Abstract
Apical membrane antigen 1 (AMA1) of the malaria parasite Plasmodium falciparum is an integral membrane protein that plays a key role in merozoite invasion of host erythrocytes. A monoclonal antibody, 4G2dc1, recognizes correctly folded AMA1 and blocks merozoite invasion. Phage display was used to identify peptides that bind to 4G2dc1 and mimic an important epitope of AMA1. Three of the highest-affinity binders—J1, J3, and J7—were chosen for antigenicity and immunogenicity studies. J1 and J7 were found to be true antigen mimics since both peptides generated inhibitory antibodies in rabbits (J. L. Casey et al., Infect. Immun. 72:1126-1134, 2004). In the present study, the solution structures of all three mimotopes were investigated by nuclear magnetic resonance spectroscopy. J1 adopted a well-defined region of structure, which can be attributed in part to the interactions of Trp11 with surrounding residues. In contrast, J3 and J7 did not adopt an ordered conformation over the majority of residues, although they share a region of local structure across their consensus sequence. Since J1 was the most structured of the peptides, it provided a template for the design of a constrained analogue, J1cc, which shares a structure similar to that of J1 and has a disulfide-stabilized conformation around the Trp11 region. J1cc binds with greater affinity to 4G2dc1 than does J1. These peptide structures provide the foundation for a better understanding of the complex conformational nature of inhibitory epitopes on AMA1. With its greater conformational stability and higher affinity for AMA1, J1cc may be a better in vitro correlate of immunity than the peptides identified by phage display.
Malaria infects 300 to 500 million people per year worldwide and causes 2 to 3 million deaths, mainly in children under 5 years of age. A considerable effort is being devoted to the development of a vaccine against malaria, and one of the leading candidates for inclusion in such a vaccine is apical membrane antigen 1 (AMA1), a type I integral membrane protein conserved throughout all Plasmodium species. AMA1 is critical for the invasion of host erythrocytes (36) and is translocated from the micronemes onto the parasite surface around the time of invasion (20). Although its precise role in invasion remains undefined, it has been postulated that AMA1 is involved in realignment of the parasite after attachment to the erythrocyte, ensuring that the apical prominence of the merozoite is in close proximity to the erythrocyte surface (8, 33). Recombinant AMA1 induced protective immune responses in mouse and monkey models of malaria (2, 10, 11, 13), and both monoclonal and polyclonal antibodies to AMA1 inhibit merozoite invasion of erythrocytes (2, 9, 11, 12, 25, 34, 42). The observation that it was not possible to obtain targeted gene disruptions of the AMA1 gene that knocked out the function of the protein further supports an important role for AMA1 in the invasion of host erythrocytes (43). Recently, a conditional knockout of Toxoplasma gondii AMA1 was created (32); the TgAMA1 deficiency had no effect on microneme secretion or initial attachment of the parasite to the host cell, but it did inhibit the secretion of the rhoptries, whose discharge is coupled to active host cell penetration, suggesting that attachment of the parasite to the host cell occurs in two stages, the second of which requires TgAMA1 and is involved in regulating rhoptry secretion.
The monoclonal antibody (MAb) 4G2dc1, which binds to correctly folded AMA1 but not to the reduced and alkylated antigen, is a useful reagent for monitoring the correct disulfide bonding of AMA1 (25). Moreover, 4G2dc1 reacted with AMA1 in 10 different isolates of P. falciparum from diverse geographical locations and consistently inhibited the invasion of P. falciparum merozoites into erythrocytes by 60 to 70% in vitro (25). The exact location of the 4G2dc1 conformational epitope is not known, but it appears to include a number of residues from a loop in domain II of AMA1 (38), which shows some conformational flexibility (15, 38). Nonetheless, it is clearly important for the generation of protective antibodies that can block merozoite invasion.
We have used phage display technology to isolate peptide sequences that mimic the conformation of the 4G2dc1 epitope of AMA1 (7). A library of random 20-residue peptides expressed as N-terminal fusions to protein III of filamentous phage M13 was selected on 4G2dc1 to develop a population of peptides that bound specifically to this antibody. Three of the highest-binding clones were selected for DNA sequencing (7). Immunization of rabbits with three of these mimotopes induced high titers of anti-peptide antibodies, which were reactive with native AMA1. In addition, using the immobilized mimotopes as immunoadsorbents, human antibodies could be affinity purified from the plasma of individuals living in regions of Papua New Guinea (PNG), where malaria is endemic. Both rabbit and human purified antibodies were able to inhibit P. falciparum invasion of erythrocytes in vitro (7). This is the first example of phage-derived peptides that mimic an important epitope of a blood-stage malaria vaccine and have the ability to induce functionally protective antibodies.
Of the phage display peptides that bound to 4G2dc1, three with the highest affinity for the antibody were chosen for further study. The sequences of these peptides—J1, J3, and J7—are shown in Fig. 1. J1 and J7 elicited antibody responses in rabbits that recognized the peptide immunogen, as well as recombinant and native AMA1. J1 antibodies were specific for particular strains of P. falciparum and could bind to 3D7, D10, HB3, and W2mef isolates of AMA1 but not to reduced and alkylated AMA1, RESA, MSP2, or MSP3 (7). In the present study we investigated the solution structures of these three peptide mimotopes. One of the most biologically active 4G2dc1 peptide mimics, J1, adopted a well-defined structure, which provided a template for the design of a constrained analogue, J1cc, that binds more tightly to 4G2dc1 than does J1.
FIG. 1.
Sequences of the J series mimotopes of apical membrane antigen 1. Identical residues are shown in boldface.
MATERIALS AND METHODS
Peptide synthesis and sample preparation.
The peptides used in the present study were synthesized by Auspep Pty., Ltd. (Parkville, Australia) and purified by high-pressure liquid chromatography to 79% for J1 and 68% for J1cc. The molecular masses of the peptides were confirmed by mass spectrometry. J1, J3, and J7 were prepared for nuclear magnetic resonance (NMR) analysis by dissolving the peptide in 500 μl of 90% H2O-10% 2H2O to a final concentration of approximately 1 mM. The pH was adjusted to 4.5 in J1 and J7 with no buffer being present, but 10 mM sodium acetate was present in the J3 sample. J1cc was dissolved in 500 μl of 95% H2O-5% 2H2O to a final concentration of approximately 1 mM. The pH of J1cc was adjusted to 4.8 in the presence of 10 mM sodium acetate buffer. 1H NMR spectra of the peptide showed a single major set of resonances, indicating that any impurities were largely nonpeptidic; minor Trp NH peaks with intensities <10% of the main peak were present in the J1cc spectrum, arising from either impurities or a minor conformer.
Generation of the J1cc phage-displayed peptide.
This was achieved by generating a “cysteine library” at positions 6, 7, 16, and 17 by incorporating degenerate oligonucleotides encoding the “wild-type” residues and cysteine simultaneously. Residue 6 (Phe), encoded by the codon TTT, was substituted by T(T/G)T, where the second base in the codon was either T or G. The product of this base change in the library would be Phe or Cys in equal proportion. Likewise, residue 7 (Thr), encoded by ACT, was substituted by (A/T)(C/G)T, producing the final amino acid proportions of 50% Ser, 25% Cys, and 25% Thr. The same process was carried out for residues 16 [Asn, encoded by AAT and mutated to (A/T)(A/G)T] and 17 [His, encoded by CAT and mutated to (C/T)(A/G)T], resulting in residue 16 having 25% Asn, 25% Tyr, 25% Ser, and 25% Cys, while residue 17 was 25% His, 25% Tyr, 25% Ser, and 25% Cys. The degenerate sequences were incorporated into two oligonucleotides with flanking regions complementary to the appropriate regions in the J1 open reading frame. In all, there were 128 different sequences represented in the library of degenerate oligonucleotides, of which four encoded cysteines at positions 6 or 7 and positions 16 or 17. Kunkel mutagenesis was then carried out by using both oligonucleotides in the same reaction to produce the J1cc library. This was followed by phage display and biopanning on MAb 4G2dc1 (7) as a mechanism for identifying the most appropriate Cys-Cys substitution while maintaining or even increasing immunoreactivity.
Reactivity of J1 and J1cc phage and peptides.
The binding activity of J1 phage mutants to 4G2dc1 was compared to the original J1 phage by enzyme-linked immunosorbent assay (ELISA) using the method described previously (9). Briefly, 100 μl of 4G2dcl/well was coated (5 μg/ml) in phosphate-buffered saline (PBS) overnight at 4°C, and wells were washed twice with PBS and blocked with 5% Blotto (skim milk powder in PBS) for 2 h. After a further two washes with PBS, dilutions of phage in PBS-0.05%Tween 20 were added in duplicate to the wells, followed by incubation for 1 h with gentle agitation. The plate was washed four times with PBS-Tween, and peroxidase-conjugated anti-M13 MAb (Amersham) was added to each well, followed by incubation for 1 h as described above. After a final four washes with PBS-Tween and one with PBS, the plate was developed with 3,3′,5,5′-tetramethylbenzidine substrate (TMB; Sigma). In order to ensure the phage peptide clones were expressed to a similar level for binding, the reactivity to the Myc epitope was measured as described previously in Coley et al. (9).
To assess the reactivity of J1 and J1cc, the peptides were coupled to peptide immobilizer plates (Exiqon, Vedbaek, Denmark). Briefly, 10 μg of peptide/ml was coated overnight at 4°C to the wells with gentle agitation, the wells were then washed three times with PBS-Tween, and dilutions of 4G2dc1 MAb in PBS-Tween were added for 1 h. After a further five washes, anti-rat immunoglobulin G (IgG)-horseradish peroxidase was added for 1 h. The wells were washed five times and detected with TMB. Human plasma ELISAs were performed as described previously in Casey et al. (7). A similar ELISA was performed as described above; plasma was diluted 1/100 in PBS containing 2% fish gelatin (Sigma), 1% Tween 20, and 1% bovine serum albumin, and peroxidase-conjugated anti-human IgG (Chemicon) was used for detection. All ELISAs were repeated to ensure consistent results.
NMR spectroscopy.
Two-dimensional homonuclear total correlation (TOCSY) spectra with a spin-lock time of 60 ms and double-quantum-filtered correlation (DQF-COSY) NMR spectra were acquired at 500 MHz on a Bruker AMX-500 spectrometer for J1, J3, and J7 and at 600 MHz on a Bruker DRX-600 for J1cc. Nuclear Overhauser effect spectroscopy (NOESY) spectra with short mixing times of 50 ms for J7 and 100 ms for J1 and a series of 1D spectra over the temperature range from 5 to 35°C at 5°C intervals were also collected for each peptide. NOESY spectra with a mixing time of 250 ms were acquired on a Bruker DRX-600 spectrometer for all four peptides. For the J1cc peptide, a NOESY spectrum with a mixing time of 250 ms was also acquired at 800 MHz on a Bruker Avance 800. Water was suppressed by using the WATERGATE pulse sequence (37). Amide exchange rates in J1, J3, and J7 were monitored by dissolving freeze-dried material in 2H2O and then recording a series of one-dimensional spectra, followed by 60-ms TOCSY and 250-ms NOESY spectra. All spectra were collected at 5°C unless otherwise stated and were referenced to an impurity peak at 0.15 ppm or to the water resonance.
Diffusion measurements were performed by using a pulsed-field gradient longitudinal eddy-current delay pulse sequence (14, 16) as implemented by Yao et al. (46). J1 at 20°C and pH 4.3 in 50 mM sodium acetate buffer was also examined by analytical ultracentrifugation.
Spectra were processed by using XWINNMR (version 3.5; Bruker Biospin) and TOPSPIN (version 1.3; Bruker Biospin) and analyzed by using XEASY (version 1.3.13) (4). A table of chemical shift assignments for each of the peptides has been deposited in the BioMagResBank (40) as entries 7212, 7213, 7214, and 7215 for J1, J1cc, J3, and J7, respectively.
Structural constraints.
3JHNHA coupling constants were measured from DQF-COSY spectra at 500 MHz for J1, J3 and J7 and at 600 MHz for J1cc and then converted to dihedral restraints as follows: 3JHNHα > 8 Hz, φ = −120 ± 40°; and 3JHNHα < 6 Hz, φ = −60 ± 30°. If a positive φ angle could be excluded on the basis of nuclear Overhauser effect (NOE) data (30), φ angles were restricted to the range −180 to 0°. A χ1 angle for Ile9 of J1 was determined based on the analysis of a short-mixing-time NOESY spectrum, but no χ1 angles were determined for J1cc. No χ1 angle restraints were included for J3 or J7. A summary of the dihedral restraints used is given in Table 1 . J1cc contained a disulfide bond, Cys6-Cys16, which was included as a structural restraint. No hydrogen bonds were included as structural restraints for any of the peptides.
TABLE 1.
Summary of experimental constraints and structural statistics for the peptides J1, J1cc, J3, and J7
| Parameter | Result for peptide
|
|||
|---|---|---|---|---|
| J1 | J1cc | J3 | J7 | |
| No. of distance restraints | 220 | 202 | 125 | 167 |
| Intraresidue (i = j) | 54 | 95 | 37 | 43 |
| Sequential (|i − j| = 1) | 104 | 72 | 69 | 105 |
| Medium range (1 < |i − j| < 5) | 59 | 32 | 19 | 19 |
| Long range (|i − j| > 4) | 3 | 3 | 0 | 0 |
| Hydrogen bond restraints | 0 | 0 | 0 | 0 |
| No. of dihedral restraints | 16 | 15 | 11 | 17 |
| Energiesa | ||||
| Mean ENOE (kcal mol−1) ± SD | 4.4 ± 1.0 | 8.0 ± 2.2 | 0.7 ± 0.6 | 3.8 ± 0.9 |
| Mean RMS deviations from expt data ± SD | ||||
| NOEs (Å) | 0.020 ± 0.002 | 0.028 ± 0.004 | 0.011 ± 0.002 | 0.021 ± 0.002 |
| Dihedrals (°) | 0.605 ± 0.357 | 0.094 ± 0.137 | 0.157 ± 0.266 | 0.018 ± 0.049 |
| Mean deviations from ideal ± SDb | ||||
| Angles (°) | 0.71 ± 0.04 | 0.72 ± 0.05 | 0.67 ± 0.04 | 0.58 ± 0.01 |
| Bonds (Å) | 0.0052 ± 0.0003 | 0.0048 ± 0.0003 | 0.0045 ± 0.0007 | 0.0022 ± 0.0001 |
| Impropers (°) | 0.45 ± 0.03 | 0.46 ± 0.05 | 0.44 ± 0.04 | 0.40 ± 0.01 |
| RMS deviations (Å)c | ||||
| Backbone atoms (global) | >2 | >2 | >2 | >2 |
| Backbone atoms [S(φ) and S(ψ) > 0.8] | 0.93 | 1.61 | ||
| Ramachandran plotd | ||||
| Most favored (%) | 44.7 | 51.8 | 46.1 | 35.6 |
| Allowed (%) | 48.7 | 45.4 | 48.9 | 58.1 |
| Additionally allowed (%) | 6.7 | 2.9 | 5.0 | 6.2 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
The values for ENOE are calculated from a square well potential with force constants of 50 kcal mol−1 Å2.
The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
The RMSD over the backbone heavy atoms (N, Cα, C) over the indicated residues.
As determined by the program PROCHECK-NMR for all residues.
Structure calculations.
Intensities of NOE cross peaks were measured in XEASY and calibrated by using the CALIBA macro of the program CYANA (version 1.0.6) (18). NOEs providing no restraint or representing fixed distances were removed. Initial structures for J1, J3, and J7 were then calculated by using torsion angle dynamics and simulated annealing protocols in CYANA (18). Structures were optimized for a low target function comprising terms for NOE and dihedral angle violations. The final constraint set was then used to calculate a new family of 100 structures using the standard simulated annealing script supplied with Xplor-NIH (39). For J1cc structure calculations, the constraint list resulting from the CALIBA macro of CYANA was used directly in Xplor-NIH to calculate a family of 100 structures using the simulated annealing script. Distance geometry routines were not used in Xplor-NIH as the flexible regions of these peptides were found not to sample the full conformational space allowed by the restraints, resulting in regions of apparently well-ordered backbone (24). The 20 lowest energy structures were selected for J1, J1cc, J3, and J7. A box of water with a periodic boundary of 18.856 Å was built around the peptide structure, and then this was energy minimized based on NOE and dihedral restraints and the geometry of the bonds, angles, and impropers. From this set of structures, final families of 20 lowest energy structures for J1, J1cc, J3, and J7 were chosen for analysis by using PROCHECK-NMR (27) and MOLMOL (26). In all cases, the final structures had no experimental distance violations greater than 0.2 Å or dihedral angle violations greater than 5°. All structural figures were prepared by using the programs MOLMOL (26) and PyMOL (13a).
RESULTS
Peptides.
Phage display has been used to identify a series of peptides (Fig. 1) that interact with the inhibitory anti-AMA1 MAb 4G2dc1 (7). All three peptides induced rabbit antibodies that inhibited parasite invasion, with anti-J1 and anti-J7 antibodies having greater potency than anti-J3 antibodies (7).
A FASTA (http://fasta.bioch.virginia.edu/fasta_www/cgi/search_frm.cgi?pgm = fa) (35) local similarity search of the J peptides against the AMA1 sequence from the P. falciparum strain 3D7 did not detect any significant sequence similarity to a common region of AMA1, although structural considerations suggest how J1 may mimic part of the 4G2dc1 epitope of AMA1 (see below).
NMR spectroscopy.
Spectra were acquired and analyzed, and structures were determined initially for the three phage-derived peptides J1, J3, and J7. Based on the structure of J1, a disulfide bridge was introduced, its exact position being optimized by phage display (see below). Since essentially identical methods were used to analyze the NMR spectra and to determine the structures of all four peptides, they are described together in this section.
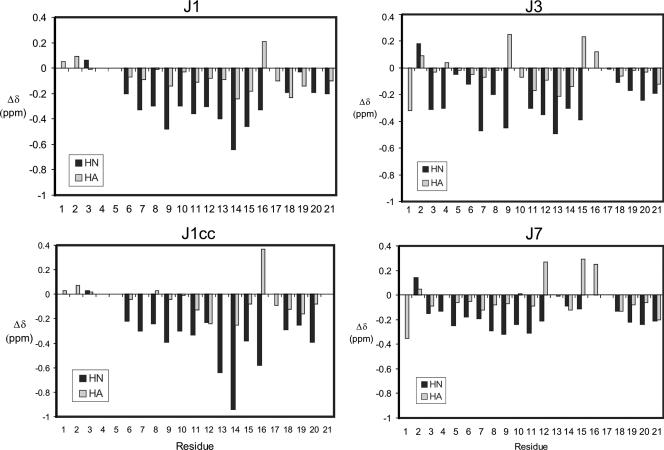
Limited chemical shift dispersion was observed for all four J peptides in aqueous solution, implying that they sample a range of conformations, as might be expected for peptides of this size that lack multiple disulfide or other cross-links. J1 and J1cc contain two Pro residues, whereas J3 and J7 each contain three. In all cases, cis-trans isomerization of the peptide bonds preceding at least one of these prolines was observed. The structural analyses described below are for the dominant trans conformation of each peptide. Minor Trp NH peaks with intensities <10% of the main peak were present in the spectrum of J1cc, possibly arising from impurities in the synthetic sample used for structure determination. 1H chemical shifts for J1, J1cc, J3, and J7 were assigned (Tables S1, S2, S3, and S4 in the supplemental material) and have been deposited in the BioMagResBank (40). All of the 1H chemical shifts were assigned except for the aromatic side chain protons of Phe1 [C(4)H] and Phe12 [C(4)H] in J3 and Phe8 [C(4)H] in J7. The deviations of the backbone NH and CαH chemical shifts from random coil values are plotted in Fig. 2. J1 shows the largest deviations around Trp11, suggesting that a well-ordered conformation may exist around this residue. The deviations for J1cc follow a pattern similar to that of J1 but are even more pronounced. There is no correlation between the deviations for J1 and J7 despite the putative consensus sequence (Fig. 1). Although there also appears to be no correlation between the deviations from random coils for J3 and J7, closer inspection reveals that the magnitude and direction of the deviations are very similar over residues 15 to 21, the consensus sequence between these two peptides.
FIG. 2.
Deviation of J peptide chemical shifts from random coil. The deviations of HN and CαH proton chemical shifts from random coil were calculated using the values of Merutka et al. (31). J1 and J1cc have been aligned to account for its two missing residues in relation to J3 and J7. For Gly residues, the two values for CαH have been averaged when necessary.
Translational diffusion coefficients of (1.09 ± 0.05) × 10−10 m2 s−1 (standard deviations [SD] over 21 resonances of the peptide), (1.54 ± 0.03) × 10−10 m2 s−1 (16 peaks), (1.07 ± 0.05) × 10−10 m2 s−1 (14 peaks), and (1.19 ± 0.05) × 10−10 m2 s−1 (13 peaks) were measured for J1, J1cc, J3 and J7, respectively, at 5°C. Diffusion coefficients of (3.48 ± 0.05) × 10−10 m2 s−1 (13 peaks) for J3 at 35°C and (1.99 ± 0.05) × 10−10 m2 s−1 (13 peaks) for J7 at 25°C were also measured. Allowing for viscosity and temperature effects, these values are similar to peptides studied by Keizer et al. (24) and Yao et al. (46), indicating that these peptides were monomeric under the solution conditions used for the present study. The higher diffusion coefficient for J1cc shows that the molecule moves more rapidly in solution compared to J1, J3, and J7, which is consistent with its more constrained structure (see below). J1 was also examined by analytical ultracentrifugation and sedimentation analysis and fitted very well to a monomeric model (data not shown).
3JHNHα coupling constants were measured from DQF-COSY spectra. Only one residue in each of J1 and J1cc had 3JHNHα values of >8 Hz, whereas several residues in each peptide had coupling constants of <6 Hz (Tables S1 to S3 in the supplemental material). These coupling constants for J1cc were from consecutive residues, Asp8 and Ile9. Initial structures were calculated by using distance and dihedral angle restraints in CYANA (18) before optimization in Xplor-NIH (39). All structures were further refined and minimized in a box of water. Summaries of the structural statistics for the final ensembles of structures are given in Table 1, and the experimental restraints are summarized in Fig. S1 in the supplemental material. For J3 and J7, the NOEs were predominantly intraresidue or sequential, with very few medium-range NOEs. In contrast, J1 and J1cc contained significantly more medium-range NOEs, particularly in the region around Trp11. There were three long-range NOEs (i − j > 4) in J1 and J1cc. No distance violations of >0.2 Å or dihedral violations of >5° were observed in the final 20 structures. In the Ramachandran plot, no residues were in the disallowed region, as determined by the program PROCHECK-NMR (27) except for Pro and Gly.
Temperature coefficients for the backbone amide protons of J1, J3, and J7 were measured to detect possible hydrogen bonds (Tables S1 to S3 in the supplemental material). Temperature coefficient magnitudes (Δδ/ΔT) of >4 ppb/K generally indicate an exposed amide proton, whereas those of <3 ppb/K indicate an amide proton that is shielded from solvent (5). In none of the three peptides were temperature coefficients of <4 ppb/K observed. However, amide exchange experiments showed that some amides were protected from solvent. After 4 h the amide protons of residues Phe6 or Asp8, Trp11 or Thr7, Ile9, Ala10, Val12, and Arg13 of J1 were still visible in a TOCSY. All of the amide protons except for those of Phe1 and Gly21 in J3 were still visible after 14 h in a TOCSY, somewhat surprisingly since this peptide does not adopt a well-defined conformation in solution (see below). In J7, resonances from Phe8, Val14, and Arg15 were visible after 4 h in a TOCSY. According to MOLMOL (26), a hydrogen bond between the amide of Trp11 and the O of Asp8 was present in 16 of the 20 structures and 14 of the 20 structures in J1 and J1cc, respectively (see below). For J3 and J7, no hydrogen bonds were present. Although the amide exchange results indicate that some backbone amide protons may be protected from solvent, no hydrogen bond restraints were used in the structure calculations because of the magnitudes of the temperature coefficients.
Structures.
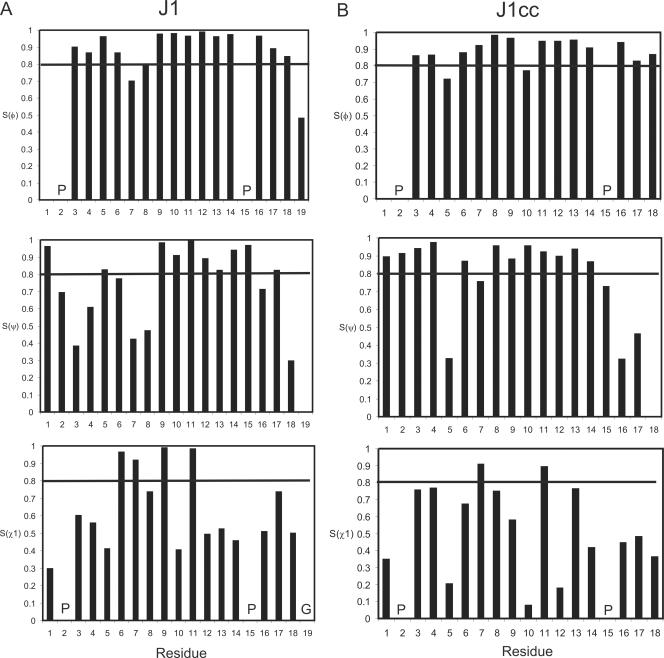
The angular order parameters (S) for the φ, ψ, and χ1 angles in the final families of structures for J1 and J1cc are plotted as a function of residue number in Fig. 3 and for J3 and J7 in Fig. S2 in the supplemental material. S values of >0.8 generally indicate that an angle is well defined within the family of structures. For J1, the φ and ψ angles were well defined, as measured by this parameter, over residues 9 to 17 with the exception of the ψ angle for Asn16, as were the side chain χ1 angles for Phe6, Thr7, Ile9, and Trp11. The backbone φ and ψ angles for J1cc were also well defined over the majority of residues, the exception being Ala5. Like J1, the side chain χ1 angles for Thr7 and Trp11 were ordered. The φ and ψ angles for J3 and J7 were mostly disordered, but in J7, the side chain χ1 angles for Asp10, Thr11, and Val14 were well defined.
FIG. 3.
Angular order parameters for J1 and J1cc. (A) J1 Sφ, SΨ, Sχ1. (B) J1cc Sφ, SΨ, Sχ1. Horizontal lines represent the cutoff for regions that are considered to be well ordered (S > 0.8). S values were measured in the program MOLMOL (26).
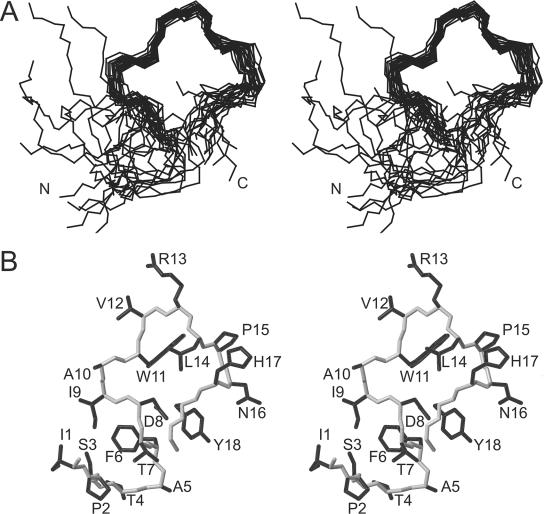
J1 appears to adopt a stable conformation over residues 9 to 17, as reflected in the φ and ψ angular order parameters and NOE patterns. This region of structure is supported by the many contacts from Asp8, Ile9, Ala10, Val12, Arg13, and Leu14 to the aromatic side chain of Trp11. In addition, J1 contains three long-range (i − j > 4) NOEs that involve residues in this region. The pairwise root mean square deviation (RMSD) over the backbone heavy atoms (N, Cα, and C) of Ile9 to His17 was 0.93 Å compared to a value of >2 Å globally. Residues Asp8-Trp11 are involved in a type I β-turn as determined by their backbone φ and ψ angles, with Ile9 and Ala10 occupying the i+1 and i+2 positions (Fig. 4). Asp at position i in a type I turn is highly favored because it potentially acts as a hydrogen bond acceptor (22). Analysis of structures predicted a hydrogen bond between the amide of Trp11 and the O of Asp8 in 16 of 20 structures, a finding consistent with the presence of such a turn. Peptides that are not constrained by disulfide or other covalent links generally sample a range of conformations in solution. As an alternate measure of how well-defined the J1 structures were, the Karplus equation (29) was used to back-calculate 3JHNHα coupling constants from the average φ angles in the final family of structures for J1. For His17, the calculated 3JHNHα coupling constant was >8 Hz, which was in good agreement with the experimental data (Table S1 in the supplemental material). There were variations in the range of coupling constants for Ala5, Ala10, and Leu14, with calculated 3JHNHα between 6 and 8 Hz compared to experimental values of <6 Hz. Thr7, Asp8, Ile9, Val12, and Arg13 had experimental 3JHNHα coupling constants between 6 and 8 Hz, but the calculated values were <6 Hz for Thr7, Asp8, and Ile9 and >8 Hz for Val12 and Arg13.
FIG. 4.
Structure of J1. (A) Stereo view of the J1 family of 20 lowest-energy structures. Structures were superimposed over the backbone heavy atoms (N, Cα, and C) of residues 9 to 17, as determined by their well-defined angular order parameters. (B) Stereo view of the closest-to-average structure in the family, showing all side chains in black and the backbone in gray.
The relatively compact structure found for J1 gave rise to the possibility that a covalently stabilized analogue of this peptide could be designed. The RCSB Protein Data Bank (6) was scanned for Cα-Cα distances across disulfides of various peptides and proteins to see where this distance could be satisfied in J1. This showed that cysteines could be introduced at positions 6 or 7 and 16 or 17 and linked to create a disulfide bridge that should stabilize the structure centered on the β-turn in J1. We therefore took a combinatorial approach to establishing which cysteine pairing was most appropriate. The phage clone selected as a result of this process had cysteine substitutions at positions 6 (Phe to Cys) and 16 (Asn to Cys).
A peptide, J1cc, was then synthesized with cysteines inserted at positions 6 and 16, and its solution structure was determined. The structure was ordered over the disulfide-bonded region, from positions 6 to 16, with a pairwise RMSD over the backbone atoms of 1.61 Å. PROCHECK revealed some secondary structure, a 310 helix between Ile9-Trp11 in 11 of 20 structures. As in J1, MOLMOL analysis of the final J1cc structures predicted a hydrogen bond between the amide of Trp11 and the O of Asp8 in 14 of 20 structures. The 3JHNHα coupling constants were also back-calculated from the Karplus equation for J1cc. For Asp8, Ile9, and Cys16 the calculated values were in good agreement with the experimentally observed 3JHNHα (Table S2 in the supplemental material). Asp8 and Ile9 had calculated values of <6 Hz, and Cys16 had calculated values of >8 Hz. Ala5, Ala10, and Val12 had calculated 3JHNHα coupling constants of < 6 Hz, and Trp11 and Leu14 had calculated 3JHNHα coupling constants of >8 Hz.
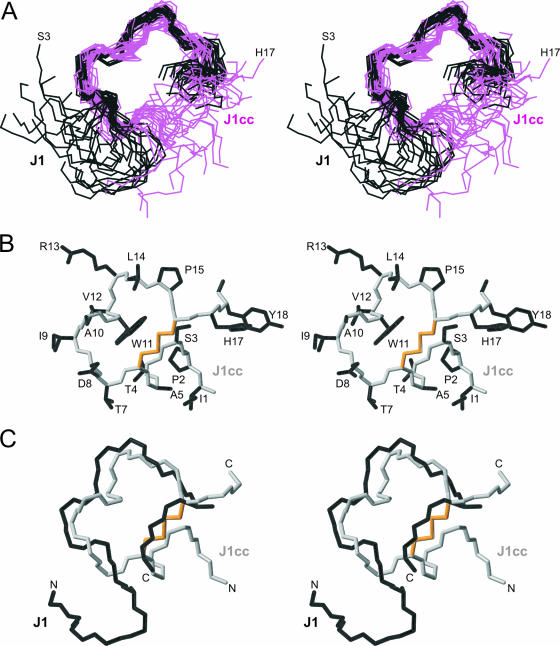
The structures for J1 and J1cc were similar, as depicted by the superposition of the families of structures with a pairwise RMSD of 1.64 Å between residues Thr7 and Pro15 (Fig. 5). It appears that the conformation of J1cc was constrained successfully, since there was no disruption in the region of local structure surrounding Trp11.
FIG. 5.
Structure of J1cc. (A) Stereo view of the superposition of the J1 and J1cc families of structures showing residues Ser3 to His17. Structures were superimposed over the backbone heavy atoms (N, Cα, C) of residues 7 to 15. J1 is shown in black; J1cc is shown in magenta. (B) Stereo view of the closest-to-average structure in the family, showing all side chains in black, the backbone in gray, and the disulfide bond (from positions 6 to 16) in orange. (C) Stereo view of a superposition of the backbones of J1cc (gray, with disulfide in orange) and J1 (black).
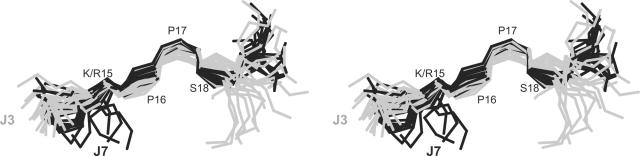
In contrast to J1, neither J3 nor J7 adopted a stable conformation over the majority of the residues. This is reflected in global backbone RMSD values of >2 Å and a lack of medium range NOEs observed for each of these peptides. Although one must be cautious in attempting to define segments of local structure in flexible peptides, it appears that both J3 and J7 contain a small region of well-defined structure over the consensus sequence from residues 15 to 18, as shown in Fig. 6. Indeed, when the J3 and J7 families of structures were superimposed over this consensus sequence, a pairwise backbone RMSD of 0.57 Å was obtained.
FIG. 6.
Stereo view of the superposition of the J3 and J7 families of structures over the backbone heavy atoms (N, Cα, and C) of residues 15 to 18, the consensus sequence. Residues 14 to 19 are shown with J3 in gray and J7 in black.
BLAST (http://www.ebi.ac.uk/blast2/index.html) (1) and FASTA (35) searches of the J peptides against the Swiss-Prot database identified several proteins with sequence similarity to these peptides for which structures are currently available in the RCSB Protein Data Bank (6). These included, for J1, an 11-residue segment from residues 76 to 86 in 6-phospho-β-galactosidase (45) and, for J7, residues 9 to 12 of a high-mobility-group-1 protein (44) and 43 to 51 of neurotoxin 6 from the Indian cobra (PDB 1LN9). It appears that J1 does not share a common structural motif with 6-phospho-β-galactosidase, but when J7 and the high-mobility-group protein were superimposed over their identical residues, RPPS, the pairwise backbone RMSD was 0.82 Å. For the neurotoxin model, the pairwise backbone RMSD was 2.19 Å from residues 9 to 17 of J7 and from residues 43 to 51 of neurotoxin 6 (see Fig. S3 in the supplemental material).
Characterization of J1 and J1cc.
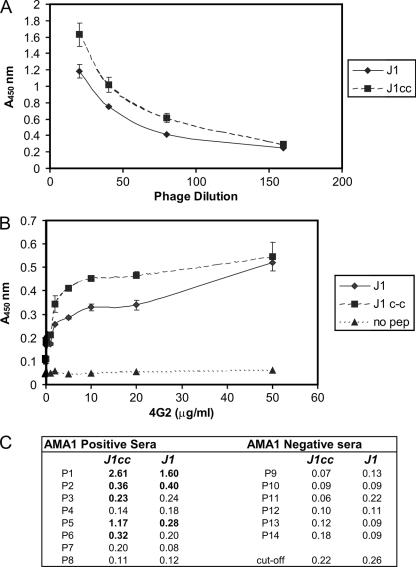
The relative binding of phage expressing J1 and J1cc was compared by ELISA (Fig. 7A). The data show an ∼1.5-fold improved binding of the J1cc phage compared to the wild-type J1 at equivalent expressed phage concentrations. Synthetic peptides J1 and J1cc were coupled to microtiter plates and found to bind to the parent MAb 4G2dc1 (Fig. 7B). It appears that there was a fivefold increase in the binding of the J1cc peptide in the linear region of the binding curve. The constrained peptide thus has a higher relative affinity than the original J1 mimotope.
FIG. 7.
J1cc has a higher relative affinity than J1. (A) ELISA showing that J1cc phage binds with a slightly higher relative affinity than J1 phage to the parent MAb 4G2dc1. The assay was performed at equivalent phage reactivity to the anti-Myc epitope tag (9) (data not shown) and repeated to ensure the data were consistent. The datum points indicate the means of duplicate wells, and error bars indicate one SD from the mean. (B) Reactivity of J1 and J1cc peptides with 4G2dc1. An ELISA was performed showing the 4G2dc1 MAb binding in a dose-dependent manner to J1 and J1cc peptides coated onto microtiter plates. The datum points indicate the means of duplicate wells, and error bars indicate one SD from the mean. (C) Detection by J1 and J1cc peptide mimotopes of IgG antibodies in human plasma samples with a high titer of AMA1 antibodies (1-8) and a low titer of AMA1 antibodies (9-14) from individuals living in an area of PNG where malaria is endemic. Samples were assayed by ELISA, and the mean absorbance level was tabulated. The cutoff level is defined as the mean of the negative population plus three SD. A positive result (shown in boldface) was defined as an absorbance reading greater than the cutoff level.
Fourteen human plasma samples from PNG blood donors were analyzed for reactivity to J1cc and J1. The AMA1 reactivity had been assessed previously, and the samples were grouped into positive and negative. J1cc detected antibodies in five of eight AMA1-positive samples, whereas J1 detected antibodies in three of eight positive samples (Fig. 7C). There were no false positives; neither J1cc nor J1 detected antibodies in the AMA1-negative samples, indicating the high specificity of the J1 epitope mimic. J1cc therefore appears to be superior to J1 for detecting antibodies in these human plasma samples. The peptides could not recognize antibodies in all of the AMA1-positive samples, and antibody titers were generally low since the peptides can only recognize antibodies to the single 4G2dc1 inhibitory epitope of AMA1.
DISCUSSION
Three peptide mimotopes of AMA1 that bind to the inhibitory MAb 4G2dc1 have been investigated by NMR in the present study in order to probe their conformations in solution. The mimotope with the highest affinity for 4G2dc1, J1, adopts a well-defined structure over residues Ile9-His17. This region involves Trp11, whose side chain forms the nucleus of a hydrophobic cluster involving the side chains of Ile9, Ala10, Val12, Arg13, and Leu14. Indeed, J1 contains a type I β-turn structure from Asp8-Trp 11, with a hydrogen bond between the amide of Trp11 and the carbonyl oxygen of Asp8 in 16 of the 20 structures. J3 and J7 do not adopt well-defined structures in aqueous solution, with only the residues in the consensus sequence, from positions 15 to 18, showing any evidence of stable structure. Inserting a tryptophan in J3 or J7 may give rise to a more ordered conformation in solution and enhance binding to 4G2dc1.
As the three J peptides mimic 4G2dc1 binding epitopes on AMA1, it was of interest to compare their sequences with that of AMA1. The three peptides show limited sequence similarity to three different regions of AMA1, predominantly in domain I. For J3 and J7, these regions of similarity occurred within domain I of AMA1, while for J1, the region of similarity overlapped the proteolytic cleavage site for production of the mature protein (21). J1 has five identical and five conservative residues in common with residues 94 to 107 of AMA1, whereas J3 has four identical and seven conservative residues with residues 134 to 148, and J7 six identical and one conservative residue with residues 168 to 184. However, the fact that 4G2dc1 binds only to native AMA1 and not to the reduced and alkylated antigen indicates that we are dealing with a conformational epitope with contributions from more than one region of the AMA1 sequence, and it is therefore unlikely that these similarities to linear segments of AMA1 are significant.
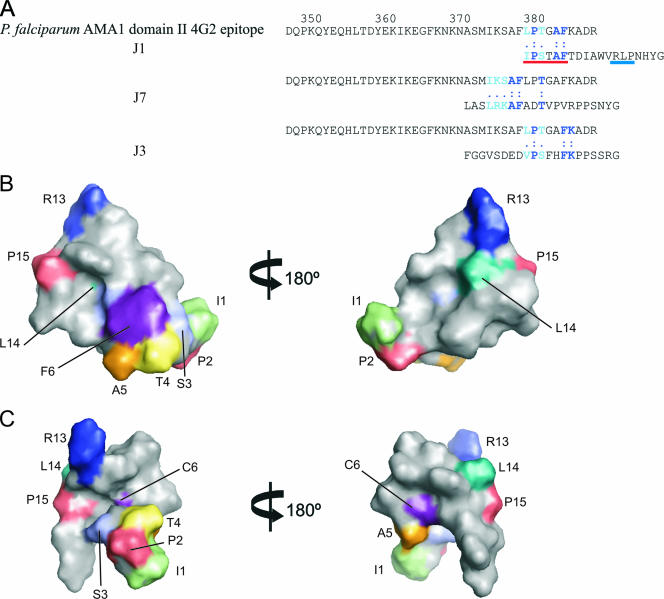
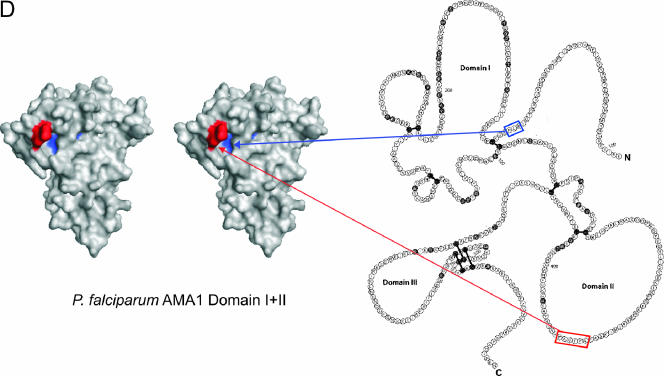
None of these sequence similarities correlates with the 4G2dc1 epitope on AMA1, which appears to be contained largely in the flexible loop region of P. falciparum AMA1 domain II between residues 348 to 389 (38). However, residues 1 to 6 at the N terminus of J1 (IPSTAF) show homology with residues 380 to 385 (LPTGAF) in domain II of AMA1, and residues 13 to 15 at the C terminus of J1 (RLP) are identical to residues 143 to 145 (RLP) in domain I of AMA1 (Fig. 8). Our structure of J1 shows that these two regions of sequence are close together in space. The two regions of AMA1 with similarity to J1 are far apart in the primary sequence and reside in different domains of the protein. Remarkably, however, when mapped onto the crystal structure of P. falciparum AMA1 domains I and II published recently (3), these regions are precisely juxtaposed, as shown in Fig. 8D. Moreover, although P. falciparum AMA1 residues 380 to 385 (LPTGAF) are not visible in the crystal structure of the ectodomain of P. vivax AMA1 because they are present on a flexible loop of domain II whose structure could not be resolved (15, 38), the closest discernible residues are indeed juxtaposed to the RLP-containing sequence in the P. vivax structure. These data support the view that the J1 peptide is acting as a scaffold to hold two regions in the appropriate spatial orientation for efficient binding of 4G2dc1. Thus, J1 appears to be mimicking structural features present on the surface of AMA1. In their mapping of the 4G2dc1 epitope, Pizzaro et al. (38) found that F385 was a key contributor, with mutation at that position abrogating 4G2dc1 binding.
FIG.8.
Possible relationship between the structure of J1 and the 4G2dc1 epitope on P. falciparum AMA1. (A) Sequence alignment of the 4G2dc1 epitope on P. falciparum AMA1 domain II and J1, J7, and J3. (B and C) Surface representations of J1 (B) and J1cc (C), highlighting the RLP motif and homology with residues in the 4G2dc1 epitope on P. falciparum AMA1 domain II. (D) Stereo view of the surface representation of the crystal structure of P. falciparum AMA1 domain I+II (3) (left) and a schematic of P. falciparum AMA1 (19) (right) highlighting the similar residues between J1 and AMA1, as shown in panel A. Residues are colored in PyMOL as follows: blue, RLP in domain I; and red, LPTGAF in the 4G2dc1 epitope in domain II.
Since J1 is the most structured of the peptides, we created a conformationally stabilized analogue by inserting a disulfide link with the aim of reducing flexibility and increasing the potency of the 4G2dc1 mimotope. Because of the constrained nature of cyclic peptides, receptor binding affinities often become more favorable (28). Cyclic peptides can be exceptional mimotopes because they have the entropic benefit of binding antibody with their structure already constrained (17, 23, 41). Cysteine residues were inserted in J1, in place of Phe6 and Asn16, such that the disulfide bond would not affect Trp11. Introduction of this disulfide succeeded in stabilizing the mean structure found for the parent J1 peptide in solution and strengthened its binding to 4G2dc1. This suggests that the solution structure resembles the structure of antibody-bound J1. Importantly, it also supports the hypothesis that the proximity of the N-terminal region of J1 and the RLP sequence creates the epitope in J1 that mimics part of the 4G2dc1 epitope on P. falciparum AMA1.
As noted above, J1, J3, and J7 do not share a consensus sequence or structural motif. J1 and J7 share a common sequence motif, AFXDXXXVRXPXXY, which correlates with both peptides eliciting antibody responses in rabbits. Asp is a shared residue, which appears in both of the well-defined regions in J1 and J7; Asp8 in J1 aligns with Asp10 in J7. When the final 20 structures for J7 were superimposed, local structure was evident from Ala7-Thr11 with a pairwise RMSD of 1.19 Å over the backbone atoms. It is possible that Asp8 may be significant for peptide activity and that mutation of Asp8 to Ala could impair binding to 4G2dc1.
It is unlikely that the epitope in either J1 or its disulfide-stabilized analogue exactly mimics the 4G2dc1 epitope on P. falciparum AMA1 in terms of structure or side chain chemistry. Nonetheless, J1cc has a higher affinity for 4G2dc1 than J1 and is likely to be a more useful mimotope than the parent peptide. We have also shown that J1cc has improved characteristics as an in vitro correlate of immunity, with the ability to detect antibodies to the 4G2dc1 epitope in the plasma of individuals exposed to malaria. Further studies could involve immunizing with the constrained peptide to produce antibodies and performing invasion inhibition studies with P. falciparum parasite cultures to assess whether J1cc is in fact a more potent peptide than J1. We know that the 4G2dcl epitope of AMA1 is important, since this MAb is one of only two characterized anti-AMA1 MAbs to date that inhibit merozoite invasion. The 4G2dc1 epitope is also present in 10 different isolates of P. falciparum AMA1. In addition, the J series of mimotopes can recognize antibodies in the plasma of individuals exposed to malaria in endemic regions of PNG, indicating the usefulness of these peptides as potential in vitro correlates of immunity. The results presented here indicate that J1cc, with its stabilized structure and tighter binding, may prove to be a more useful second-generation 4G2dc1 mimotope than J1.
Supplementary Material
Acknowledgments
We thank Geoff Howlett for performing analytical ultracentrifugation analyses and Robin Anders, Mark Hinds, and Shenggen Yao for valuable discussions.
This study was supported in part by grants from the Australian National Health and Medical Research Council.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 23 October 2006.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunization with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 3.Bai, T., M. Becker, A. Gupta, P. Strike, V. J. Murphy, R. F. Anders, and A. H. Batchelor. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. USA 102:12736-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartels, C., T. H. Xia, M. Billeter, P. Güntert, and K. Wüthrich. 1995. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR 6:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, N. J., and M. P. Williamson. 1997. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 9:359-369. [DOI] [PubMed] [Google Scholar]
- 6.Berman, H. M., T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne, K. Burkhardt, Z. Feng, G. L. Gilliland, L. Iype, S. Jain, P. Fagan, J. Marvin, D. Padilla, V. Ravichandran, B. Schneider, N. Thanki, H. Weissig, J. D. Westbrook, and C. Zardecki. 2002. The protein data bank. Acta Crystallogr. D. Biol. Crystallogr. 58:899-907. [DOI] [PubMed] [Google Scholar]
- 7.Casey, J. L., A. M. Coley, R. F. Anders, V. J. Murphy, K. S. Humberstone, A. W. Thomas, and M. Foley. 2004. Antibodies to malaria peptide mimics inhibit Plasmodium falciparum invasion of erythrocytes. Infect. Immun. 72:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis, C. E., and M. J. Blackman. 2000. Host cell invasion by malaria parasites. Parasitol. Today 16:411-415. [DOI] [PubMed] [Google Scholar]
- 9.Coley, A. M., K. Parisi, R. Masciantonio, J. Hoeck, J. L. Casey, V. J. Murphy, K. S. Harris, A. H. Batchelor, R. F. Anders, and M. Foley. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 74:2628-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, and J. S. Sullivan. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 11.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deans, J. A., T. Alderson, A. W. Thomas, G. H. Mitchell, E. S. Lennox, and S. Cohen. 1982. Rat monoclonal antibodies which inhibit the in vitro multiplication of Plasmodium knowlesi. Clin. Exp. Immunol. 49:297-309. [PMC free article] [PubMed] [Google Scholar]
- 13.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66-kDa merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 13a.Delano, W. L. 2002. The PyMOL Molecular Graphics System. Delano Scientific, San Carlos, CA. [Online.] http://www.pymol.org.
- 14.Dingley, A. J., J. P. Mackay, B. E. Chapman, M. B. Morris, P. W. Kuchel, B. D. Hambly, and G. F. King. 1995. Measuring protein self-association using pulsed-field-gradient NMR spectroscopy: application to myosin light chain 2. J. Biomol. NMR 6:321-328. [DOI] [PubMed] [Google Scholar]
- 15.Feng, Z. P., D. W. Keizer, R. A. Stevenson, S. Yao, J. J. Babon, V. J. Murphy, R. F. Anders, and R. S. Norton. 2005. Structure and inter-domain interactions of domain II from the blood-stage malarial protein, apical membrane antigen 1. J. Mol. Biol. 350:641-656. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs, S. J., and C. S. Johnson. 1991. A PEG NMR experiment for accurate diffusion and flow studies in the presence of eddy currents. J. Magn. Reson. 93:395-402. [Google Scholar]
- 17.Giebel, L. B., R. T. Cass, D. L. Milligan, D. C. Young, R. Arze, and C. R. Johnson. 1995. Screening of cyclic peptide phage libraries identifies ligands that bind streptavidin with high affinities. Biochemistry 34:15430-15435. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209-227. [DOI] [PubMed] [Google Scholar]
- 19.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 20.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 21.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 22.Hutchinson, E. G., and J. M. Thornton. 1994. A revised set of potentials for β-turn formation in proteins. Protein Sci. 3:2207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz, B. A. 1995. Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry 34:15421-15429. [DOI] [PubMed] [Google Scholar]
- 24.Keizer, D. W., L. A. Miles, F. Li, M. Nair, R. F. Anders, A. M. Coley, M. Foley, and R. S. Norton. 2003. Structures of phage-display peptides that bind to the malarial surface protein, apical membrane antigen 1, and block erythrocyte invasion. Biochemistry 42:9915-9923. [DOI] [PubMed] [Google Scholar]
- 25.Kocken, C. H., E. Hundt, B. Knapp, D. Brazel, B. Enders, D. L. Narum, J. A. Wubben, and A. W. Thomas. 1998. Immunization of Aotus monkeys with recombinant Plasmodium falciparum hybrid proteins does not reproducibly result in protection from malaria infection. Infect. Immun. 66:373-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph 14:29-32; 51-55. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 28.Li, P., and P. P. Roller. 2002. Cyclization strategies in peptide derived drug design. Curr. Top. Med. Chem. 2:325-341. [DOI] [PubMed] [Google Scholar]
- 29.Ludvigsen, S., K. V. Andersen, and F. M. Poulsen. 1991. Accurate measurements of coupling constants from two-dimensional nuclear magnetic resonance spectra of proteins and determination of ϕ-angles. J. Mol. Biol. 217:731-736. [DOI] [PubMed] [Google Scholar]
- 30.Ludvigsen, S., and F. M. Poulsen. 1992. Positive ϕ-angles in proteins by nuclear magnetic resonance spectroscopy. J. Biomol. NMR 2:227-233. [DOI] [PubMed] [Google Scholar]
- 31.Merutka, G., H. J. Dyson, and P. E. Wright. 1995. “Random coil” 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J. Biomol. NMR 5:14-24. [DOI] [PubMed] [Google Scholar]
- 32.Mital, J., M. Meissner, D. Soldati, and G. E. Ward. 2005. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol. Biol. Cell 16:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, W. R. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132:185-219. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piotto, M., V. Saudek, and V. Sklenář. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2:661-665. [DOI] [PubMed] [Google Scholar]
- 38.Pizarro, J. C., B. Vulliez-Le Normand, M. L. Chesne-Seck, C. R. Collins, C. Withers-Martinez, F. Hackett, M. J. Blackman, B. W. Faber, E. J. Remarque, C. H. Kocken, A. W. Thomas, and G. A. Bentley. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308:408-411. [DOI] [PubMed] [Google Scholar]
- 39.Schwieters, C. D., J. J. Kuszewski, N. Tjandra, and G. M. Clore. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65-73. [DOI] [PubMed] [Google Scholar]
- 40.Seavey, B. R., E. A. Farr, W. M. Westler, and J. L. Markley. 1991. A relational database for sequence-specific protein NMR data. J. Biomol. NMR 1:217-236. [DOI] [PubMed] [Google Scholar]
- 41.Sem, D. S., B. L. Baker, E. J. Victoria, D. S. Jones, D. Marquis, L. Yu, J. Parks, and S. M. Coutts. 1998. Structural characterization and optimization of antibody-selected phage library mimotopes of an antigen associated with autoimmune recurrent thrombosis. Biochemistry 37:16069-16081. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, A. W., J. A. Deans, G. H. Mitchell, T. Alderson, and S. Cohen. 1984. The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol. 13:187-199. [DOI] [PubMed] [Google Scholar]
- 43.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 44.Weir, H. M., P. J. Kraulis, C. S. Hill, A. R. Raine, E. D. Laue, and J. O. Thomas. 1993. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 12:1311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiesmann, C., W. Hengstenberg, and G. E. Schulz. 1997. Crystal structures and mechanism of 6-phospho-β-galactosidase from Lactococcus lactis. J. Mol. Biol. 269:851-860. [DOI] [PubMed] [Google Scholar]
- 46.Yao, S., G. J. Howlett, and R. S. Norton. 2000. Peptide self-association in aqueous trifluoroethanol monitored by pulsed field gradient NMR diffusion measurements. J. Biomol. NMR 16:109-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.