Abstract
Subtilase cytotoxin (SubAB) is a recently identified AB5 subunit toxin produced by Shiga-toxigenic Escherichia coli. The A subunit is thought to be a subtilase-like, serine protease, whereas the B subunit binds to the toxin receptor on the cell surface. We cloned the genes from a clinical isolate; the toxin was produced as His-tagged proteins. SubAB induced vacuolation at concentrations greater than 1 μg/ml after 8 h, in addition to the reported cytotoxicity induced at a ng/ml level after 48 h. Vacuolation was induced with the B, but not the A, subunit and was dependent on V-type ATPase. The cytotoxicity of SubAB at low concentrations was associated with the inhibition of protein synthesis; the 50% inhibitory dose was ∼1 ng/ml. The A subunit, containing serine 272, which is thought to be a part of the catalytic triad of a subtilase-like serine protease, plus the B subunit was necessary for this activity, both in vivo and in vitro. SubAB did not cleave azocasein, bovine serum albumin, ovalbumin, or synthetic peptides. These data suggest that SubAB is a unique AB toxin: first, the B subunit alone can induce vacuolation; second, the A subunit containing serine 272 plus the B subunit inhibited protein synthesis, both in vivo and in vitro; and third, the A subunit proteolytic activity may have a strict range of substrate specificity.
Shiga toxins belong to a large family of bacterial toxins. Shiga toxin (Stx) produced by Shigella dysenteriae and Shiga toxin 1 (Stx1 or VT1) produced by Escherichia coli are almost identical: Shiga toxin 2 (Stx2 or VT2) produced by E. coli has a similar structure but differs in sequence. This family of Shiga toxins belongs to the AB family of protein toxins; the A subunit has enzymatic activity, and the B subunits bind to the cell surface. In the case of Shiga toxin, the B moiety consists of five identical B subunits and binds to glycolipid receptors at the cell surface (13, 35). The A subunit contains internal disulfide bonds, and proteolytic cleavage releases A1 and A2 fragments (8, 29). The A1 fragment inhibits protein synthesis by removing adenine from 28S rRNA, thereby inhibiting the binding of amino-acyl-tRNA (5). Shiga toxin acts only on cells that have specific receptors (usually glycolipid Gb3). The toxin is internalized by endocytosis into the endosome; transport through the Golgi apparatus and endoplasmic reticulum (ER) is required for intoxication (28).
Infections with Shiga toxin-producing bacteria are responsible for diseases such as hemolytic uremic syndrome (HUS), which is characterized by thrombocytopenia, microangiopathic hemolytic anemia, and renal failure. In some cases, severe neurological manifestations are observed (10, 11, 12, 33). These infections are a great threat to human health, not only in developing but also in developed countries. However, whether Shiga toxin is responsible for the clinical presentations is still not clear. Putative accessory virulence factors have been reported, including enterohemolysin, serine protease, and heat-stable enterotoxin. There may be additional factors responsible for disease severity (2, 3, 17, 24, 30, 31).
A new member of the AB5 toxin family, named subtilase cytotoxin (SubAB), was identified by Paton et al. (23) from E. coli O113:H21 strain 98NK2, which produced Stx2 and was responsible for an outbreak of HUS (21). The SubAB A subunit (SubA), with a molecular weight of 35 kDa, has a structure similar to that of a subtilase-like serine protease and is distantly related to the BA 2875 gene product of Bacillus anthracis. The B subunit (SubB) consists of a pentamer of 13-kDa monomers, which are related to a putative exported protein from Yersinia pestis. SubAB was shown to be cytotoxic to Vero cells and lethal for mice, causing extensive microvascular thrombosis as well as necrosis in the brain, kidney, and liver (23).
We wanted to elucidate the presently unknown molecular mechanism of action of this novel cytotoxin. Paton et al. showed the existence of the subA gene in 11% of E. coli producing Stx (22). In a survey of 11 strains of Stx-producing E. coli, we found the subAB region by PCR with specific primers for subAB in one strain of E. coli O29 which produced Stx2. Recombinant SubAB protein at concentrations of >1 μg/ml induced vacuolation in Vero cells after 8 h, an observation not reported earlier (23). Lower concentrations of toxin (<1 μg/ml) inhibited growth; after 48 h, rounded cells detached from the plate, as had been noted previously (23). We found that SubAB cytotoxicity was associated with the inhibition of protein synthesis, as described for Shiga toxin, but occurred by a different mechanism, most likely the putative protease activity of SubA. This activity was not found in the SubAB(S272A) mutant toxin, which still had vacuolation activity. We report here that SubAB is a novel toxin; its activities include the inhibition of protein synthesis and vacuole formation. The inhibition of protein synthesis by SubA required serine at position 272 and SubB, whereas SubB alone induced vacuolation.
MATERIALS AND METHODS
Bacteria and cells.
E. coli O29 was isolated from a patient hospitalized with acute intestinal infection. It produced VT2 but not VT1, as found by using an immunodetection kit (Wako Pure Chemical Industries, Ltd.). Vero cells were cultured in Eagle's minimum essential medium (MEM) (Sigma) containing 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere.
Chemicals.
Bafilomycin A1 was purchased from Wako Pure Chemical Industries, Ltd. His-Select Nickel Affinity Gel was purchased from Sigma. Anti-β-COP antibodies were from Affinity Bioreagents, and anti-EEA1 antibodies were from Transduction Laboratories. Alexa488-conjugated anti-rabbit and anti-mouse immunoglobulin G (IgG) antibodies were from Molecular Probes. l-[U-14C]Leucine (50 μCi/ml) was from Amersham Pharmacia Biotech. Synthetic peptides Z-Ala-Ala-Leu-pNA, Boc-Arg-Val-Arg-Arg-MCA (where Boc is t-butyloxycarbonyl, and MCA is 4-methylcoumarin), and MOCAc-Ser-Glu-Val-Asp-Leu-Asp-Ala-Glu-Phe-Arg-Lys(Dnp)-Arg-Arg-NH2 [where MOCAc is (7-methoxycoumarin-4-yl)acetyl, and Dnp is 2,4-dinitrophenol] were purchased from Peptide Institute, Inc. Cholera toxin B subunit was purchased from Calbiochem, and Stx1 was prepared by the method reported previously (15).
Cloning and expression of SubAB from E. coli O29.
DNA was extracted from E. coli O29. PCR amplification using forward primer 5′-GCTGGATCCGATGCTTAAGATTTTATGGACG-3′ and reverse primer 3′-GATTATCTCGAGTGAGTTCTTTTTCCTGTCAGG-3′ (underlined sequences are restriction sites for BamH1 and Xho1, respectively) resulted in a product of the expected size, which was subcloned into pCR-TOPO vector by TOPO TA cloning (Invitrogen). DNA was digested by BamH1 and Xho1 and ligated into pET23b(+) (Novagen), which had been digested with BamH1 and Xho1; the vector was used for the transformation of E. coli DH5α. The plasmid was sequenced and then used for the transformation of E. coli BL21(DE3). Expressed recombinant six-His-tagged protein was purified under native conditions using Ni-nitrilotriacetic acid (NTA) agarose (23). SubA and SubB were produced using a method similar to that for SubAB, using forward primer 5′-GCTGGATCCGATGCTTAAGATTTTATGGACG-3′ and reverse primer 5′-GGCTCGAGCAGTTCTTCACTCATCCTTC-3′ (underlined sequences are restriction sites for BamH1 and Xho1, respectively) for SubA and forward primer 5′-GCGGATCCGATGACGATTAAGCGTTTTTTTG-3′ and reverse primer 5′-GATTATCTCGAGTGAGTTCTTTTTCCTGTCAGG for SubB (underlined sequences are restriction sites for BamH1 and Xho1, respectively).
Site-directed mutagenesis of SubAB.
To replace active site serine (S272) with alanine in SubA, a QuikChange site-directed mutagenesis kit (Stratagene) was used. Primers were 5′-CTGGTAGCGGAACGGCAGAAGCAACAGCTATAG-3′ and 5′-CTATAGCTGTTGCTTCTGCCGTTCCGCTACCA-3′ (mutated bases are underlined). The sequence was confirmed with an ABI 377 automatic sequencer. The mutant, SubAB(S272A), was purified using the same method as described for the wild-type SubAB.
Assay of protein synthesis in vivo.
Vero cells were seeded at 2 × 105 cells/24-well plate (each well, 1 ml) and grown overnight. Medium was replaced with 0.5 ml of MEM-10% FBS, containing 0.625 μCi/ml of [14C]leucine. After the addition of SubAB (50 μl), the plate was incubated at 37°C for 2 h in a water bath. Protein synthesis was stopped by the addition of 0.25 ml of 30% trichloroacetic acid (TCA); cells were washed three times with 1 ml of 10% TCA and lysed in 0.25 ml of 0.5 M KOH for 10 min at 37°C. The lysate was neutralized with 0.25 ml of 0.5 M acetate, and protein synthesis was quantified by a radioassay of 14C (36).
Assay of protein synthesis in vitro.
Protein synthesis in a cell-free system was investigated using a Retic lysate IVT in vitro translation kit (Ambion Inc.) following the manufacturer's instructions. Various amounts of toxin were incubated at 30°C for 60 min with [14C]leucine in a reaction mixture (company supplied) having a total volume of 12.5 μl. The reaction mixture was then placed on ice, and 3-μl samples were placed into new tubes with 0.5 ml distilled water and incubated at 30°C for 10 min after the addition of a 0.5 ml decolorizing solution (1 N NaOH, 1 mM leucine, 1.5% H2O2). Cold 25% TCA was added, and after 5 min on ice, the precipitate was collected by vacuum filtration on GFC glass filters, and the 14C radioactivity was determined.
Assay of cell vacuolation.
Cells (2 × 104 in 90 μl) were seeded into a 96-well plate; indicated concentrations of toxin were added (final volume, 100 μl), followed by incubation at 37°C for 18 to 20 h. Medium was removed, and 50 μl of 0.025% neutral red in phosphate-buffered saline (PBS) was added, followed by incubation for 7 min. Cells were washed with PBS twice, and then 100 μl of solution (0.4% 1 N HCl in 70% ethanol) was added, followed by the determination of A540.
Assay of viable cells.
Various concentrations of SubAB were added to 1 × 105 cells/24-well plates (500 μl/well), which were incubated at 37°C in a 5% CO2 atmosphere. At each time point, cells were detached from the substratum by trypsin and collected in a microtube by centrifugation (100 × g, 5 min). A solution of 0.04% trypan blue-PBS was added, and viable, unstained cells were counted.
Internalization of SubAB.
Internalization of SubAB was determined as described previously (16). Briefly, Vero cells (2 × 104 cells/well) were cultured in Eagle's MEM with 10% fetal calf serum in 96-well plates overnight. Biotin-labeled SubAB was prepared according to the supplier's instructions, using EZ-Link sulfo-N-hydoroxylsulfosuccinimide-biotin (sulfo-NHS-SS-biotin) (Pierce). Cells were washed with cold Hanks' balanced salt solution with 0.1% bovine serum albumin (BSA) (HBSS-BSA) three times before the addition of 100 μl of SubAB (2 μg/ml) in HBSS-BSA, incubated at 4°C for 30 min, and then washed three times with cold HBSS to remove unbound toxin. Cells were then incubated at 37°C for the indicated time before washing immediately with cold HBSS, fixation with 0.25% glutaraldehyde in PBS, and the addition of 0.5 M 2-mercaptoethanesulfonic acid (MESNA), which does not enter cells, to reduce disulfide bonds and release biotin from SubAB bound to the cell surface. After additional incubation at room temperature for 30 min, cells were permeabilized with 1% Triton X-100 for 15 min and fixed for 20 min. After blocking with 3% BSA in PBS for 1 h, cells were incubated with horseradish peroxidase (HRP)-conjugated streptavidin for 1 h, and HRP was detected with BM blue substrate (Roche Diagnostic Corp.). After a 20-min incubation, the reaction was stopped with 1 M H2SO4, and color was measured at 450 nm. Background absorbance was subtracted. Between each step, cells were washed with PBS three times.
Localization of fluorescein-labeled SubAB.
SubAB was labeled with Cy3 using a Fluorolink-Ab Cy3 labeling kit (Amersham Pharmacia Biotech, Ltd.). Cells (2 × 105/well) were seeded in six-well plates containing coverslips and incubated at 37°C overnight. After washing, cells were incubated at 4°C for 30 min in HBSS-BSA containing Cy3-labeled SubAB. Then, cells were washed to remove unbound toxin and incubated at 37°C for the indicated times. Cells were then fixed with 3% paraformaldehyde for 20 min, quenched with 50 mM NH4Cl for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 4 min. Then, cells were blocked with 3% BSA in PBS for 1 h. Cells were incubated with mouse anti-EEA1 antibody (1:100) or rabbit anti-β-COP (1:500) for 1 h and then for 1 h with Alexa488-conjugated anti-mouse IgG (1:500) or Alexa488-conjugated anti-rabbit IgG (1:500) secondary antibody, respectively. Colocalization was determined by confocal microscopy.
Protease activity with azocasein.
Sample was added to a reaction mixture containing 0.4% azocasein and 10 mM Tris-HCl buffer (pH 7.5) in a final volume of 500 μl, followed by incubation at 37°C for 1 h. The reaction was terminated by the addition of 10% TCA, and the solution was centrifuged at 13,000 × g for 10 min. The supernatant was mixed with an equal volume of 1 N NaOH, and the optical density of the solution was measured at 450 nm (38).
RESULTS
We found the subAB construct by PCR with specific primers for the subAB region in a strain of E. coli O29 that produced VT2 but not VT1 (data not shown). The PCR product was subcloned into a pCR-TOPO cloning vector and sequenced. The sequence of the gene differed by three bases from the subAB construct (nucleotides 13,725 to 14,768 of GenBank accession no. AF399919.3) (23), which resulted in a two-amino-acid difference: alanine 263 of SubA was replaced with glycine, and lysine 102 of SubB was replaced with glutamic acid. The subAB construct was subcloned into pET23b using the BamHI and XhoI sites and transformed into strain BL21(DE3). Recombinant SubAB protein was purified under native conditions to more than 95% purity using a Ni-NTA agarose column (data not shown).
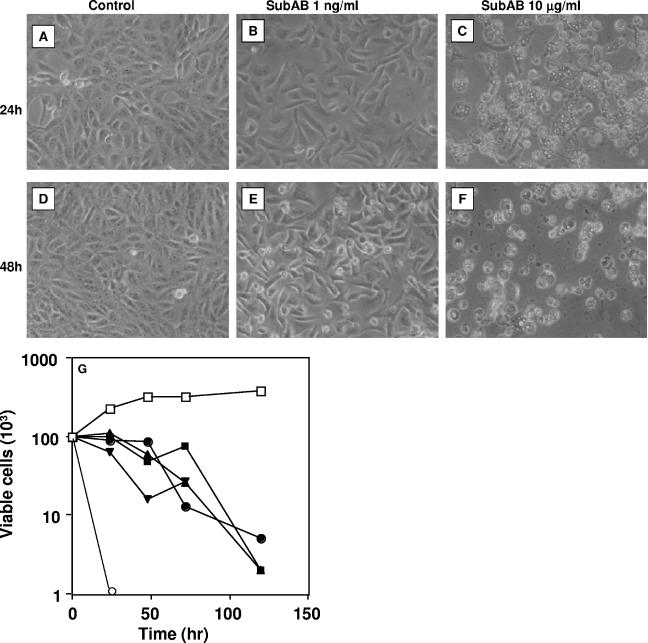
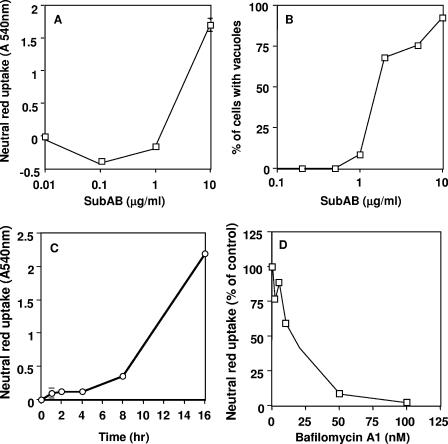
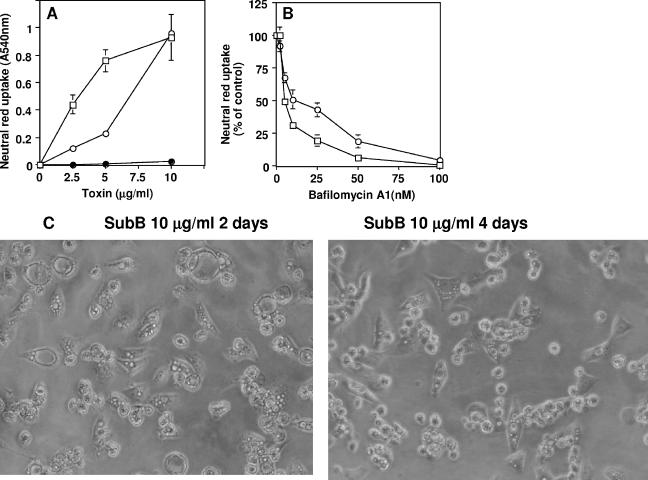
SubAB produced large vacuoles in Vero cells (Fig. 1C). Vacuolation was induced at concentrations of SubAB greater than 1 μg/ml when assessed by direct count (phase-contrast microscopy) or neutral red incorporation (Fig. 2A and B) and appeared around 8 h after the addition of SubAB to Vero cells (Fig. 2C). Cells were disrupted at 48 h (Fig. 1F). To determine whether SubAB-induced vacuolation is dependent on the function of vacuole-type ATPase proton pumps, we used bafilomycin A1, which targets V-ATPase protein pumps (1, 6, 14). Bafilomycin A1 inhibited SubAB-induced vacuolation in a concentration-dependent manner (Fig. 2D).
FIG. 1.
Cytotoxicity of SubAB for Vero cells (A to F). Cells were treated with the indicated amounts of SubAB for 24 (A to C) and 48 h (D to F). Phase-contrast microscopy pictures are shown. (G) Viable cells were counted at the indicated times using trypan blue exclusion. Control (□), 0.001 μg/ml (▪), 0.01 μg/ml (•), 0.1 μg/ml (▴), 1 μg/ml (▾), and 10 μg/ml (○). Experiments were repeated twice with similar results.
FIG. 2.
Vacuolation by SubAB in Vero cells. (A) Monolayers in 96-well plates were treated with the indicated concentrations of SubAB for 18 h, and neutral red incorporation at A540 was determined. A540 in control cells (1.3) was subtracted. (B) Cells (1 × 105) were seeded in 24-well plates and incubated with various concentrations of SubAB for 18 h; the percentage of cells with vacuolation was calculated by direct microscopic visualization. (C) Monolayers in 96-well plates were treated with 10 μg/ml of SubAB for the indicated times, and neutral red incorporation was evaluated. A540 in control cells (1.46) was subtracted. (D) Monolayers in 96-well plates were treated with the indicated concentrations of bafilomycin A1 and 5 μg/ml of SubAB for 18 h, and neutral red incorporation was evaluated at A540. Absorbance in control cells (2.2) was subtracted.
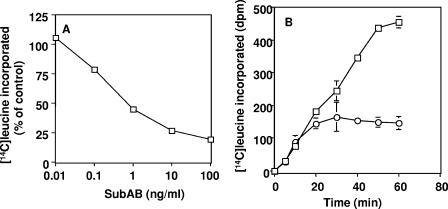
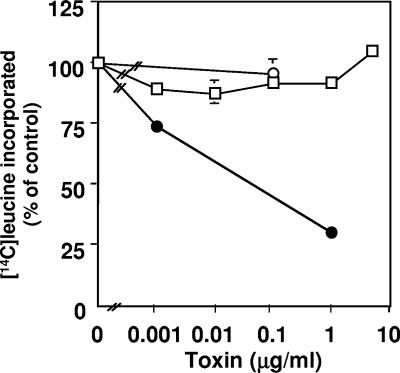
Lesser amounts of toxin (0.1 ng/ml to ∼1 μg/ml) induced growth inhibition (Fig. 1B and E), and at more than 48 h, round cells appeared, which were detached from the substratum (Fig. 1E). These round cells were stained with trypan blue, suggesting that they were not viable. A similar cell-damaging effect of SubAB was reported by Paton et al. (23). Toxin (10 μg/ml) induced vacuolation, and viable cells were not seen after 1 day (Fig. 1G). Toxin concentrations less than 1 μg/ml did not induce vacuolation, as reported above; however, cell growth was suppressed, and gradually, viable cells were lost. As this cell damage took more than 2 days, we investigated whether protein synthesis of Vero cells was suppressed by SubAB. Leucine incorporation into a TCA-precipitated fraction of Vero cells was suppressed by SubAB in a dose-dependent manner: a 50% inhibitory dose was ∼1 ng/ml (Fig. 3A). Leucine incorporation was suppressed at 20 min (P = 0.14) and 30 min (P = 0.02) with 10 μg/ml of toxin (Fig. 3B).
FIG. 3.
Effect of SubAB on [14C]leucine incorporation into Vero cells. (A) Vero cells (2 × 105 cells) were seeded in a 24-well plate and incubated overnight. [14C]Leucine and the indicated concentrations of SubAB were added to cells, which were then incubated for 2 h. After the addition of TCA (5%) to stop the reaction, [14C]leucine incorporation was determined and expressed as a percentage of control. The disintegrations per minute (dpm) value of control samples was 4,093 ± 744. Data are the means ± standard deviations (SD) of values from four samples. (B) Vero cells were incubated with (○) or without (□) 10 μg/ml of SubAB for the indicated times, and [14C]leucine incorporation was determined. Basal dpm (43.7) was subtracted. Data are the means ± SD of values from four samples. Experiments were repeated three times with similar results. P values for the comparison of control and SubAB-treated cells at 20 and 30 min were 0.14 and 0.02, respectively.
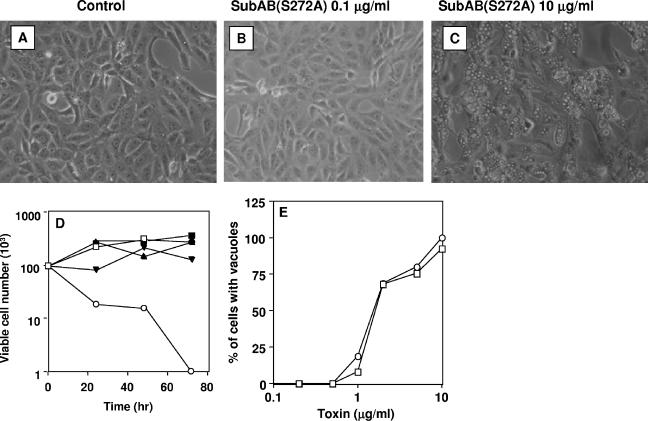
Paton et al. reported that the cytotoxicity of SubAB was abolished by replacing the active site serine residue of SubA with alanine (S272A) (23). We investigated whether this replacement also abolished the inhibition of protein synthesis. The SubAB(S272A) mutant did not inhibit cell growth at a low dose (Fig. 4B and D), and the protein synthesis inhibitory activity of SubAB(S272A) was completely lost, even at 10 μg/ml (data not shown). The mutant, however, did induce vacuolation (Fig. 4C and E) at concentrations of more than 1 μg/ml, similar to wild-type SubAB.
FIG. 4.
Effect of mutant SubAB on Vero cells. (A to C) SubAB(S272A) was added to monolayers of Vero cells for 24 h, and cell morphology was observed with a phase-contrast microscope. (D) SubAB(S272A) was added to monolayers of Vero cells, and viable cells were counted by trypan blue dye exclusion. Control (□), 0.001 μg/ml (▪), 0.01 μg/ml (•), 0.1 μg/ml (▴), 1 μg/ml (▾), and 10 μg/ml (○) of SubAB(S272A). (E) Vacuolating activity of SubAB(S272A) was evaluated by direct microscopic visualization and compared to that of SubAB, as shown in Fig. 2B. SubAB (□) and SubAB(S272A) (○).
We prepared recombinant SubA and SubB to determine the subunit responsible for vacuolation or inhibition of protein synthesis; both subunits had a C-terminal His tag. SubB, but not SubA, caused vacuolation (Fig. 5A), and the vacuolation was suppressed by bafilomycin A1 (Fig. 5B). Cells containing vacuoles continued to attach to the substratum of the plate for at least 4 days (Fig. 5C). They did not proliferate, even when the medium was changed 1 day after treatment, and were stained by trypan blue, suggesting that the vacuole-containing cells were no longer viable. SubA alone did not inhibit cell growth (data not shown), and consistent with that, SubA alone did not inhibit protein synthesis (Fig. 6).
FIG. 5.
Effect of A and B subunits on vacuolation. (A) Monolayers in a 96-well plate were treated with various concentrations of SubA (•), SubB (□), and SubAB (○) for 18 h, and neutral red incorporation was evaluated at A540. Data are the means ± standard deviations (SD) of values from three samples. (B) Monolayers in 96-well plates were treated with the indicated concentrations of bafilomycin A1 and 5 μg/ml SubB (□) or 10 μg/ml SubAB (○) for 18 h. Neutral red incorporation was evaluated at A540. Data are the means ± SD of values from three samples. (C) Phase-contrast microscopy pictures of vacuoles formed in Vero cells treated with 10 μg/ml SubB for 2 and 4 days.
FIG. 6.
Effect of A and B subunits on protein synthesis in Vero cells. The indicated concentrations of SubA (□), SubB (○), or SubAB (•) and [14C]leucine were added to monolayers of Vero cells as described in the legend to Fig. 3A. [14C]Leucine incorporation was determined and expressed as a percentage of control. The disintegrations per minute value of control samples was 1,426 ± 396. Data are the means ± SD of values from three samples. Experiments were repeated twice with similar results.
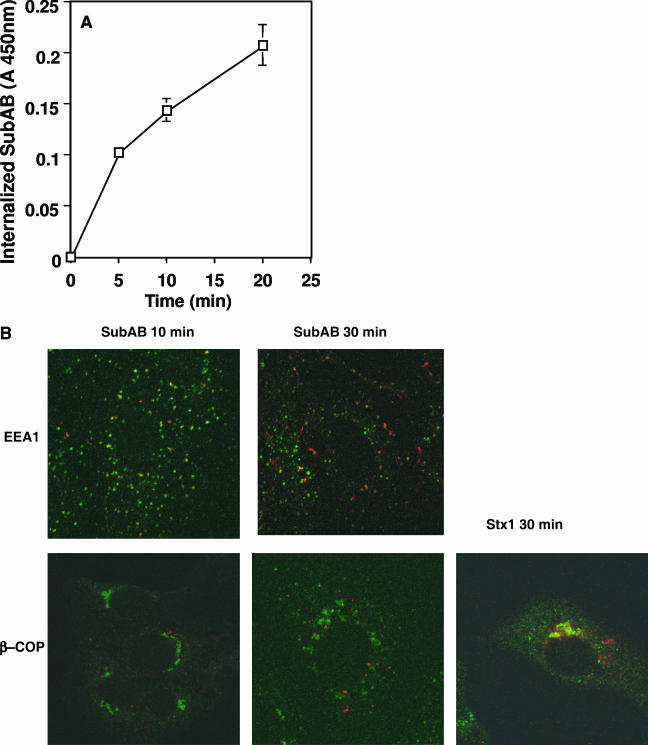
We assessed whether SubAB was internalized into Vero cells and how it trafficked inside cells. Internalization of SubAB was investigated using SubAB labeled with EZ-Link sulfo-NHS-SS-biotin (biotin-SubAB). As biotin-SubAB binding to Vero cells was maximal at 2 μg/ml (data not shown), we used this concentration. Cells were incubated with 2 μg/ml biotin-SubAB at 4°C for 30 min, and after washing to remove unbound toxin, the cells were incubated at 37°C for the indicated times. SubAB internalization was quantified after removing biotin from the cell surface with MESNA. SubAB was internalized into cells in a time-dependent manner (Fig. 7A). We then examined SubAB delivery inside the cells using indirect immunostaining with Cy3-labeled SubAB. Following a 10-min incubation at 37°C, SubAB colocalized with EEA1, an early endosome marker. Colocalization with EEA1 was significantly less at 30 min. SubAB did not colocalize with β-COP, a Golgi marker, at both 10 and 30 min (Fig. 7B), even after incubation for 60 min (data not shown). In contrast, Stx1 colocalized with β-COP at 30 min (Fig. 7B). These data suggest that SubAB was endocytosed into early endosomes and then may move from endosomes into cytosol without the involvement of the retrograde pathway to the Golgi or the ER.
FIG. 7.
Internalization of SubAB in Vero cells. (A) Time course of internalization of SubAB. Biotin-labeled SubAB was bound to monolayers of Vero cells in a 96-well plate at 4°C for 30 min and then incubated at 37°C for the indicated times after washing to remove unbound toxin. At the indicated time, cells were fixed and biotin bound to the cell surface was removed with MESNA; internalized biotin was quantified as in Materials and Methods. The total binding to Vero cells was 0.871 ± 0.041. Data are the means ± SD of values from four samples. (B) Colocalization of SubAB with EEA1 and β-COP. Cy3-labeled SubAB or Stx1 was bound to Vero cells at 4°C for 30 min, which were then incubated at 37°C for the indicated times after washing to remove unbound toxin. Cells were fixed and incubated with anti-EEA1 or anti-β-COP antibodies, after treatment with 0.1% Triton X-100, followed by Alexa488-conjugated secondary antibodies, as described in Materials and Methods.
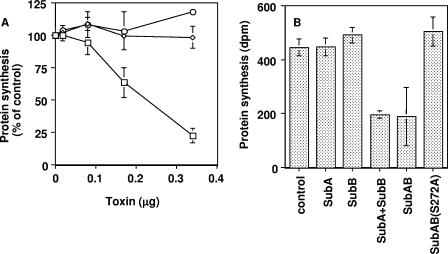
We further examined the effect of SubAB on protein synthesis in a cell-free system. In vitro translation of mRNA (capped RNA for Xenopus elongation factor 1) was performed using a company-supplied kit. As expected, protein synthesis in vitro was also suppressed by wild-type SubAB but not by the SubAB(S272A) mutant; however, unexpectedly, SubA alone did not inhibit protein synthesis (Fig. 8A). The addition of SubB to SubA was required; thus, both subunits are necessary for the inhibition of protein synthesis (Fig. 8B).
FIG. 8.
Effect of SubAB on the protein synthesis in a cell-free system. (A) The indicated concentrations of SubAB (□), SubAB(S272A) (⋄), and SubA (○) were incubated with [14C]leucine in a reaction mixture (company supplied) (total volume of 12.5 μl) at 30°C for 60 min, and [14C] radioactivity of TCA-precipitated samples on GFC glass filters was determined as described in Materials and Methods. Disintegration per minute of control sample without toxin was 751.5 ± 25.5. Data are expressed as a percentage of the control. Data are the means ± standard deviations (SD) of values from four samples. (B) Effect of A and B subunits on protein synthesis in a cell-free system. SubA (80 ng), SubB (90 ng), SubA+SubB (90 ng), SubAB (340 ng), and SubAB(S272A) (340 ng) were incubated with [14C]leucine in a reaction mixture at 30°C for 60 min. Data are the means ± SD of values from four samples of SubAB and three samples of the others.
Serine at the 272 position was important for the inhibition of protein synthesis by SubAB. Paton et al. showed that this serine formed the catalytic triad of the protease (23). We examined the protease activity of SubAB using azocasein, BSA, and ovalbumin as substrates; however, no proteolytic activity was observed (data not shown). We also used synthetic peptides Z-Ala-Ala-Leu-pNA (a subtilase substrate), Boc-Arg-Val-Arg-Arg-MCA (a substrate for furin, which cleaves at the carboxyl side of paired basic residues), and MOCAc-Ser-Glu-Val-Asp-Leu-Asp-Ala-Glu-Phe-Arg-Lys(Dnp)-Arg-Arg-NH2. However, no digestion was observed (data not shown).
DISCUSSION
In this report, we show that SubAB produced by E. coli O29 had strong vacuolation activity. Vacuolation was induced by a SubAB mutant without subtilase-like serine protease activity, suggesting that there is a functional site for vacuolation, distinct from the serine protease-active site. Recombinant SubA was not necessary for vacuolation. Recombinant SubB alone induced vacuolation. One structural difference from that reported by Paton et al. is that glutamic acid 102 of our B subunit was replaced with lysine. As this amino acid substitution may be critical to the vacuolating activity of SubB, we made a point mutation in subB resulting in the recombinant SubB(E102K) mutant. SubB(E102K) (8 μg/ml) induced vacuolation in ∼80% of cells (data not shown); thus, the amino acid difference was not responsible for the vacuolating activity. Vacuolation was not found with another AB5 toxin (e.g., cholera toxin B subunit did not induce vacuolation when added to Vero cells at 20 μg/ml [data not shown]); therefore, vacuolation of SubB is not a general phenomenon of AB5 toxins. Vacuolation induced by bacterial toxins was recently examined using VacA produced by Helicobacter pylori (4, 7, 18, 19). Sequence similarity between the functional vacuolating domain of VacA (37) and that of SubB was not found. However, similar to VacA, V-ATPase was necessary to induce vacuolation by SubB, suggesting that vacuoles may be formed by a similar mechanism. Vacuolation resulted in cell damage; however, a larger amount was required compared to that required for protein synthesis inhibition.
In this report, we also show that cell cytotoxicity induced by ng/ml amounts of toxin was associated with protein synthesis inhibition. About 20 min (Fig. 3B) was required to abolish protein synthesis, consistent with the time necessary for the delivery of toxin. This was also suggested by the experiment using biotin- or Cy3-labeled toxin (Fig. 7). Some other AB subunit toxins also suppress protein synthesis. One is Stx, a member of bacterial toxins produced by Shigella dysenteriae and Stx-producing E. coli. The Stx A subunit has RNA N-glycosidase activity (5), which is responsible for inhibiting protein synthesis; the B5 subunits of Stx bind to the receptor (e.g., globotriaosyl ceramide [Gb3]) (13, 35). Other toxins which inhibit protein synthesis include diphtheria toxin and pseudomonal exotoxin A, which inactivate elongation factor 2 through ADP ribosylation (9, 20). In both cases, toxins are endocytosed after binding to their respective cell surface receptors and are transported to different intracellular destinations. Stx internalizes by endocytosis into early endosomes, traffics by retrograde movement from the Golgi to the ER, and then crosses the vesicle membrane into cytosol (27, 29). By contrast, diphtheria toxin is internalized by endocytosis and moves across endosome membranes into cytosol (26, 34). The toxins then exert their effects in the cytosol. This trafficking caused a delay in toxin action, similar to that seen with the inhibition of protein synthesis by SubAB. Confocal microscopic observation (Fig. 5B) suggested that SubAB did not traffic in a retrograde manner and might instead traffic directly into cytosol from endosomal membranes.
SubAB is unique because A and B subunits are necessary for the inhibition of protein synthesis both in vivo and in vitro. SubA alone did not inhibit protein synthesis in vivo (Fig. 6) and did not cause cell damage. SubA delivery into cells using Chariot (Active Motif) also failed to induce cytotoxicity (data not shown), suggesting that SubB is necessary not only to internalize SubA into cells but perhaps also to stabilize SubA activity; alternatively, SubB may be necessary for SubA to interact with its target protein or SubA may not be freely accessible to the target protein without SubB.
Subtilases are a superfamily of subtilisin-like serine proteases produced by archaea, bacteria, fungi, yeast, and higher eukaryotes. They are quite common in gram-positive bacteria (e.g., Bacillus species) (25, 32). They have highly similar arrangements of His, Asp, and Ser residues, which form a catalytic triad. SubA has a putative catalytic triad, and serine replacement with alanine in the triad resulted in the loss of protein synthesis inhibitory activity. We tried to detect the protease activity of the toxin using azocasein and some synthetic peptides. However, we failed to find a substrate for this toxin. Serine protease inhibitors, phenylmethylsulfonyl fluoride, aprotinin, and leupeptin, did not block the effects of SubAB on protein synthesis (data not shown). We are trying to determine whether this toxin can digest proteins in microsomal or cytosolic fractions. However, no differences in proteins were found between the samples with and those without toxin. If SubA digestion cleaves only a small peptide from the target protein or if the protein was present at low concentrations, it might be difficult to detect the difference. Paton et al. also examined the protease activity of SubAB using collagen or fibronectin, but the toxin did not cleave them (23). They showed that all three of the amino acids in the catalytic triad were important for the cytotoxic activity, so SubAB might be a subtilase-like serine protease. Based on our data, SubA proteolytic activity might have a very limited substrate specificity.
Acknowledgments
This work was supported by Grants-in-Aid of Scientific Research from the Ministry of Education, Science and Culture of Japan. J. Moss was supported by the International Research Program, NIH/NHLBI.
The authors thank I. Kato for useful discussions and critical review of the manuscript. We thank K. Okamoto, Okayama University, for his advice on the assay of protease activity. We thank S. Yamazaki, Osaka Prefecture University, for the supply of Stx-producing strains. We thank R. Komine and A. Kiuchi for technical assistance.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Arai, H., G. Terres, S. Pink, and M. Forgac. 1988. Topography and subunit stoichiometry of the coated vesicle proton pump. J. Biol. Chem. 263:8796-8802. [PubMed] [Google Scholar]
- 2.Beutin, L., M. A. Montenegro, I. Ørskov, F. Ørskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 4.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 5.Endo, Y., K. Mitsui, M. Motizuki, and K. Tsurugi. 1987. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 262:5908-5912. [PubMed] [Google Scholar]
- 6.Forgac, M. 1992. Structure and properties of the coated vesicle proton pump. Ann. N. Y. Acad. Sci. 671:273-283. [DOI] [PubMed] [Google Scholar]
- 7.Fujikawa, A., D. Shirasaka, S. Yamamoto, H. Ota, K. Yahiro, M. Fukada, T. Shintani, A. Wada, N. Aoyama, T. Hirayama, H. Fukamachi, and M. Noda. 2003. Mice deficient in protein tyrosine phosphatase receptor type Z are resistant to gastric ulcer induction by VacA of Helicobacter pylori. Nat. Genet. 33:375-381. [DOI] [PubMed] [Google Scholar]
- 8.Garred, O., B. van Deurs, and K. Sandvig. 1995. Furin-induced cleavage and activation of Shiga toxin. J. Biol. Chem. 270:10817-10821. [DOI] [PubMed] [Google Scholar]
- 9.Iglewski, B. H., and D. Kabat. 1975. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc. Natl. Acad. Sci. USA 72:2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inward, C. D., A. J. Howie, M. M. Fitzpatrick, F. Rafaat, D. V. Milford, and C. M. Taylor for the British Association for Paediatric Nephrology. 1997. Renal histopathology in fatal cases of diarrhoea-associated haemolytic uraemic syndrome. Pediatr. Nephrol. 11:556-559. [DOI] [PubMed] [Google Scholar]
- 11.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, and B. T. Steele. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet 2:1299-1300. [DOI] [PubMed] [Google Scholar]
- 13.Lingwood, C. A., H. Law, S. Richardson, M. Petric, J. L. Brunton, S. De Grandis, and M. Karmali. 1987. Glycolipid binding of purified and recombinant Escherichia coli produced verotoxin in vitro. J. Biol. Chem. 262:8834-8839. [PubMed] [Google Scholar]
- 14.Marquez-Sterling, N., I. M. Herman, T. Pesacreta, H. Arai, G. Terres, and M. Forgac. 1991. Immunolocalization of the vacuolar-type (H+)-ATPase from clathrin-coated vesicles. Eur. J. Cell Biol. 56:19-33. [PubMed] [Google Scholar]
- 15.Miyake, M., E. Utsuno, and M. Noda. 2000. Binding of avian ovomucoid to Shiga-like toxin type 1 and its utilization for receptor analog affinity chromatography. Anal. Biochem. 281:202-208. [DOI] [PubMed] [Google Scholar]
- 16.Morinaga, N., Y. Iwamaru, K. Yahiro, M. Tagashira, J. Moss, and M. Noda. 2005. Differential activities of plant polyphenols on the binding and internalization of cholera toxin in vero cells. J. Biol. Chem. 280:23303-23309. [DOI] [PubMed] [Google Scholar]
- 17.O'Brien, A. D., and R. K. Holmes. 1987. Shiga and Shiga-like toxins. Microbiol. Rev. 51:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papini, E., E. Gottardi, B. Satin, M. de Bernard, P. Massari, J. Telford, R. Rappuoli, S. B. Sato, and C. Montecucco. 1996. The vacuolar ATPase proton pump is present on intracellular vacuoles induced by Helicobacter pylori. J. Med. Microbiol. 45:84-89. [DOI] [PubMed] [Google Scholar]
- 19.Papini, E., B. Satin, C. Bucci, M. de Bernard, J. L. Telford, R. Manetti, R. Rappuoli, M. Zerial, and C. Montecucco. 1997. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J. 16:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pappenheimer, A. M., Jr. 1977. Diphtheria toxin. Annu. Rev. Biochem. 46:69-94. [DOI] [PubMed] [Google Scholar]
- 21.Paton, A. W., and J. C. Paton. 1999. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect. Immun. 67:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, A. W., and J. C. Paton. 2005. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J. Clin. Microbiol. 43:2944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polgar, L. 2005. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62:2161-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratts, R., H. Zeng, E. A. Berg, C. Blue, M. E. McComb, C. E. Costello, J. C. vanderSpek, and J. R. Murphy. 2003. The cytosolic entry of diphtheria toxin catalytic domain requires a host cell cytosolic translocation factor complex. J. Cell Biol. 160:1139-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saleh, M. T., J. Ferguson, J. M. Boggs, and J. Gariepy. 1996. Insertion and orientation of a synthetic peptide representing the C-terminus of the A1 domain of Shiga toxin into phospholipid membranes. Biochemistry 35:9325-9334. [DOI] [PubMed] [Google Scholar]
- 28.Sandvig, K., O. Garred, K. Prydz, J. V. Kozlov, S. H. Hansen, and B. van Deurs. 1992. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature 358:510-512. [DOI] [PubMed] [Google Scholar]
- 29.Sandvig, K., and B. van Deurs. 1996. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 76:949-966. [DOI] [PubMed] [Google Scholar]
- 30.Savarino, S. J., A. Fasano, D. C. Robertson, and M. M. Levine. 1991. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J. Clin. Investig. 87:1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, H., J. Scheef, H. I. Huppertz, M. Frosch, and H. Karch. 1999. Escherichia coli O157:H7 and O157:H(−) strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siezen, R. J., and J. A. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 5:1817-1822. [DOI] [PubMed] [Google Scholar]
- 34.Umata, T., and E. Mekada. 1998. Diphtheria toxin translocation across endosome membranes. A novel cell permeabilization assay reveals new diphtheria toxin fragments in endocytic vesicles. J. Biol. Chem. 273:8351-8359. [DOI] [PubMed] [Google Scholar]
- 35.Waddell, T., S. Head, M. Petric, A. Cohen, and C. Lingwood. 1988. Globotriaosyl ceramide is specifically recognized by the Escherichia coli verocytotoxin 2. Biochem. Biophys. Res. Commun. 152:674-679. [DOI] [PubMed] [Google Scholar]
- 36.Williams, J. M., N. Lea, J. M. Lord, L. M. Roberts, D. V. Milford, and C. M. Taylor. 1997. Comparison of ribosome-inactivating proteins in the induction of apoptosis. