Abstract
The progression of Legionella pneumophila infection in macrophages is controlled by the Lgn1 gene locus, which expresses the nonpermissive phenotype in cells from BALB/c mice but the permissive phenotype in cells from A/J mice. Activation of dendritic cells and macrophages by L. pneumophila is mediated by the pathogen recognition receptor Toll-like receptor 2 (TLR2); furthermore, Legionella induces innate and adaptive immune cytokines by the MyD88-dependent pathway. TLR9 is coupled to MyD88 and mediates the production of interleukin-12 (IL-12) in dendritic cells infected with other facultatively intracellular pathogens. In the current study, L. pneumophila growth in dendritic cells from BALB/c and A/J mice was examined along with the role of TLR9 in the induction of IL-12 in these cells. Dendritic cells from both strains were nonpermissive for L. pneumophila intracellular growth, suggesting that the products of the Lgn1 gene locus that control intracellular growth in macrophages do not control the growth of Legionella in dendritic cells. In addition, chloroquine treatment suppressed IL-12 p40 production in response to Legionella treatment in dendritic cells and macrophages from BALB/c and A/J mice. Furthermore, the TLR9 inhibitor ODN2088 suppressed the Legionella-induced IL-12 production in dendritic cells from both mouse strains. These results suggest that L. pneumophila is similar to other intracellular bacteria in that it stimulates the production of immune-transitioning cytokines, such as IL-12, through activation of TLR9 and that this receptor provides a common mechanism for sensing these types of microbes and inducing innate and adaptive immunity.
Legionella pneumophila is the causative agent of Legionnaires' disease and Pontiac fever (26) and replicates intracellularly by evading phagosome-lysosome fusion (4) within human monocytes/macrophages (22) and in permissive macrophages from A/J mice (43). This ability to replicate within A/J macrophages is due to allelic variation in a single gene on chromosome 13 called Naip5 (Birc1e) (neuronal apoptosis inhibitory protein 5, also known as Birc1e, for baculovirus inhibitor of apoptosis repeat-containing protein 1) (12, 15), located within the Lgn1 gene locus (44). Following sublethal infection with L. pneumophila, BALB/c mice develop Th1 adaptive immunity, complete with antigen-specific gamma interferon production by T cells and anti-L. pneumophila immunoglobulin G2a (IgG2a) antibodies, which protect them following a secondary challenge infection (36). Protection from primary infection in the mouse, however, results from innate immunity rather than adaptive immunity and does not require Th1 responses (28).
Innate immune responses are mediated by pattern recognition receptors that recognize pathogen-associated patterns (3). Toll-like receptors (TLRs) serve as pattern recognition receptors for mammals and respond to various pathogen-associated antigens (3, 42). For example, TLR4 responds to lipopolysaccharides (LPS) (14, 37), and TLR2 responds to microbial peptidoglycans, lipoproteins, and lipopeptides (42). Studies with L. pneumophila have shown that LPS and viable organisms function through TLR2, not TLR4 (2, 7, 16). TLR5 is activated by flagellin protein (20), and in humans a common polymorphism in the TLR5 gene causes a deficiency in mediating signals from flagellin and increased susceptibility to Legionnaires' disease (19). TLR9 is activated by DNA or synthetic oligonucleotides, such as murine ODN1826, that contain unmethylated CpG (21), and the activity of these receptors can be blocked by inhibitory ligands, such as murine ODN2088 (41), or by treatment with the antimalarial chloroquine, which through its buffering capacity interferes with acidification or maturation of the endosomes (39, 46). Chloroquine has been reported to suppress the antigen-presenting function of L. pneumophila-infected macrophages due to neutralization of endocytic compartments (8, 17, 33). Regarding dendritic cells (DCs), L. pneumophila is internalized by these cells, similar to case for macrophages, but the bacterium does not grow intracellularly (34). In addition, infection induces interleukin-12 (IL-12) in DCs (27, 30); however, the TLR involved has not been determined. In the current study, we demonstrate the requirement for the TLR9 pathway in IL-12 p40 production by bone marrow-derived DCs and macrophages infected with L. pneumophila.
MATERIALS AND METHODS
Mice.
