Abstract
Alternate sigma factors have been implicated in the survival of mycobacteria in response to specific stresses. To characterize the role of SigM in Mycobacterium tuberculosis, a sigM deletion mutant was generated by allelic exchange in the virulent CDC1551 strain. Comparing the wild-type and ΔsigM strains by complete genomic microarray, we observed a low level of baseline expression of sigM in wild-type M. tuberculosis and no significant differences in the gene expression patterns between these two strains. Alternatively, a SigM-overexpressing M. tuberculosis strain was constructed and microarray profiling revealed SigM-dependent expression of a relatively small group of genes, which included four esat-6 homologues: esxE, esxF, esxT, and esxU. An assessment of SigM-dependent promoters from the microarray analysis revealed a putative consensus sequence for M. tuberculosis SigM of −35 GGAAC and −10 CGTCR. In vitro expression studies showed that M. tuberculosis sigM transcripts accumulate slightly in stationary phase and following heat shock. To understand the role of SigM in pathogenesis, the M. tuberculosis sigM deletion strain was compared with the isogenic wild-type strain and the complemented mutant strain for survival in murine macrophages and in the mouse model. The mutant was found to have similar abilities to survive in both the resting and activated J774A.1 macrophages. Mouse organ bacterial burdens indicated that the mutant proliferated and persisted at the same level as that of the wild-type and complemented strains in lung and spleen tissues. In time-to-death experiments in the mouse model, the ΔsigM mutant exhibited lethality times comparable to those observed for the wild-type and complemented strains. These data indicate that M. tuberculosis SigM governs the expression of a small set of genes, including four esat-6 homologues, and that the loss of sigM does not confer a detectable virulence defect in the macrophages and mouse models of infection.
Tuberculosis remains a leading cause of human deaths throughout the world (10). Mycobacterium tuberculosis, the microbial cause of the disease, is a successful pathogen in part because of its high adaptability for survival in different host environments. Differential coordinate gene expression is one such adaptive mechanism, and it is clear that numerous response regulons are present in M. tuberculosis (23). In prokaryotes, gene sets are frequently regulated at the level of transcription, and alternate sigma factors frequently govern conditionally expressed responses, such as heat shock responses, sporulation, motility, and extracytoplasmic functions (ECFs), which include transport, cell wall modifications, or the secretion of proteins (14). The genome sequence of M. tuberculosis reveals the presence of 13 sigma factor genes, including 10 that have been classified as belonging to the ECF subfamily (9, 12).
Several M. tuberculosis ECF sigma factors have already been studied in detail. The M. tuberculosis sigE gene encodes a member of the ECF family of sigma factors, and the sigE mutant of M. tuberculosis, characterized by Manganelli et al. (25), demonstrated greater susceptibility to heat shock, sodium dodecyl sulfate (SDS), and various oxidative stresses than did the wild type. This mutant was also defective in its ability to grow inside both human and murine resting macrophages and was more susceptible to the killing activity of activated murine macrophages. In mouse studies, M. tuberculosis ΔsigE mutants proliferated and survived in mouse tissues but were less lethal by time-to-death analysis (1, 22). Kaushal et al. showed M. tuberculosis sigH to be essential for the production of immunopathology and lethality in mice (19). The M. tuberculosis ΔsigH mutant also exhibited a higher susceptibility to heat and oxidative stress in vitro (24, 29). These studies suggested that the ECF sigma factor SigH plays a central role in a regulatory network involved in stress responses. Another ECF sigma factor, SigD, has been shown to be involved in the virulence of M. tuberculosis (6, 27). By microarray analysis, it was shown that SigD regulates the expression of several physiologically significant genes. In a mouse model of infection, the ΔsigD mutant strain was found to be attenuated, with differences in survival and the inflammatory response in the lung between mice infected with the mutant and those infected with the wild type (27). In a similar observation by Calamita et al., SigD was reported to be required for virulence in mice and was found to play a major role in controlling the expression of genes required for protein synthesis during late stationary phase (6).
A fourth ECF sigma factor, SigC, has been evaluated by Sun et al., who observed a dramatic delay in median time to death in mice infected with the M. tuberculosis ΔsigC mutant compared to that in mice infected with the wild type (>300 days versus 170 days), without a significant difference in bacterial organ CFU counts (31). By whole-genome microarray experiments, it was observed that SigC regulates the expression of several important genes involved in virulence, such as senX3 and mtrA, a two-component sensor kinase and a two-component response regulator, respectively, as well as hspX, encoding an α-crystalline homologue (31). Recently, by using M. tuberculosis H37Rv mutants lacking sigma factor SigC, SigF, or SigM, it was observed that all of these mutants exhibited comparable growth levels in Middlebrook 7H9 broth as well as in the human monocytic cell line THP-1. Moreover, following low-dose aerosol infection of guinea pigs, the SigM mutant-infected animals did not exhibit differences in lung and spleen granuloma formation relative to animals infected with wild-type M. tuberculosis H37Rv. In contrast, a SigF mutant was partially attenuated, with the induction of necrotic spleen granulomas and poorly defined lung granulomas, while the SigC mutant exhibited attenuation in the lung and spleen and did not induce granuloma formation (18). In another report, it has been shown that an ECF sigma factor of M. tuberculosis, SigL, affects virulence in mice. By overexpressing sigL in M. tuberculosis and conducting microarray profiling with the sigL-overexpressing strain, it was observed that SigL controls the expression of several genes of M. tuberculosis, including those encoding polyketide synthases and secreted or membrane proteins (13).
In the present study, we have characterized SigM, a member of the ECF class of mycobacterial sigma factors. There is little information regarding the function of SigM in mycobacteria. Previous studies indicated that the expression levels of sigM were nearly half those of sigA in the exponential phase of mycobacterial growth and that sigM expression was not changed under several stress conditions except in the presence of 0.05% SDS, which led to a 3.5-fold reduction in its expression (21). However, in a study by Arraiz et al., it was observed that in both M. smegmatis and Mycobacterium bovis BCG, the expression of sigM was induced at a high temperature and in stationary phase (2), and in this study, we observed a similar expression pattern with induction in stationary phase as well as under heat shock. To assess the role of SigM in M. tuberculosis and its contribution to virulence, a ΔsigM mutant strain was constructed in M. tuberculosis CDC1551 by allelic exchange. We investigated the SigM regulon using microarray and quantitative reverse transcription-PCR (RT-PCR) analysis and identified a putative consensus promoter sequence. We also assessed the virulence of the ΔsigM mutant and persistence in the mouse and macrophage tuberculosis models.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and chemicals.
