including differential modulation induced by transcription factors, are currently being investigated. Elevated T-cell responsiveness to TprK (Fig. 2) in animals with long-term infections suggests that this antigen may be frequently (or continuously) expressed during the course of infection. We were unable to examine tprK expression directly in the long-term-infected rabbits used for the T-cell assays because the numbers of treponemes sufficient for RNA analysis are not available after long-term infection. Thus, the measured immune responses reflect the antigens preferentially offered to the immune system over several months of untreated disease, while the quantitative real-time data indicate transcription only during early active infection.
tprA was shown to be scarcely transcribed in treponemes harvested at the peak of testicular infection; furthermore, no humoral response toward the recombinant TprA peptide was detected at the same time point in Nichols, Chicago, and Bal 73-1 strain-infected rabbits (14). The absence of humoral and T-cell responses could, however, be due to the fact that the recombinant TprA peptide used for the studies refers to the tprA ORF downstream of the frameshift present in this gene, which would not allow translation in vivo of the peptide used herein. Sequence analysis showed that the Chicago and Bal 73-1 strains also carry a frameshift mutation within the tprA ORF identical to the one described in Nichols. In contrast, in the Sea 81-4 tprA, the frameshift is reverted by a fourth CT dinucleotide repeat. In our studies, the T-cell reactivity seen toward the TprA peptide by Sea 81-4-infected rabbits predicted the presence of a functional tprA ORF and induced us to sequence that locus.
Similarly, the lack of T-cell reactivity toward T. pallidum TprJ peptide in Sea 81-4-infected rabbits is consistent with the fact that the tprJ locus contains a tprG/J hybrid sequence (9). These studies have demonstrated how comparison of immune response and expression data can lead to identification of new gene sequence information. The Sea 81-4 strain harbors considerable differences in its tpr architecture compared to Nichols, which confirms this isolate as a valuable instrument for further comparative studies.
Three of the strains analyzed in the present study express tprB at a similar low level, whereas in the Sea 81-4 strain expression is almost absent. T-lymphocyte proliferation data suggest, however, that tprB transcription does occur during Sea 81-4 infection, since a significant cellular immune response was noted in long-term-infected rabbits. Future studies should examine the temporal modulation of tprB expression in T. pallidum strains.
tprH expression, significantly higher in Bal 73-1 with respect to Nichols, Chicago, and Sea 81-4 is not matched by enhanced T-cell proliferation to TprH peptide in Bal73-1-infected rabbits. TprH could, however, constitute a preferential target for the humoral response, as previously shown by the higher antibody reactivity to this antigen seen at day 17 postinfection in Bal 73-1-infected animals (14).
The expression of tprC/D was seen in all of the strains examined here. Because these two loci contain identical ORFs in Nichols, Chicago, and Bal73-1, it is not possible to know from our studies whether one or both loci are being transcribed. However, in Sea 81-4, which contains the D2 allele at the tprD locus, it is possible to identify the transcribed locus as tprC. In fact, tprC is expressed at significantly higher levels than the other tpr genes in this strain. An important role of TprC during infection with this strain is also suggested by the proliferation values obtained toward the TprC/D peptide; unfortunately, the T-cell responses to TprD2 were not measured. High T-cell reactivity toward the NH2-SubI peptide (containing the amino terminus of subfamily I members) is an indicator of the expression of any subfamily I member. As a further confirmation, Sun et al. (22), analyzing the humoral and cellular response toward subfamily I antigens in Sea 81-4-infected rabbits at peak orchitis, showed a remarkable reactivity toward the amino-conserved region of TprC, suggesting that this region is a common target for both the humoral and the cellular immune response even early in infection. The revised PSORT prediction of TprC and -D to outer membrane localization, as well as the now-recognized sequence diversity in the tprC and tprD loci in many T. pallidum strains, is consistent with a role for these proteins in pathogenesis.
There are several factors that could influence the results and interpretation of the present study, including different growth rates of individual strains of T. pallidum and the effects of possible animal-to-animal variation on the disease. Different T. pallidum strains are often characterized by distinct growth rates, which influence optimal harvest time and yield in terms of the number of organisms available for experimental procedures. We chose, however, despite the differences in time elapsed between infection and harvest of the Nichols and Chicago strains compared to the Sea 81-4 and Bal 73-1 strains, to measure expression of the tpr genes when each animal exhibited the same stage of experimental infection (peak orchitis), before the immune system could clear the majority of bacteria. The present study was conducted using treponemes from a single rabbit infected with each strain of T. pallidum; thus, differences in tpr expression potentially induced by the host cannot be determined. Unpublished new data from our laboratory (not shown), however, suggest only a limited degree of rabbit-to-rabbit variability in tpr gene expression during the experimental infection of multiple rabbits by a given strain.
It is well acknowledged that individuals with syphilis can have very different clinical courses of infection with differential involvement of the central nervous system and other sites. Although host susceptibility factors undoubtedly play an important role in influencing clinical course, the heterogeneity of T. pallidum strains in terms of tpr sequence, as well as in gene expression, suggests that T. pallidum strains may differ in pathogenic potential. Further studies will be needed to examine the correlation between tpr gene sequence and expression with clinical disease.
Acknowledgments
We thank Rebecca LaFond, Karin Hevner, and Troy Leader for help and suggestions to improve the quantification assay described here and Heidi Pecoraro for manuscript preparation.