Female BALB/c (National Cancer Institute-Harlan, Frederick, MD) and A/J (Jackson Laboratory, Bar Harbor, ME) mice were used at 8 to 12 weeks of age. The mice were housed and cared for in the animal facility of University of South Florida Health Sciences Center, which is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Bacteria.
Legionella pneumophila M124, a serogroup 1 isolate from a case of Legionnaires' disease (Tampa General Hospital, Tampa, FL), was grown on buffered charcoal yeast extract (BD-Difco, Sparks, MD) plates for 48 h from a passage 3 stock stored at −80°C. For L. pneumophila infection, DCs and macrophages were centrifuged, resuspended in medium (0.5 ml), and infected at a 10:1 ratio (bacteria to cells) for 30 min at 37°C in a CO2 incubator. Following the 30 min of uptake, cells were washed twice to remove noninternalized bacteria.
Bone marrow-derived dendritic cells and macrophages.
The leg bones were removed, cleaned, and sterilized. The bone marrow was flushed from femurs and tibias by use of a syringe containing culture medium. For DC isolation, the bone marrow cells were washed and cultured overnight with RPMI 1640 medium (Sigma, St. Louis, MO) containing 10% bovine growth serum (HyClone, Logan, UT) plus granulocyte-macrophage colony-stimulating factor (GM-CSF [5 ng/ml]; BD Pharmingen, San Diego, CA), antibiotic/antimycotic solution, and 2-mercaptoethanol (0.05%; Sigma). On the following day, nonadherent cells were discarded, fresh GM-CSF (10 ng/ml)-containing medium was added to the adherent population, and the cells were cultured for an additional 7 to 9 days. Bone marrow-derived macrophages were generated using L929 conditioned (30%) medium containing M-CSF in place of GM-CSF, following a previously described protocol (9). These macrophages were examined by flow cytometric analysis and found to be >95% positive for CD11b, >85% positive for F4/80, and <5% positive for CD11c (data not shown).
RT-PCR.
RNAs were extracted with TRI reagent (Sigma) from 3-h-infected DCs as previously described (35), and DNA was removed with a DNA-free kit from Ambion (Austin, TX). Reverse transcription (RT) of RNAs was performed with avian myeloblastosis virus reverse transcriptase (Promega, San Diego, CA), and the cDNA products were PCR amplified as previously described, using a MasterGradient thermocycler (Eppendorf, Westbury, NY) (40). The gene products were amplified using Taq DNA polymerase from Takara Mirus Bio Corporation (Madison, WI) and primer pairs specific for β-actin, IL-12 p40 (40), and TLR9 (10). PCR products for β-actin and IL-12 p40 were amplified by duplex reactions, i.e., both sets of primers were run in the same reaction mix. All products were visualized with ethidium bromide in a 2% agarose gel.
Flow cytometry analysis.
ODN1826-fluorescein isothiocyanate (ODN1826-FITC) and ODN2088 (InvivoGen, San Diego, CA), an inhibitor of TLR9 (41), were used to detect intracellular TLR9 by flow cytometry. Cells were pretreated without or with ODN2088 (5 μM) for 0.5 to 3 h, followed by treatment with ODN1826-FITC (0.5 μM) for 3 h. The cells were centrifuged, and Fc Block (BD-Pharmingen) was added for 5 min, followed by the addition of anti-CD11c-allophycocyanin and anti-CD11b-phycoerythrin for 30 min at 4°C. Cells were then washed and fixed in 1% paraformaldehyde. The percentage of positive cells was determined with a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) equipped with lasers tuned to 488 nm and 635 nm.
In vitro cultures.
The cells were incubated in 5% fetal calf serum (HyClone)-RPMI plus 2-mercaptoethanol at a concentration of 106 cells/ml unless stated otherwise. For chloroquine treatment, the drug (100 μM; MP Biomedicals, Aurora, OH) was added for 30 min prior to stimulation with ODN1826 (25) (0.5 μM; InvivoGen), Escherichia coli O111:B4 LPS (100 ng/ml; Sigma), or formalin-killed L. pneumophila cells (107 bacteria/ml) or, as previously stated, prior to infection with L. pneumophila at a 10:1 ratio. In some experiments, ODN2088 (5 to 10 μM; InvivoGen), an inhibitor of TLR9 (41), was added 0.5 to 3 h prior to ODN1826 or formalin-killed L. pneumophila or prior to infection with L. pneumophila. All supernatants were collected at 24 h of culture and analyzed for IL-12 p40/70 by an enzyme-linked immunosorbent assay (ELISA).