In this study, we used Escherichia coli strain DH5α {F′ endA1 hsdR17(rk− mk+) glnV44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR [φ80dlac Δ(lacZ) M15]}, procured from Stratagene, and HB101 [F−, thi-1, hsdS20(rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 galK2 rpsL20 (Strr) xyl-5 mtl-1], purchased from Promega. Mycobacterium tuberculosis CDC1551 and Mycobacterium smegmatis mc2155 were obtained from Colorado State University, Fort Collins, CO. Both the M. smegmatis and M. tuberculosis (the wild type and its mutants) cells were cultured in Middlebrook 7H9 containing 0.05% Tween 80 or Middlebrook 7H10 without Tween 80. Both types of medium were supplemented with 1× oleic acid-albumin-dextrose-catalase and 0.5% glycerol. Liquid cultures were grown in roller bottles, and plates were incubated at 37°C. All tissue culture media were purchased from Sigma unless otherwise specified. The following antibiotics were added when necessary: ampicillin (50 μg/ml), kanamycin (25 μg/ml for E. coli and 15 μg/ml for mycobacteria), and hygromycin (150 μg/ml for E. coli and 50 μg/ml for mycobacteria). M. tuberculosis was cultured under different stress conditions as described earlier (11). Stress conditions were as follows: 5 mM diamide (oxidative stress); 1 mM cumene hydroperoxide (CHP) (oxidative stress); pH 4.2 (acid stress); phosphate-buffered saline (PBS) (nutrient starvation); 1 mM DETA/NO (dimethylenetriamine nitric oxide adduct, nitrosative stress); 4°C water bath (cold shock); and 42°C (heat shock). The effect of acid stress was examined by washing the cultures twice with PBS and resuspending cells in complete 7H9 medium at pH 4.2 and incubating them for 3 h.
DNA techniques.
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs, and Taq polymerase was purchased from the Invitrogen Corporation. Protocols for DNA manipulations, including plasmid DNA preparation, restriction endonuclease digestion, agarose gel electrophoresis, and isolation and ligation of DNA fragments, are described elsewhere (30). E. coli DH5α was transformed by the standard protocol (30), and M. tuberculosis was transformed by electroporation. PCR amplifications were carried out according to the manufacturer's specifications (Applied Biosystems). Each of the 30 cycles was carried out at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. DNA fragments used for cloning and labeling reactions were purified by using a QIAGEN gel extraction kit (QIAGEN) as per the manufacturer's specifications.
sigM gene disruption and complementation.
An M. tuberculosis ΔsigM::hyg mutant was constructed by targeted mutagenesis as described previously (3). Briefly, DNA fragments flanking sigM were amplified by PCR from M. tuberculosis genomic DNA and cloned into the cosmid vector pYUB854 (a kind gift from William R. Jacobs, Jr., Albert Einstein College of Medicine, NY), at the sites flanking the hygromycin resistance cassette. The resulting plasmid (AES; allelic exchange substrate) was sequenced and ligated into the temperature-sensitive shuttle plasmid phAE87 (a kind gift from William R. Jacobs, Jr., Albert Einstein College of Medicine, NY) after digestion with PacI. The recombinant vector phAE87::AES was packaged in vitro using an in vitro packaging kit (Gigapack Gold; Stratagene), and the packaged bacteriophages harboring the recombinant vector phAE87::AES were used to transduce E. coli HB101 cells. Recombinants were selected on hygromycin plates, and plasmid DNA was isolated. This sigM-disrupting construct in phAE87 was electroporated into M. smegmatis mc2155, cells were mixed with top agar after revival at 30°C and plated on 7H10 plates, and plaques were obtained after 3 days of incubation at 30°C. Transducing phage isolation and the transduction of M. tuberculosis were performed as described previously (3). The resulting hygromycin-resistant colonies of M. tuberculosis were screened for the disruption of the sigM by gene-specific PCR and further confirmed by Southern blotting. To complement the ΔsigM::hyg mutant, a 1,032-bp DNA fragment, including the coding sequence of the sigM gene and 166 bp of 5′ sequence (including its native promoter) and a 197-bp sequence downstream of the sigM gene, was amplified by PCR with the primers sigM1 (5′-GGCCACAACACCATCTCG-3′) and sigM2 (5′-CGTACCCGGTTCAACGC-3′) and cloned into the TA cloning vector (Invitrogen). The resulting PCR amplicon was released from the recombinant vector by restriction digestion with SpeI and EcoRV and subsequently cloned in integrating vector pMH94 (20) at the Klenow end-filled blunt EcoRI site and cohesive XbaI site. The recombinant vector pMH94-sigM was subjected to nucleotide sequencing by using the forward primer sigM1 and subsequently used to transform the M. tuberculosis ΔsigM::hyg mutant strain. Candidate Hygr Kanr colonies were selected, and the presence of the intact complementing sigM allele was identified by PCR and Southern blotting.
Oxidative stress and detergent stress phenotypes.
Susceptibility of the ΔsigM::hyg mutant to diamide (1 M), cumene hydroperoxide (20 mM), H2O2 (100 mM), and sodium dodecyl sulfate (1%) was determined by using a disk diffusion assay as previously described (33). For the assay, a 10-μl solution of each of these oxidizing agents was loaded on 4.5-mm Whatman paper disks. Since the cumene hydroperoxide is prepared as a dimethyl sulfoxide (DMSO) solution, a negative control with 10 μl of DMSO was used.
RNA extraction.
Total RNA was isolated from M. tuberculosis by using the TRIzol method according to the instructions provided by the supplier (Invitrogen Corporation, Carlsbad, CA).
Quantitative RT-PCR.
RNA isolated from the bacterial cultures was subjected to 30 cycles of PCR to ensure that all DNA had been removed by treatment with RNase-free DNase I (Ambion) as evaluated by ethidium-bromide-stained agarose gel analysis before proceeding with RT of the RNAs. After RT, the relative amount of cDNA obtained from each transcript was measured by 35 cycles of RT-PCR. The relative change (n-fold) of mRNA of the genes was measured by normalizing the PCR product of the gene of interest to that of sigA, the level of which was the same in wild-type and sigM-overexpressing strains (attB::Pace-sigM) of M. tuberculosis by microarray analysis. The change of specific mRNA levels in M. tuberculosis CDC1551 attB::Pace-sigM strain relative to the wild-type strain was calculated as the ratio of the final PCR product change (n-fold) in the M. tuberculosis CDC1551 attB::Pace-sigM to that of sigA and change (n-fold) of PCR product to sigA of the gene of interest in the wild type. Transcript levels of sigM at different growth phases and following different stress conditions were calculated by using the sigA transcript for normalization for RNA amounts as described earlier (11).
Microarray preparation and data analysis.