This study was supported by NIH grants AI 42143, AI 63940, and AI 34616.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Baker-Zander, S. A., M. J. Fohn, and S. A. Lukehart. 1988. Development of cellular immunity to individual soluble antigens of Treponema pallidum during experimental syphilis. J. Immunol. 141:4363-4369. [PubMed] [Google Scholar]
- 2.Centurion-Lara, A., T. Arroll, R. Castillo, J. M. Shaffer, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 1997. Conservation of the 15-kilodalton lipoprotein among Treponema pallidum subspecies and strains and other pathogenic treponemes: genetic and antigenic analyses. Infect. Immun. 65:1440-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centurion-Lara, A., C. Castro, L. Barrett, C. Cameron, M. Mostowfi, W. C. Van Voorhis, and S. A. Lukehart. 1999. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centurion-Lara, A., C. Godornes, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect. Immun. 68:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centurion-Lara, A., R. E. LaFond, K. Hevner, C. Godornes, B. J. Molini, W. C. Van Voorhis, and S. A. Lukehart. 2004. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 52:1579-1596. [DOI] [PubMed] [Google Scholar]
- 6.Centurion-Lara, A., E. S. Sun, L. K. Barrett, C. Castro, S. A. Lukehart, and W. C. Van Voorhis. 2000. Multiple alleles of Treponema pallidum repeat gene D in Treponema pallidum isolates. J. Bacteriol. 182:2332-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelkens, H. J., F. J. ten Kate, J. Judanarso, V. D. Vuzevski, J. B. van Lier, J. C. Godschalk, J. J. van der Sluis, and E. Stolz. 1993. The localization of treponemes and characterization of the inflammatory infiltrate in skin biopsies from patients with primary or secondary syphilis, or early infectious yaws. Genitourin. Med. 69:102-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, J. C. Venter, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 9.Giacani, L., K. Hevner, and A. Centurion-Lara. 2005. Gene organization and transcriptional analysis of the tprJ, tprI, tprG, and tprF loci in the Nichols and Sea 81-4 Treponema pallidum isolates. J. Bacteriol. 187:6084-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giacani, L., V. Sambri, A. Marangoni, F. Cavrini, E. Storni, M. Donati, S. Corona, P. Lanzarini, and R. Cevenini. 2005. Immunological evaluation and cellular location analysis of the TprI antigen of Treponema pallidum subsp. pallidum. Infect. Immun. 73:3817-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giacani, L., E. S. Sun, K. Hevner, B. J. Molini, W. C. Van Voorhis, S. A. Lukehart, and A. Centurion-Lara. 2004. Tpr homologs in Treponema paraluiscuniculi Cuniculi A strain. Infect. Immun. 72:6561-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazlett, K. R. O., T. J. Sellati, T. T. Nguyen, D. L. Cox, M. L. Clawson, M. J. Caimano, and J. D. Radolf. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaFond, R. E., A. Centurion-Lara, C. Godornes, A. M. Rompalo, W. C. Van Voorhis, and S. A. Lukehart. 2003. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J. Bacteriol. 185:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leader, B. T., K. Hevner, B. J. Molini, L. K. Barrett, W. C. Van Voorhis, and S. A. Lukehart. 2003. Antibody responses elicited against the Treponema pallidum repeat proteins differ during infection with different isolates of Treponema pallidum subsp. pallidum. Infect. Immun. 71:6054-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukehart, S. A., S. A. Baker-Zander, R. M. Lloyd, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. II. Nature of cellular infiltration and Treponema pallidum distribution in testicular lesions. J. Immunol. 124:461-467. [PubMed] [Google Scholar]
- 16.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Morgan, C. A., S. A. Lukehart, and W. C. Van Voorhis. 2002. Immunization with the N-terminal portion of Treponema pallidum repeat protein K attenuates syphilitic lesion development in the rabbit model. Infect. Immun. 70:6811-6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan, C. A., S. A. Lukehart, and W. C. Van Voorhis. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect. Immun. 71:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, C. A., B. J. Molini, S. A. Lukehart, and W. C. Van Voorhis. 2002. Segregation of B and T-cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J. Immunol. 169:952-957. [DOI] [PubMed] [Google Scholar]
- 20.Smajs, D., M. McKevitt, J. K. Howell, S. J. Norris, W. W. Cai, T. Palzkill, and G. M. Weinstock. 2005. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J. Bacteriol. 187:1866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamm, L. V., S. R. Greene, H. L. Bergen, J. M. Hardham, and N. Y. Barnes. 1998. Identification and sequence analysis of Treponema pallidum tprJ, a member of a polymorphic multigene family. FEMS Microbiol. Lett. 169:155-163. [DOI] [PubMed] [Google Scholar]
- 22.Sun, E. S., B. J. Molini, L. K. Barrett, A. Centurion-Lara, S. A. Lukehart, and W. C. Van Voorhis. 2004. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 6:725-737. [DOI] [PubMed] [Google Scholar]
- 23.Tantalo, L. C., S. A. Lukehart, and C. M. Marra. 2005. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J. Infect. Dis. 191:75-80. [DOI] [PubMed] [Google Scholar]
- 24.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.
- 25.Van Voorhis, W. C., L. K. Barrett, D. M. Koelle, J. M. Nasio, F. A. Plummer, and S. A. Lukehart. 1996. Primary and secondary syphilis lesions contain mRNA for Th1 cytokines. J. Infect. Dis. 173:491-495. [DOI] [PubMed] [Google Scholar]