Cytokine ELISA.
Supernatants and standards were tested for IL-12 p40 activity by sandwich ELISAs using antibody pairs from BD-Pharmingen (San Diego, CA) following previously described protocols (29). The plates were read at 450 nm on an Emax microplate reader (Molecular Devices, Menlo Park, CA), and units were calculated from the standard curve performed for each plate. The low-end sensitivity was 100 pg/ml for IL-12 p40.
CFU determination.
DC and macrophage cultures were infected with L. pneumophila at a 10:1 ratio (bacteria to cells) for 30 min at 37°C and washed to remove noninternalized bacteria. Uptake by all cell groups was similar, ranging from 1 × 104 to 3 × 104 CFU per culture. The infected cultures were incubated for 0 to 48 h and centrifuged prior to being lysed with 0.2% saponin. The lysates were diluted and incubated on buffered charcoal yeast extract plates. Bacterial CFU were determined on an AutoCount counter (Dynatech Labs, Chantilly, VA).
Statistical analysis.
Data were analyzed by one-way analysis of variance with Dunnett's test for comparing groups, using SigmaStat (Jandel Scientific, San Rafael, CA).
RESULTS AND DISCUSSION
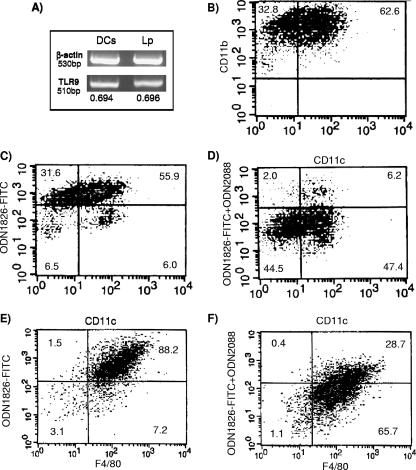
DCs are known to be important in sensing and responding to microbial products and are vital for induction of innate and adaptive immunity (24). DCs have been reported to respond to TLR9 ligands with the production of IL-12, and this production is augmented by costimulation of the cells with ligands for other TLRs, such as LPS (32). To characterize the expression of TLR9 in DCs and macrophages from BALB/c and A/J mice, bone marrow cells were cultured with GM-CSF- or M-CSF-containing L929 supernatant and analyzed for various markers by flow cytometry and RT-PCR. Figure 1A shows a robust expression of TLR9 mRNA by DCs from BALB/c mice which was not attenuated by L. pneumophila infection. Figure 1B shows that these cells were also over 95% positive for CD11b and 62% positive for CD11c, as determined by flow cytometry, and that the CD11c cells were approximately 90% positive for TLR9, as determined by specific binding of the TLR9 ligand ODN1826-FITC (Fig. 1C and D). Macrophages from BALB/c mice were >95% positive for the F4/80 marker and were approximately 90% positive for TLR9, as determined by specific ligand binding (Fig. 1E and F). Similar results were obtained with DC- and macrophage-enriched populations isolated from A/J mice: the mean fluorescence intensities (± standard errors of the means [SEM]) of the two populations were 454.7 ± 62 and 672.3 ± 83, respectively, for mice treated with ODN1826-FITC and 122.3 ± 24 and 139.3 ± 30, respectively, for mice treated with ODN1826-FITC and the ODN 2088 inhibitor. The fluorescence intensity was significantly decreased by TLR9 inhibitor treatment.
FIG. 1.
TLR9 expression in BALB/c bone marrow cells incubated with either GM-CSF (A to D) or L929 conditioned medium containing M-CSF (E and F). (A) RT-PCR for TLR9 mRNA in GM-CSF-matured cells uninfected or infected for 2 h with L. pneumophila. (B) Flow cytometry results for GM-CSF-matured cells analyzed for CD11b and CD11c. (C and D) Flow cytometry results for GM-CSF-matured cells analyzed for CD11c and the binding of the TLR9 ligand ODN1826-FITC in either the absence (C) or presence (D) of the ligand inhibitor ODN2088. (E and F) Flow cytometry results for L929 supernatant-matured cells analyzed for F4/80 and the binding of the TLR9 ligand ODN1826-FITC in either the absence (E) or presence (F) of the ligand inhibitor ODN2088. Data are representative of three repeats.