Microarray slides were prepared as described earlier (19). Briefly, a set of 4,100 oligonucleotides (70-mer) encompassing the complete genome sequence of M. tuberculosis (Operon Technologies) was dissolved in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and printed in duplicate onto poly-l-lysine-coated glass slides using an OmniGrid arrayer (GeneMachines). For probe preparation, cDNA was prepared from 8 μg of total RNA from the test and control strains of M. tuberculosis. For each hybridization, the cDNA probes labeled with Cy3 or Cy5 (Amersham Biosciences) were used in pairs. Duplicate RNA samples were prepared, and each RNA sample was hybridized twice through reverse labeling of the respective cDNAs. Slides were scanned using a GenePix 4000A microarray scanner (Axon Instruments), and the resulting images were analyzed using GenePix Pro 4.0 software (Axon Instruments). A total of three hybridizations were performed, and six hybridization intensity values were obtained because of double printing for each gene. The fluorescence intensity of Cy3 and Cy5 was normalized against the overall intensity. The expression ratio for the test and the control genes was determined from the normalized fluorescence intensity. The expression ratio was calculated as the average change (n-fold) of the six data points. Student's t test was used to determine the significance of the change (n-fold). The P value was calculated from the one-tailed distribution with a two-sample equal-variance t test.
Mouse virulence and persistence testing.
For persistence testing, three groups of C57BL/6 mice were infected with ΔsigM, ΔsigM-complemented, and wild-type CDC1551 strains, respectively, by the aerosol route with an inoculum that implanted ∼102 CFU in lung. Six mice from each group were subsequently sacrificed at day 1 and at weeks 1, 2, 4, and 8 postinfection to determine the organ CFU count. Lung and spleen tissues were homogenized in their entirety in PBS, and colonies were enumerated on 7H10 plates after 3 weeks of incubation at 37°C. Time-to-death experiments were conducted using 5-week-old C57BL/6 mice. Twenty mice in three groups were infected by different strains via the aerosol route with similar numbers of bacilli. Three mice from each group were sacrificed at day 1 postinfection to obtain the bacillary load in lung.
Infection of murine macrophages.
The mouse macrophage J774A.1 cell line was cultured in RPMI 1640 medium containing 10% fetal bovine serum in 24-well tissue culture plates at a concentration of ∼106 cells/ml. For activation, cells were stimulated with gamma interferon (IFN-γ) (500 U ml−1) overnight, followed by lipopolysaccharide (LPS) (200 ng ml−1) for 3 h. Cells were subsequently infected with wild-type, ΔsigM, and ΔsigM-complemented M. tuberculosis at a multiplicity of infection of 1:1. After 2 h, cells were washed three times with PBS, and fresh culture medium was added. At designated time points, three wells of cells were washed with PBS and then lysed in 0.1% Triton X-100 for 5 min to release intracellular bacteria. The bacteria were serially diluted in PBS and plated on 7H10 for CFU analysis.
RESULTS
Expression of sigM in M. tuberculosis in vitro.
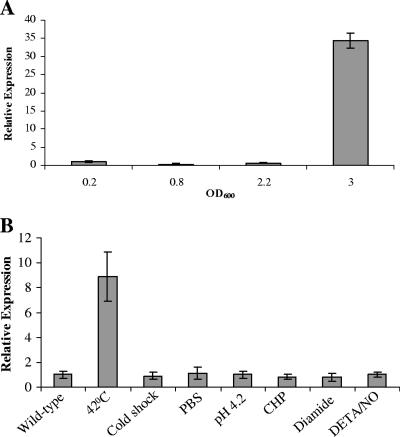
SigM is one of the ECF mycobacterial sigma factors. To assess the signal(s) for sigM expression in M. tuberculosis, we measured sigM transcript levels at different growth phases as well as under several stress conditions in vitro by quantitative RT-PCR. We examined the expression of the sigM gene from the early exponential to late stationary phases of growth. sigM expression was evaluated in M. tuberculosis by comparing its mRNA transcript levels with those of a housekeeping gene, sigA. The mRNA copy numbers of sigM and sigA were calculated from a standard curve prepared by performing quantitative RT-PCR using different known amounts of genomic DNA as a template, and it was observed that sigA mRNA levels were comparable at the different stages of mycobacterial growth (optical density at 600 nm [OD600], 0.2 to 3.0) (data not shown) as has been reported earlier (15). Transcript levels for sigM were found to be constant at an OD600 of 0.2 to 2.2 (Fig. 1A) and were at levels approximately 1,000-fold below that of sigA (data not shown). In late stationary phase (OD600 = 3.0), an ∼30-fold increase in sigM transcript level was observed compared with that in early exponential phase (Fig. 1A); however, sigM transcript relative abundance remained low, with levels approximately 30-fold below that of sigA (data not shown).
FIG. 1.
Expression of sigM in M. tuberculosis. (A) Analysis of the expression of the sigM gene at different growth phases in M. tuberculosis. Total RNA was isolated from M. tuberculosis CDC1551 cultures grown to the OD600 shown. Phases of growth were identified as described earlier (11). Quantitative RT-PCR was performed, and the relative expression of sigM was calculated using sigA transcript as normalization for RNA amounts and a 0.2 OD growth phase as a control. The graph shown represents an average of three experiments. (B) Analysis of the expression of the sigM gene under various stress conditions in M. tuberculosis. Total RNA was isolated from M. tuberculosis CDC1551 cultures grown to an OD600 of 1.0 and treated with various stresses for 3.0 h as described in Materials and Methods. Quantitative RT-PCR was performed, and the relative expression of sigM was calculated using normalization to sigA transcript levels, which remained constant under these conditions. The graph shown represents an average of three experiments. Error bars indicate standard deviations.
Moreover, we analyzed sigM expression levels during in vitro stress conditions that a mycobacterium may encounter during its life cycle, as discussed previously (11). As shown in Fig. 1B, sigM expression remained uniform under most of these stresses, except during heat shock at 42°C, where an ∼9-fold induction was observed compared with baseline expression (Fig. 1B).
Deletion of sigM gene and complementation of a ΔsigM mutant in M. tuberculosis CDC1551: effects on stress phenotypes.
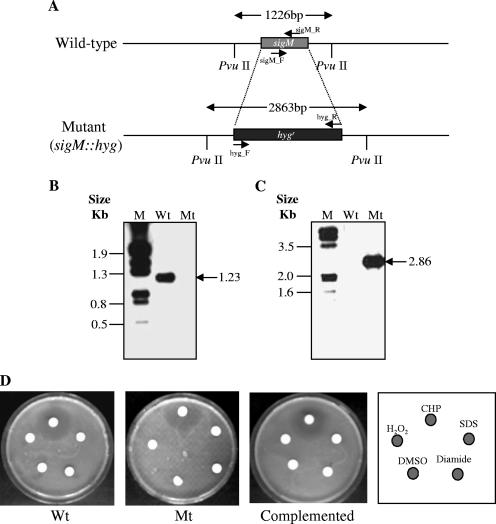
The sigM gene was deleted by allelic exchange as previously described (3). The gene replacement construct was generated by cloning 5′ and 3′ sigM flanking sequences (0.86 and 1.0 kb, respectively) into cosmid vector pYUB854 (3), thereby deleting the complete sigM open reading frame which was replaced by a hygromycin resistance cassette (Fig. 2A). The ΔsigM strain was obtained by selecting Hygr and Kans clones and subsequently confirmed by PvuII digestion of genomic DNA and Southern blot analysis using sigM and hyg gene-specific probes. As shown in Fig. 2B, the sigM gene-specific probe hybridized to a 1,226-bp PvuII fragment from wild-type genomic DNA, but there was no signal with DNA from the sigM::hyg insertion-deletion mutant (Fig. 2B). On the other hand, by using a hyg gene-specific probe, a band of ∼2.9 kb was obtained with the genomic DNA from the sigM::hyg insertion-deletion mutant, but there was no signal with genomic DNA from the wild-type strain (Fig. 2C). The complementation of the ΔsigM strain was obtained by transforming the mutant strain with a single-copy, chromosomally integrating vector containing the kanamycin resistance gene and a copy of the wild-type sigM gene expressed by its own promoter. The complemented strain exhibited Hygr and Kanr phenotypes (data not shown).
FIG. 2.
Strategy for construction of sigM deletion and ΔsigM complementation strains and the resulting effects on the growth phenotype of M. tuberculosis CDC1551. (A) The M. tuberculosis sigM locus and strategy for deletion mutant construction. (B and C) Southern blot analysis of wild-type M. tuberculosis CDC1551 (Wt) and sigM deletion mutant (Mt). Genomic DNA of each strain was digested with PvuII, electrophoresed in a 0.8% agarose gel, and transferred to a nylon membrane. The resulting blot was UV cross-linked and subsequently probed with either the sigM-specific PCR amplicon or a hyg gene-specific PCR amplicon. The sigM-specific probe was prepared by amplifying the DNA fragment using the primers sigM_F (5′-GCAGCGGGCACTGATGCG-3′ and sigM_R (5′-TAGCCCAGCAGCCGCGC-3′) and digoxigenin-labeled dUTP (Roche); similarly, the hyg gene-specific probe was prepared by amplifying the DNA fragment using the primers hyg_F (5′-GGGAATTCCATATGACACAAGAATCCCTG-3′) and hyg_R (5′-CCTTAATTAATCAGGCGCCGGGGGCGGT-3′) and digoxigenin-labeled dUTP (Roche). Expected sizes for the PvuII fragments from the wild-type and sigM mutant strains were 1.2 and 2.9 kb, respectively. (D) Susceptibility testing of different mycobacterial strains to various oxidative stresses and detergent stress (each 10 μl) by disk diffusion assay using 4.5-mm Whatman paper disks. The photographs demonstrate the effects of the oxidative stresses, cumene hydroperoxide (20 mM), diamide (1 M), and hydrogen peroxide (H2O2; 100 mM), and the detergent, SDS (1%), on the survival of wild-type (Wt), ΔsigM mutant (Mt), and ΔsigM-complemented strains of M. tuberculosis. The effects of these stress agents were determined by measuring the diameter of the zone of inhibition. These photographs were obtained with three individual sets of experiments and a representative photograph is shown in the figure. Since the cumene hydroperoxide is prepared as a DMSO solution, a negative control with 10 μl of DMSO was used.
The wild-type, mutant, and complemented mutant strains of M. tuberculosis had identical in vitro growth rates in Middlebrook 7H9 medium supplemented with oleic acid-albumin-dextrose-catalase, glycerol, and Tween 80, and no clear differences were observed in the colony morphologies of these strains when grown on solid medium (data not shown). To examine stress phenotypes of the sigM mutant strains, we compared the susceptibility of the ΔsigM strains to that of wild-type and ΔsigM-complemented strains by using a disk diffusion assay (33). As shown in Fig. 2D, the deletion of sigM did not affect the lethality of several oxidative agents (H2O2, cumene hydroperoxide, and diamide) and the detergent sodium dodecyl sulfate on the survival of M. tuberculosis on solid medium (Fig. 2D).
Identification of SigM-regulated genes by microarray.
To identify SigM-regulated genes, we performed global gene expression analysis using microarrays. As described above, sigM transcription was low at an OD600 of 0.2 to 2.2, and in late stationary phase (OD600 = 3.0), a modest sigM transcript accumulation was observed. Thus, we compared the expression pattern of genes in the ΔsigM strain with that of wild-type M. tuberculosis CDC1551 at an OD600 of 3.0. The cDNA from bacterial transcripts was labeled and hybridized to a 4,100-spot microarray comprised of 70-mers specific for M. tuberculosis genes. It was observed that the sigM mutant exhibited a gene expression profile similar to that observed with wild-type M. tuberculosis with no transcripts achieving statistically significant differences (data not shown). We attributed this failure to identify reductions in certain SigM-dependent transcripts in the mutant to the relatively low levels of sigM expression in the wild type under the conditions tested. As an alternative approach, we used “knock-in” technology to conditionally overexpress sigM prior to microarray profiling. To accomplish this, sigM was overexpressed in M. tuberculosis under the control of an acetamide-inducible promoter. The coding sequence of the sigM gene was cloned under the control of the acetamide-inducible promoter (Pace) (26, 32) in an integrating vector, pSCW38, derived from pMH94 (20) to create a strain with a single extra copy of sigM (M. tuberculosis CDC1551 attB::Pace-sigM). The acetamide promoter-containing vector pSCW38 was introduced into M. tuberculosis CDC1551, and this empty vector-containing clone served as a control strain (M. tuberculosis CDC1551 attB::Pace) for microarray analyses. Both strains M. tuberculosis CDC1551 attB::Pace-sigM and M. tuberculosis CDC1551 attB::Pace were grown to an OD600 of 0.5, induced for 6 h by the addition of acetamide to a final concentration of 0.2%, and subjected to RNA extraction prior to gene expression analysis using oligonucleotide microarrays. We performed the microarray experiments three times, each in duplicate with three independent RNA preparations, and the results were analyzed statistically. The genes that were differentially expressed (P < 0.05; ≥1.9-fold difference) in the sigM-overexpressing strain are listed in Table 1. The differential expression of several genes in the overexpression strain was confirmed by quantitative RT-PCR, as described in the next section.
TABLE 1.
Differentially expressed genes in the sigM-overexpressing strain relative to wild-typea
| Gene productb | Locus | Change in expression (fold) | SD (fold) | P value |
|---|---|---|---|---|
| FadD34 | Rv0035 | −2.2 | 0.015 | 8.44E-05 |
| Conserved hypothetical proteins* | Rv0313 | −2.2 | 0.042 | 3.11E-04 |
| CMP* | Rv0479c | −2.0 | 0.069 | 2.54E-03 |
| Tuf | Rv0685 | −2.0 | 0.057 | 1.25E-03 |
| CMP* | Rv3092c | 2.1 | 0.040 | 1.83E-02 |
| Hypothetical oxidoreductase | Rv3093c | 4.1 | 0.063 | 2.94E-02 |
| Conserved hypothetical proteins* | Rv3094c | 2.7 | 0.036 | 2.72E-02 |
| Hypothetical transcriptional regulatory protein* | Rv3095 | 1.9 | 0.039 | 3.24E-02 |
| Conserved hypothetical proteins* (alanine and proline rich) | Rv3439c | 2.2 | 0.042 | 3.98E-02 |
| HP* | Rv3440c | 3.2 | 0.042 | 3.89E-03 |
| EsxT | Rv3444c | 13.0 | 0.035 | 4.60E-04 |
| EsxU | Rv3445c | 3.2 | 0.082 | 4.61E-02 |
| CMP* | Rv3447c | 8.4 | 0.022 | 2.50E-02 |
| MycP4 | Rv3449 | 5.4 | 0.025 | 6.67E-03 |
| EsxE | Rv3904c | 2.2 | 0.014 | 9.36E-03 |
| EsxF | Rv3905c | 2.4 | 0.003 | 2.70E-02 |
| Conserved hypothetical proteins* | Rv3906c | 6.3 | 0.044 | 1.30E-02 |
| SigM | Rv3911 | 19.7 | 0.057 | 4.20E-04 |
Shown is a summary of genes over-/underexpressed >1.9-fold in the SigM conditionally overexpressing strain of M. tuberculosis. Results shown are relative to wild-type strain containing the empty vector, pSCW38. SD, standard deviation.
The asterisks denote gene products annotated as either conserved hypothetical proteins, conserved membrane proteins (CMP), or hypothetical proteins (HP).
As shown in Table 1, the treatment of the M. tuberculosis CDC1551 attB::Pace-sigM strain with 0.2% acetamide led to an ∼20-fold induction in sigM transcript level compared to its level in the M. tuberculosis CDC1551 attB::Pace strain. Our microarray results indicated that sigM overexpression also resulted in an ≥1.9-fold induction of genes Rv3092c, Rv3093c, Rv3094c, Rv3095, Rv3439c, Rv3440c, Rv3444c (esxT), Rv3445c (esxU), Rv3447c, Rv3449 (mycP4), Rv3904c (esxE), Rv3905c (esxF), and Rv3906c (Table 1). Analysis of genomic organization suggests that these genes could be part of four putative operons, Rv3092c-Rv3094c; Rv3439c-Rv3447c; Rv3448-Rv3449, and Rv3904c-Rv3906c. Two of these putative operons, Rv3439-Rv3447c and Rv3904-Rv39006c, contain tandem esat-6-like genes, esxT-esxU and esxE-esxF, respectively, which were significantly induced by sigM overexpression (esxT, 13.0-fold; esxU, 3.2-fold; esxE, 2.2-fold, and esxF, 2.4-fold). The other genes which were upregulated in sigM-overexpressed strains included Rv3092c (2.1-fold,), Rv3093c (4.1-fold), Rv3094c (2.7-fold), Rv3095 (1.9-fold), Rv3439c (2.2-fold), Rv3440c (3.2-fold), Rv3447c (8.4-fold), Rv3449 (5.4-fold), and Rv3906c (6.3-fold). We also observed a few genes which were significantly downregulated by sigM overexpression (Table 1). Those genes included Rv0035 (fadD34; 2.2-fold), Rv0313 (2.2-fold), Rv0479c (2.0-fold), and Rv0685 (tuf, encoding a probable iron-regulated elongation factor; 2.0-fold). These results indicate that SigM regulates the expression of various genes and thus may be considered a global regulator in M. tuberculosis.
Quantitative RT-PCR analysis.
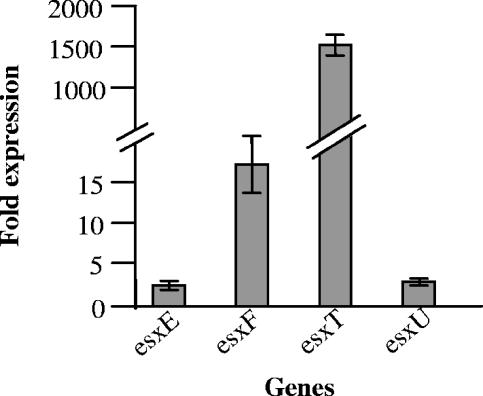
The microarray results indicate that SigM specifically regulates the expression of M. tuberculosis esat-6 homologues. In order to confirm the microarray results, we conducted RT-PCR experiments to analyze the differential expression of these genes under the influence of SigM overexpression. We compared the expression levels of the esxT, esxU, esxE, and esxF genes in the SigM-overexpressed and vector-containing wild-type M. tuberculosis strains. The amount of each respective transcript was normalized to the expression level of an essential sigma factor gene, sigA, which was not altered in either of these strains. As may be seen from Fig. 3, each of the esat-6-like genes, esxT, esxU, esxE, and esxF, exhibited significantly elevated expression levels in the SigM-overexpressed strain, thus confirming the microarray results (Fig. 3).
FIG. 3.
RT-PCR confirmation of altered gene expression levels in the M. tuberculosis sigM-overexpressing strain. Gene expression levels observed to be upregulated in the sigM-overexpressing strain relative to the wild-type strain by microarray analysis were evaluated by quantitative RT-PCR. M. tuberculosis expression levels of the genes of interest in these two strains were measured as described in Materials and Methods and normalized to sigA, an essential sigma factor gene whose expression was not altered in the sigM-overexpressing strain (data not shown). The expression (n-fold) indicates the ratio of normalized gene expression levels in the sigM-overexpressing strain relative to those in the wild type. Error bars indicate standard deviations.
Identification of a putative SigM promoter consensus sequence.
From the microarray analyses, several genes were up-regulated in the M. tuberculosis CDC1551 attB::Pace-sigM strain. It is possible that some of these genes are directly transcribed by the SigM-containing RNA polymerase holoenzyme, while other genes may be indirectly regulated by SigM through the intermediate effects of genes under direct SigM transcriptional control. The sigW alternate sigma factor gene from Bacillus subtilis, like M. tuberculosis sigM, encodes an ECF sigma factor induced in stationary phase (16). Because of their structural and functional similarity, we assumed that B. subtilis SigW and M. tuberculosis SigM might share similar promoter consensus sequences. Using the search pattern function in Tuberculist (http://genolist.pasteur.fr/TubercuList/index.html) and the known B. subtilis SigW consensus recognition sequence, TGAAAC-N16-CGTCww (7, 16, 17), we identified several SigM-controlled genes which are preceded by sequences similar to that of the SigW consensus sequence. Moreover, we restricted our analysis to genes with a distinct 5′ untranslated region rather than those that are downstream members of a gene cluster. We included in our search the genes overexpressed by ≥1.4-fold in the M. tuberculosis CDC1551 attB::Pace-sigM strain relative to the wild type. Putative promoter sequences that had no more than three mismatches in any of the hexamers with a spacer size ranging from 16 to 18 nucleotides were found in intergenic regions up to 350 bp from the translational start site. Our analysis revealed 10 genes that exhibited overexpression in the M. tuberculosis CDC1551 attB::Pace-sigM strain with no more than three mismatches in the −35 and −10 regions relative to the sigW promoter consensus template. Analysis of these genes revealed a putative consensus sequence for M. tuberculosis SigM of −35 GGAAC and −10 CGTCR (Table 2).
TABLE 2.
Putative M. tuberculosis SigM promoter consensus sequences
| Locus (MT/Rv/annotation)a | Distance from ATG (no. of base pairs) | Putative −35 consensus | Spacer (no. of nucleotides) | Putative −10 consensus | Overexpression level (fold) |
|---|---|---|---|---|---|
| MT0680/Rv0651/rplJ | −315 | GGCGAC | 17 | CGTCGC | 1.4 |
| MT1736/Rv2009 | −294 | TGTAGC | 16 | CGGCGG | 1.7 |
| MT2113/Rv2053c | −95 | TCGATC | 16 | CGTCAT | 1.5 |
| MT2973/Rv2905/lppW | −193 | AGGAAA | 18 | CGTCGA | 1.4 |
| MT3067/Rv2989 | −217 | TCGAAC | 16 | CCTCAT | 1.4 |
| MT3178/Rv3094c* | −40 | GGTAAC | 16 | CGTCAT | 2.7 |
| MT3179/Rv3095 | −330 | CGACAC | 17 | CGTCAC | 1.9 |
| MT3553/Rv3447c* | −89 | CGGAAC | 17 | CGCCCG | 8.4 |
| MT3554/Rv3448* | −298 | AGAAAC | 17 | CGGCGA | 1.7 |
| MT4026/Rv3907c/pcnA* | −95 | CGACAC | 18 | CGGCAA | 1.6 |
| Consensus | GGAAC | CGTCR |
Shown is the identification of the consensus promoter sequence for SigM by aligning the promoter regions of the overexpressed genes obtained by microarray. Asterisks represent genes which appear to be arranged in an operon-like fashion, and for these genes only, the sequence upstream of the first gene of the operon was considered for the identification of putative SigM promoter consensus sequences. R indicates A or G.
Effects of sigM deletion on the survival of M. tuberculosis in murine macrophages.
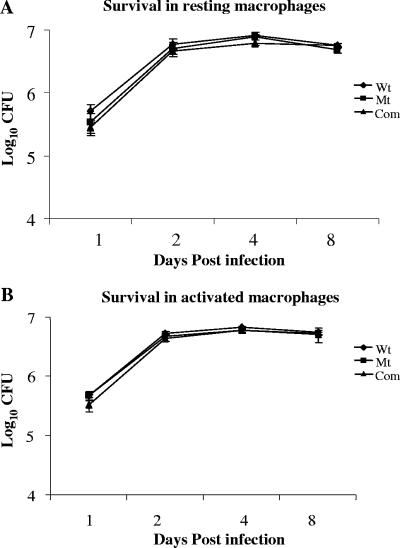
To assess the effects of sigM deletion on the survival of M. tuberculosis in macrophages, ΔsigM, wild-type, and ΔsigM-complemented M. tuberculosis strains were used to infect both resting and activated J774A.1 mouse macrophages. As may be seen in Fig. 4A, all three strains exhibited comparable survival rates in resting J774A.1 macrophages over a period of 8 days, indicating that the loss of sigM did not impair the ability of M. tuberculosis to survive in this murine macrophage line. To determine whether the activation of macrophages might reveal a defect in intracellular survival of the M. tuberculosis ΔsigM strain, macrophages were activated before infection by all three strains. For activation, macrophages were incubated overnight with IFN-γ, followed by incubation for 3 h with LPS as described in Materials and Methods. Our results indicated that similar to resting macrophages, IFN-γ/LPS-activated macrophages did not alter the survival of the ΔsigM mutant strain of M. tuberculosis in comparison with that of the wild-type and complemented strains (Fig. 4B).
FIG. 4.
Survival profile of M. tuberculosis CDC1551, the ΔsigM mutant, and the ΔsigM-complemented strains in resting and activated J774A.1 murine macrophages. Macrophages were infected with wild-type (Wt), ΔsigM mutant (Mt), and ΔsigM-complemented (Com) strains. Macrophages were activated with IFN-γ and LPS before infection. The results are expressed as CFU/well in a 24-well plate, and data represent the average of three experiments. (A) Macrophages resting prior to infection. (B) Macrophages activated prior to infection. Error bars indicate standard deviations.
Effects of sigM deletion on the virulence of M. tuberculosis in the mouse model of infection.
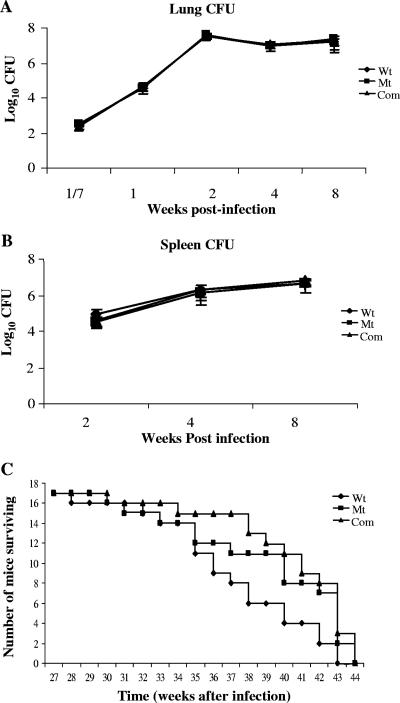
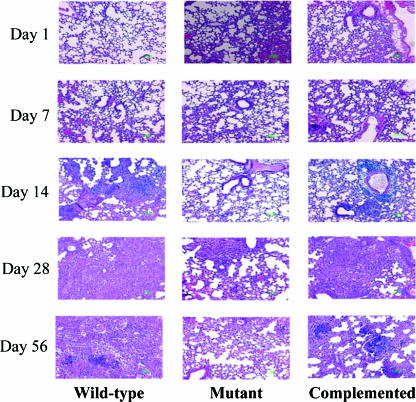
In order to evaluate the role of sigM in virulence, we investigated the in vivo persistence of the ΔsigM mutant strain of M. tuberculosis in the mouse model of tuberculosis. For this, C57BL/6 mice were infected in groups with the wild-type, ΔsigM, and complemented strains of M. tuberculosis CDC1551 by the aerosol route. At different time points, the mice were euthanized and the bacilli were enumerated in the spleen and lung tissues by plating serially diluted samples of the infected tissue homogenates. The day 1 lung CFU counts were determined, which revealed that the numbers of bacilli implanted in the mice were essentially similar for the ΔsigM (2.3 × 102 ± 0.3 × 102), wild-type (2.8 × 102 ± 0.2 × 102), and complemented (3.0 × 102 ± 0.2 × 102) strains. The lung bacillary count of the ΔsigM strain-infected mice showed a characteristic increase up to 2 weeks postinfection, followed by persistence in the plateau phase, and it remained comparable to the bacillary loads for both the wild-type and complemented strains over the course of the 16-week experiment (Fig. 5A). A similar pattern was obtained for the bacillary loads in spleen, with all three strains showing ∼106 CFU counts in the spleens at 4 weeks after aerosol infection (Fig. 5B). Hematoxylin-and-eosin-stained lung tissues from infected C57BL/6 mice were compared for histologic changes in tissue pathology, and significant variations in the inflammatory responses in the lungs of mice infected with the wild-type and ΔsigM strains were not detected (Fig. 6).
FIG. 5.
Effects of sigM deletion on the virulence of M. tuberculosis in the mouse model of infection. Lung (A) and spleen (B) CFU counts and time-to-death analysis (C) in C57BL/6 mice infected with wild-type (Wt), ΔsigM mutant (Mt), and ΔsigM-complemented (Com) strains of M. tuberculosis by the aerosol route. Groups of six infected mice were evaluated for CFU counts at each time point. Groups of 20 mice were infected for time-to-death analysis; of these, three from each group of infected mice were sacrificed on day 1 (shown as week 1/7) to determine the initial lung CFU counts. Error bars indicate standard deviations.
FIG. 6.
Tissue histopathology of lungs of mice infected with wild-type, ΔsigM, and ΔsigM-complemented strains of M. tuberculosis. The ΔsigM mutant produces tissue pathology in lungs of infected C57BL/6 mice similar to that with wild-type and complemented strains. C57BL/6 mice infected with ∼102 bacilli by the aerosol route were analyzed at day 1 and at 7, 14, 28, and 56 days (magnification, ×200; hematoxylin and eosin staining).
The role of SigM in the virulence of M. tuberculosis was also evaluated by monitoring the median time-to-death value of C57BL/6 strains of mice infected with the ΔsigM, ΔsigM-complemented, and wild-type strains of M. tuberculosis. Mice were infected in groups with all three strains of M. tuberculosis CDC1551 by the aerosol route, and the day 1 counts demonstrated essentially similar bacillary loads (200 ± 50 CFU) in lungs of mice infected with these strains. As shown in Fig. 5C, mice infected with all three strains of M. tuberculosis exhibited comparable times to death (median times to death were 248 days for wild-type-, 262 days for ΔsigM-complemented-strain-, and 259 days for ΔsigM strain-infected mice), and these median time-to-death values did not differ statistically using Kaplan-Meier analysis.
DISCUSSION
Resistance to various environmental stresses has often been associated with the ability of pathogenic bacteria to survive in their hosts, and alternative σ factors have been implicated in such survival (23). In this paper, we characterize a mutant of M. tuberculosis CDC1551 in which the gene encoding the ECF σ factor σM was disrupted. Contrary to previous observations with the sigM mutant strain of M. smegmatis, where it was shown that ΔsigM mutants of M. smegmatis were susceptible to oxidative stress (2), we observed no change in the susceptibility of the ΔsigM strain of M. tuberculosis to various environmental stresses, such as oxidative stress and SDS, in comparison to that of the wild type (Fig. 2D). Thus, our results indicate that M. tuberculosis SigM is not required for oxidative and detergent stress survival. These results were further corroborated by the observations that sigM expression remained unaltered in M. tuberculosis following exposure to several stress conditions, including oxidative stresses (Fig. 1B), thus suggesting that the roles of the SigM orthologues in M. tuberculosis and M. smegmatis may differ from each other. However, similar to previous observations with other mycobacterial strains, we observed that sigM is induced in M. tuberculosis in late stationary phase and under heat shock (Fig. 1). Though sigM transcription was substantially induced in our study under these experimental conditions, it remained at low levels relative to that of sigA. Therefore, future studies are needed to examine the significance of the minor inductions in sigM transcript levels under these experimental conditions. Moreover, the differences in the expression patterns of sigM at the various growth phases in our study and the previously published report by Manganelli et al. (21) could be due to differences in the experimental procedures between the two studies. In their report, by using molecular beacons, Manganelli et al. used the total amount of RNA as starting material for the normalization since they observed differences in sigA transcript levels during the different growth phases. However, a previous report by Hu and Coates suggests that sigA levels remain constant in M. tuberculosis at several growth stages and under several environmental stresses (15). Similarly, we also observed no significant change in sigA transcript levels under different growth stages and various stress conditions in our quantitative RT-PCR experiments and thus used this for the normalization of sigM RNA amounts.
Our gene profiling experiments comparing the M. tuberculosis ΔsigM mutant to its wild-type parent grown in vitro in rich medium failed to identify significant expression differences by using complete genome microarrays. The simplest explanation for this is our related finding that sigM expression levels in wild-type bacilli are low during axenic growth in Middlebrook medium, and hence, the SigM regulon is likely inactive even in the wild-type strain. Using an alternative approach, chemical-dependent “knock-in” expression of the sigM gene in a wild-type strain background, we were able to detect differential expression of a relatively small set of genes, as identified in Table 1, that were up-regulated upon conditional sigM expression. The majority of these putative SigM-dependent gene products are annotated as hypothetical or conserved hypothetical proteins. However, subanalysis of these hypothetical proteins reveals some potential clues as to the physiology of the SigM regulon. Four putative SigM-dependent genes were esat-6 homologues (esxT, esxU, esxE, and esxF). While the precise role of ESAT-6 is currently unknown, ESAT-6 is a secreted protein that has been postulated to modulate host cellular responses during infection (5). Other genes which were found to be upregulated in a sigM-overexpressed strain included Rv3092 (exhibiting similarity to a putative ABC transport system membrane protein in Corynebacterium diphtheriae), Rv3447c (encoding a probable conserved membrane protein related to the FtsK/SpoIIIE family), and Rv3449 (encoding a probable membrane-anchored mycosin Mycp4, a serine protease). The preponderance of genes encoding cell membrane-associated proteins and the presence of four esat-6 homologues which are likely to encode secreted polypeptides suggests that the SigM regulon may be associated with the production and export of secreted gene products, a hypothesis consistent with the fact that SigM belongs to the extracytoplasmic class of sigma factors. Despite the small number of genes in the putative regulon, we were able to identify a likely SigM promoter consensus sequence of −35 GGAAC and −10 CGTCR by searching the 5′ untranslated regions of these genes (Table 2).
Recently (during the review of this paper), Raman et al. (28) characterized the sigM regulon in M. tuberculosis, identifying a number of the same genes found in our analysis, such as the four esat-6 homologues mentioned above. Interestingly, Raman et al. also found that SigM down-regulates several genes involved in lipid metabolism, including the kasA-kasB operon. The apparent differences between the microarray list of Raman et al. and our microarray experiments may be due to the differences in technique, arrays, or the parent mycobacterial strains, M. tuberculosis CDC1551 versus M. tuberculosis H37Rv (which are known to differ in genetic composition as well as virulence phenotypes), used in these two studies (4).
To study the importance of sigM in M. tuberculosis survival under the stresses encountered inside the macrophage, we used the sigM mutant to infect J774A.1 murine macrophages. Unactivated J774A.1 macrophages are permissive for the growth of M. tuberculosis, and when activated with gamma interferon and LPS, they are bactericidal by means of an NO-dependent mechanism (8). Similar to the previously published reports with other sigma factor mutants of M. tuberculosis, sigC and sigD (6, 31), we found that the deletion of sigM in M. tuberculosis CDC1551 does not bear any effect on its survival in unactivated as well as activated J774A.1 macrophages. We also studied the effect of sigM deletion on the virulence of M. tuberculosis in the mouse model of infection, and similarly, we did not observe attenuation of the ΔsigM strain (Fig. 4 and Fig. 5) by both the organ CFU and time-to-death experiments. Despite this apparent lack of virulence of the ΔsigM mutant strain in the mouse model of tuberculosis, it remains possible that sigM is required for survival in other animal models which more closely mimic the human. In a recent report by Karls et al. using the low-dose aerosol infection of guinea pigs with different sigma factor mutants of M. tuberculosis, the sigM mutant was the least attenuated (18). It resulted in necrotic granulomas in the lungs and spleen and the calcification of lung granulomas at 20 weeks postinfection comparable to those observed in H37Rv-infected animals. Similar to our observations with a mouse model of infection, the authors suggested that SigM is not essential for virulence in the guinea pig host (18).
In conclusion, our study reveals that M. tuberculosis SigM governs the expression of a small set of genes, including four esat-6 homologues and several genes encoding membrane proteins, and it defines a putative SigM consensus promoter recognition sequence. Although the loss of sigM did not result in a loss of virulence in the macrophage or mouse models of M. tuberculosis infection, it remains possible that sigM is required for conditional survival and pathogenicity in aspects of the M. tuberculosis disease cycle, such as persistence, survival in necrotic granulomas, or transmission among the hosts via droplet nuclei which are not measurable by these experimental models. Further studies are needed to clarify the precise microenvironment in which this M. tuberculosis ECF sigma factor and its regulon are significantly activated.
Acknowledgments
The support of NIH grants AI36973, AI37856, AI43846, and AI51668 is gratefully acknowledged.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Ando, M., T. Yoshimatsu, C. Ko, P. J. Converse, and W. R. Bishai. 2003. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect. Immun. 71:7170-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arraiz, N., L. Salazar, G. Lopez, R. Rodriguez, Y. Casart, and H. Takiff. 2001. Characterization of the expression and function of SigM an ECF sigma factor in mycobacteria. Acta Cient. Venez. 52(Suppl. 1):40-41. (In Spanish.) [PubMed] [Google Scholar]
- 3.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 4.Bishai, W. R., A. M. Dannenberg, Jr., N. Parrish, R. Ruiz, P. Chen, B. C. Zook, W. Johnson, J. W. Boles, and M. L. Pitt. 1999. Virulence of Mycobacterium tuberculosis CDC1551 and H37Rv in rabbits evaluated by Lurie's pulmonary tubercle count method. Infect. Immun. 67:4931-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin, P., L. Majlessi, L. Marsollier, M. I. de Jonge, D. Bottai, C. Demangel, J. Hinds, O. Neyrolles, P. D. Butcher, C. Leclerc, S. T. Cole, and R. Brosch. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calamita, H., C. Ko, S. Tyagi, T. Yoshimatsu, N. E. Morrison, and W. R. Bishai. 2005. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell. Microbiol. 7:233-244. [DOI] [PubMed] [Google Scholar]
- 7.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 8.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doukhan, L., M. Predich, G. Nair, O. Dussurget, I. Mandic-Mulec, S. T. Cole, D. R. Smith, and I. Smith. 1995. Genomic organization of the mycobacterial sigma gene cluster. Gene 165:67-70. [DOI] [PubMed] [Google Scholar]
- 10.Dye, C., S. Scheele, P. Dolin, V. Pathania, M. C. Raviglione, et al. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 11.Geiman, D. E., T. R. Raghunand, N. Agarwal, and W. R. Bishai. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez, J. E., J. M. Chen, and W. R. Bishai. 1997. Sigma factors of Mycobacterium tuberculosis. Tuber. Lung Dis. 78:175-183. [DOI] [PubMed] [Google Scholar]
- 13.Hahn, M. Y., S. Raman, M. Anaya, and R. N. Husson. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 187:7062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 15.Hu, Y., and A. R. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 18.Karls, R. K., J. Guarner, D. N. McMurray, K. A. Birkness, and F. D. Quinn. 2006. Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerosol infection of guinea pigs suggests a role for SigC in pathogenesis. Microbiology 152:1591-1600. [DOI] [PubMed] [Google Scholar]
- 19.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 22.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extra cytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 25.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 26.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267-7226. [DOI] [PubMed] [Google Scholar]
- 27.Raman, S., R. Hazra, C. C. Dascher, and R. N. Husson. 2004. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186:6605-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raman, S., X. Puyang, T. Y. Cheng, D. C. Young, D. B. Moody, and R. N. Husson. 2006. Mycobacterium tuberculosis SigM positively regulates Esx secreted protein and nonribosomal peptide synthesis genes and down regulates virulence-associated surface lipid synthesis. J. Bacteriol. 188:8460-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor SigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 32.Triccas, J. A., T. Parish, W. J. Britton, and B. Gicquel. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:151-156. [DOI] [PubMed] [Google Scholar]
- 33.Wallace, R. J., Jr., J. R. Dalovisio, and G. A. Pankey. 1979. Disk diffusion testing of susceptibility of Mycobacterium fortuitum and Mycobacterium chelonei to antibacterial agents. Antimicrob. Agents Chemother. 16:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]