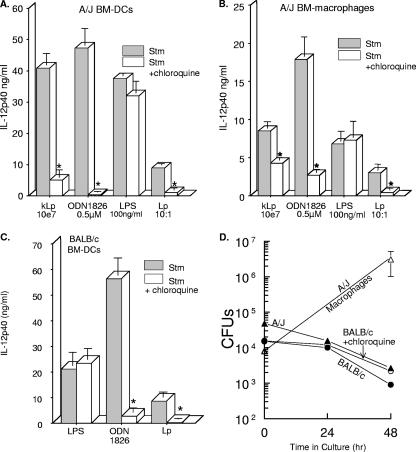
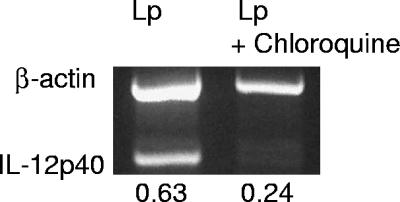
A requirement for TLR9 activation is endosomal maturation and/or acidification, which is inhibited by chloroquine pretreatment (39, 46). To see if L. pneumophila was stimulating IL-12 production through TLR9, DCs (Fig. 2A) and macrophages (Fig. 2B) from A/J mice were stimulated with formalin-killed L. pneumophila (kLp), E. coli LPS, or ODN1826 or were infected with virulent L. pneumophila (Lp) in either the presence or absence of chloroquine. The results show that chloroquine attenuated ODN1826 stimulation of the IL-12 p40 level (Fig. 2A and B), as previously reported, and also suppressed IL-12 p40 in response to L. pneumophila cells (killed or living) (Fig. 2A and B), suggesting that the bacteria are activated through TLR9. The effect of LPS, as previously reported, was not suppressed by chloroquine. Similar results with chloroquine were obtained with DCs from BALB/c mice (Fig. 2C). To ensure that chloroquine treatment was not affecting the uptake and intracellular survival of L. pneumophila, CFU were measured in the DC cultures from both mouse strains and in macrophages from A/J mice. As reported by others (5, 33, 45), macrophages from A/J mice supported the replication of L. pneumophila, as evidenced by an increase in the number of CFU (Fig. 2D); however, intracellular growth was not seen in DCs from both mouse strains, and furthermore, chloroquine treatment had no effect on the number of CFU (Fig. 2D). It should also be noted that the uptake of L. pneumophila by all groups of cells was similar, ranging from 104 to 3 × 104 CFU. In addition to IL-12 p40 protein, chloroquine also suppressed the mRNA in infected DCs, as measured by RT-PCR (Fig. 3).
FIG. 2.
Chloroquine attenuates IL-12 p40 production in DCs from A/J mice (A), macrophages from A/J mice (B), and DCs from BALB/c mice (C). Cells were pretreated with chloroquine (100 μM) for 30 min prior to stimulation (Stm) with killed L. pneumophila (kLp), ODN1826, living L. pneumophila (Lp), or LPS for 24 h. Culture supernatants were analyzed for IL-12 p40 by ELISA, and the data are means ± SEM for three to six experiments. Panel D shows the numbers of L. pneumophila CFU in DC cultures from A/J and BALB/c mice treated with chloroquine and also in A/J macrophages. BM, bone marrow. *, P < 0.05.
FIG. 3.
Chloroquine inhibits IL-12 p40 mRNA in L. pneumophila-infected (Lp) DCs from BALB/c mice. DCs were pretreated with chloroquine for 30 min, followed by a 30-min L. pneumophila infection and RNA extraction at 3 h postinfection. Semiquantitative RT-PCR was performed as described in Materials and Methods. The data are representative of three repeats.
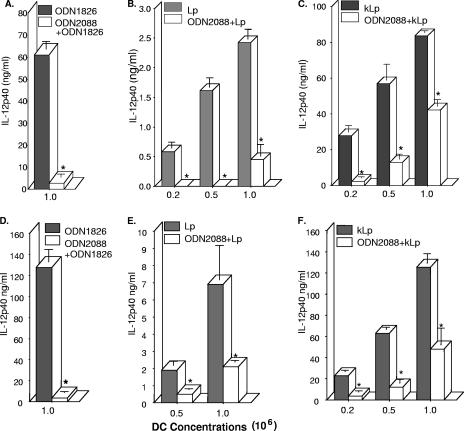
In addition to suppression by chloroquine, the activation of TLR9 can also be assessed by treatment with the ligand inhibitor ODN2088 (41). As expected, pretreatment (0.5 to 3 h) with the inhibitor completely attenuated IL-12 p40 production in response to the TLR9 ligand ODN1826 in cells from both BALB/c and A/J mice (Fig. 4A and D). It is interesting that ODN2088 pretreatment also significantly inhibited cytokine production in response to stimulation with either living or killed L. pneumophila cells (Fig. 4B to F), suggesting a role for TLR9. Although cells from A/J mice (Fig. 4D and E) appeared to produce more IL-12 p40 in response to infection, cells from both strains were equally inhibited by ODN2088. Interestingly, attenuation by the inhibitor was lower at higher concentrations of L. pneumophila, suggesting that moieties other than TLR9 are involved in the response.
FIG. 4.
ODN2088 inhibits IL-12 p40 production in DC cultures from BALB/c (A to C) and A/J (D to F) mice. Cell cultures were pretreated or not with ODN2088 (5 to 10 μM) for 0.5 to 3 h, followed by stimulation with ODN1826 (0.5 μM), infection with L. pneumophila (Lp; 10:1 [bacteria:DC]), or stimulation with killed L. pneumophila (kLp; 107 bacteria/ml) for 24 h and analysis of IL-12 p40 by ELISA. Data are means ± SEM for three to five experiments. *, P < 0.05.
The molecular and intracellular mechanisms surrounding L. pneumophila infection of macrophages have been studied extensively, but less is known concerning the mechanisms in infected DCs. Macrophages from A/J mice are permissive for L. pneumophila growth (43) because of low activity of the Naip5 (Birc1e) protein transcribed from the Lgn1 gene locus (11, 45). These proteins contain domains that interact with caspases and also contain Nod and LRR domains, which are important in sensing pathogen-associated antigens in the cytosol. Recently, it was shown that Birc1e proteins in the cytosol controlled the intracellular replication of L. pneumophila in macrophages through the activation of caspase-1 (IL-1β converting enzyme-protease-activating factor) (45). Others have shown that caspase-3 activation and cell death are important in controlling L. pneumophila growth in macrophages, although apoptosis was delayed following caspase-3 activation (1, 11, 13). TLR stimulation on macrophages also appears to be involved in resistance to L. pneumophila infection. For example, intracellular growth inhibition was dependent on TLR2 but not on TLR4 (2), and IL-12 and IL-10 production was attenuated in macrophages from TLR2-deficient mice. Furthermore, mice deficient in the TLR adapter protein MyD88 were more susceptible to L. pneumophila infection, and macrophages from these mice were deficient in IL-12, IL-6, and tumor necrosis factor alpha production following infection with L. pneumophila (5). Flagellin filaments by a TLR5-independent but Naip5-dependent mechanism were shown to induce defense against L. pneumophila replication in macrophages by activating caspase-1 and causing rapid cell death (38). This mouse macrophage resistance mechanism also required, in addition to the activation of caspase-1, a type IV secretion system and was postulated to contain features of pyroptosis and autophagy (31).
From these studies, it is clear that various innate molecular mechanisms are activated in macrophages following L. pneumophila infection, including Nod-LRR-sensing proteins, TLRs, caspase activation, and inflammatory cytokine production. Besides macrophages, DCs are also infected by L. pneumophila. DCs from nonpermissive C57BL/6 mice were shown in culture to minimally support the growth of L. pneumophila (2), and the growth was not affected in cells from TLR2- and TLR4-deficient mice. In the current report, we studied the growth of L. pneumophila in bone marrow-derived DCs from both nonpermissive BALB/c mice and permissive A/J mice. Our results show that, unlike the case in macrophages, there is a gradual decline in the number of intracellular CFU in DCs from permissive as well as nonpermissive strains. Thus, it appears that the innate mechanisms that restrict L. pneumophila growth in DCs are dependent upon factors other than the Naip5 proteins in that the diminished functioning of these proteins in A/J mice does not affect the overall restrictive capacity of the cell.
Cytokine production has also been studied in DC cultures exposed to L. pneumophila. For example, cells from A/J mice cultured with formalin- or heat-killed L. pneumophila cells produced more IL-12 p40, tumor necrosis factor alpha, and IL-6 than cells stimulated with living bacteria; furthermore, the stimulation of IL-12 p40 by heat-killed, but not formalin-killed, bacteria was shown to require, in part, TLR4 ligation (27). Also, bone marrow DCs from TLR2-deficient mice but not from TLR4-deficient mice showed lowered responses to purified L. pneumophila LPS as well as to viable or formalin-killed L. pneumophila (7). These findings, coupled with our own preliminary results showing that DCs from TLR4-deficient mice display a robust IL-12 p40 response following infection with live L. pneumophila (data not shown), led us to speculate that TLRs other than TLR2 and TLR4 play a significant role in mediating cytokine production in response to living bacteria. Our results show that cells from both permissive and nonpermissive strains are potent producers of IL-12, with L. pneumophila causing cytokine production in the ng/ml range. Furthermore, although the A/J cells appeared to be slightly more responsive in this regard, both groups of cells were equally inhibited by the TLR9 inhibitor, suggesting equivalent responsiveness to L. pneumophila infection. Bone marrow macrophages from the two strains are also equivalent regarding IL-12 p40 production (data not shown), consistent with the view that some immune functions are independent of the mechanisms controlled by the Naip5 proteins.
Regarding the involvement of TLRs, our results with chloroquine and ODN2088 treatment strongly support a role for TLR9 ligation in the stimulation of IL-12 p40 by L. pneumophila. This is also supported by previous findings linking the TLR9 adaptor protein MyD88 to L. pneumophila resistance (5, 18). Although this is the first report showing an association between TLR9 and Legionella, other facultatively intracellular bacteria have been shown to induce IL-12 through this receptor in DCs and macrophages. TLR9-deficient mice displayed defective IL-12 production in response to infection with Mycobacterium tuberculosis, and a reported cooperation was observed between TLR9 and TLR2 in the overall cytokine response and resistance to infection (6). In addition, the DC production of IL-12 in response to treatment with heat-killed Brucella abortus was shown to be dependent on TLR9 as well as the production of Th1 cytokines in whole-animal studies (23). In total, these results demonstrate that the induction of immunity by L. pneumophila is mediated through a number of cellular receptor systems in macrophages and DCs. The array of bacterial antigens stimulates not only several different TLRs but also members of the Nod-LRR recognition system, resulting in restriction of bacterial growth, destruction of the host cell, and induction of cytokines important in innate and adaptive immunity.
Acknowledgments
This work was supported by grants DA03646 and AI45169 from the National Institutes of Health.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 23 October 2006.
REFERENCES
- 1.Abu-Zant, A., M. Santic, M. Molmeret, S. Jones, J. Helbig, and Y. Abu Kwaik. 2005. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect. Immun. 73:5339-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamine, M., F. Higa, N. Arakaki, K. Kawakami, K. Takeda, S. Akira, and A. Saito. 2005. Differential roles of Toll-like receptors 2 and 4 in in vitro responses of macrophages to Legionella pneumophila. Infect. Immun. 73:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archer, K. A., and C. R. Roy. 2006. MyD88-dependent responses involving Toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires' disease. Infect. Immun. 74:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bafica, A., C. A. Scanga, C. G. Feng, C. Leifer, A. Cheever, and A. Sher. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braedel-Ruoff, S., M. Faigle, N. Hilf, B. Neumeister, and H. Schild. 2005. Legionella pneumophila mediated activation of dendritic cells involves CD14 and TLR2. J. Endotoxin Res. 11:89-96. [DOI] [PubMed] [Google Scholar]
- 8.Byrd, T. F., and M. A. Horwitz. 1991. Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. A potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. J. Clin. Investig. 88:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celada, A., P. W. Gray, E. Rinderknecht, and R. D. Schreiber. 1984. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 160:55-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta, S. K., V. Redecke, K. R. Prilliman, K. Takabayashi, M. Corr, T. Tallant, J. DiDonato, R. Dziarski, S. Akira, S. P. Schoenberger, and E. Raz. 2003. A subset of Toll-like receptor ligands induces cross-presentation by bone marrow-derived dendritic cells. J. Immunol. 170:4102-4110. [DOI] [PubMed] [Google Scholar]
- 11.Derre, I., and R. R. Isberg. 2004. Macrophages from mice with the restrictive Lgn1 allele exhibit multifactorial resistance to Legionella pneumophila. Infect. Immun. 72:6221-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez, E., S. H. Lee, S. Gauthier, Z. Yaraghi, M. Tremblay, S. Vidal, and P. Gros. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat. Genet. 33:55-60. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, S. F., J. Vier, C. Muller-Thomas, and G. Hacker. 2006. Induction of apoptosis by Legionella pneumophila in mammalian cells requires the mitochondrial pathway for caspase activation. Microbes Infect. 8:662-669. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, K. A., D. C. Rowe, and D. T. Golenbock. 2004. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. 6:1361-1367. [DOI] [PubMed] [Google Scholar]
- 15.Fortier, A., E. Diez, and P. Gros. 2005. Naip5/Birc1e and susceptibility to Legionella pneumophila. Trends Microbiol. 13:328-335. [DOI] [PubMed] [Google Scholar]
- 16.Girard, R., T. Pedron, S. Uematsu, V. Balloy, M. Chignard, S. Akira, and R. Chaby. 2003. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J. Cell Sci. 116:293-302. [DOI] [PubMed] [Google Scholar]
- 17.Harley, V. S., and B. S. Drasar. 1989. Interaction of Legionella pneumophila with tissue culture cells. Biochem. Soc. Trans. 17:1118-1119. [DOI] [PubMed] [Google Scholar]
- 18.Hawn, T. R., K. D. Smith, A. Aderem, and S. J. Skerrett. 2006. Myeloid differentiation primary response gene (88)- and Toll-like receptor 2-deficient mice are susceptible to infection with aerosolized Legionella pneumophila. J. Infect. Dis. 193:1693-1702. [DOI] [PubMed] [Google Scholar]
- 19.Hawn, T. R., A. Verbon, K. D. Lettinga, L. P. Zhao, S. S. Li, R. J. Laws, S. J. Skerrett, B. Beutler, L. Schroeder, A. Nachman, A. Ozinsky, K. D. Smith, and A. Aderem. 2003. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to Legionnaires' disease. J. Exp. Med. 198:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 21.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, L. Y., K. J. Ishii, S. Akira, J. Aliberti, and B. Golding. 2005. Th1-like cytokine induction by heat-killed Brucella abortus is dependent on triggering of TLR9. J. Immunol. 175:3964-3970. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 25.Jakob, T., P. S. Walker, A. M. Krieg, E. von Stebut, M. C. Udey, and J. C. Vogel. 1999. Bacterial DNA and CpG-containing oligodeoxynucleotides activate cutaneous dendritic cells and induce IL-12 production: implications for the augmentation of Th1 responses. Int. Arch. Allergy Immunol. 118:457-461. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. H., and B. A. Cunha. 1993. Atypical pneumonias. Clinical and extrapulmonary features of Chlamydia, Mycoplasma, and Legionella infections. Postgrad. Med. 93:69-72, 75-76, 79-82. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi, T., T. Kobayashi, K. Gomi, T. Suzuki, Y. Tokue, A. Watanabe, and T. Nukiwa. 2004. Dendritic cells pulsed with live and dead Legionella pneumophila elicit distinct immune responses. J. Immunol. 172:1727-1734. [DOI] [PubMed] [Google Scholar]
- 28.Klein, T. W., C. Newton, R. Widen, and H. Friedman. 1993. Delta 9-tetrahydrocannabinol injection induces cytokine-mediated mortality of mice infected with Legionella pneumophila. J. Pharmacol. Exp. Ther. 267:635-640. [PubMed] [Google Scholar]
- 29.Klein, T. W., C. A. Newton, N. Nakachi, and H. Friedman. 2000. Δ9-Tetrahydrocannabinol treatment suppresses immunity and early IFNγ, IL-12, and IL-12 receptor β2 responses to Legionella pneumophila infection. J. Immunol. 164:6461-6466. [DOI] [PubMed] [Google Scholar]
- 30.Lu, T., C. Newton, I. Perkins, H. Friedman, and T. W. Klein. 2006. Role of cannabinoid receptors in delta-9-tetrahydrocannabinol suppression of IL-12p40 in mouse bone marrow-derived dendritic cells infected with Legionella pneumophila. Eur. J. Pharmacol. 532:170-177. [DOI] [PubMed] [Google Scholar]
- 31.Molofsky, A. B., B. G. Byrne, N. N. Whitfield, C. A. Madigan, E. T. Fuse, K. Tateda, and M. S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napolitani, G., A. Rinaldi, F. Bertoni, F. Sallusto, and A. Lanzavecchia. 2005. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neild, A., T. Murata, and C. R. Roy. 2005. Processing and major histocompatibility complex class II presentation of Legionella pneumophila antigens by infected macrophages. Infect. Immun. 73:2336-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neild, A. L., C. R. Roy, and E. R. Unanue. 2003. Legionella reveal dendritic cell functions that facilitate selection of antigens for MHC class II presentation. Immunity 18:813-823. [DOI] [PubMed] [Google Scholar]
- 35.Newton, C., S. McHugh, R. Widen, N. Nakachi, T. Klein, and H. Friedman. 2000. Induction of interleukin-4 (IL-4) by Legionella pneumophila infection in BALB/c mice and regulation of tumor necrosis factor alpha, IL-6, and IL-1beta. Infect. Immun. 68:5234-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newton, C. A., T. W. Klein, and H. Friedman. 1994. Secondary immunity to Legionella pneumophila and Th1 activity are suppressed by delta-9-tetrahydrocannabinol injection. Infect. Immun. 62:4015-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palsson-McDermott, E. M., and L. A. O'Neill. 2004. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 113:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutz, M., J. Metzger, T. Gellert, P. Luppa, G. B. Lipford, H. Wagner, and S. Bauer. 2004. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur. J. Immunol. 34:2541-2550. [DOI] [PubMed] [Google Scholar]
- 40.Salins, S., C. Newton, R. Widen, T. W. Klein, and H. Friedman. 2001. Differential induction of gamma interferon in Legionella pneumophila-infected macrophages from BALB/c and A/J mice. Infect. Immun. 69:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stunz, L. L., P. Lenert, D. Peckham, A. K. Yi, S. Haxhinasto, M. Chang, A. M. Krieg, and R. F. Ashman. 2002. Inhibitory oligonucleotides specifically block effects of stimulatory CpG oligonucleotides in B cells. Eur. J. Immunol. 32:1212-1222. [DOI] [PubMed] [Google Scholar]
- 42.Ulevitch, R. J., J. C. Mathison, and J. da Silva Correiaqq. 2004. Innate immune responses during infection. Vaccine 22(Suppl. 1):S25-S30. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, Y., T. W. Klein, C. A. Newton, R. Widen, and H. Friedman. 1988. Growth of Legionella pneumophila in thioglycolate-elicited peritoneal macrophages from A/J mice. Infect. Immun. 56:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida, S., Y. Goto, Y. Mizuguchi, K. Nomoto, and E. Skamene. 1991. Genetic control of natural resistance in mouse macrophages regulating intracellular Legionella pneumophila multiplication in vitro. Infect. Immun. 59:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamboni, D. S., K. S. Kobayashi, T. Kohlsdorf, Y. Ogura, E. M. Long, R. E. Vance, K. Kuida, S. Mariathasan, V. M. Dixit, R. A. Flavell, W. F. Dietrich, and C. R. Roy. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318-325. [DOI] [PubMed] [Google Scholar]
- 46.Zou, W., A. Amcheslavsky, and Z. Bar-Shavit. 2003. CpG oligodeoxynucleotides modulate the osteoclastogenic activity of osteoblasts via Toll-like receptor 9. J. Biol. Chem. 278:16732-16740. [DOI] [PubMed] [Google Scholar]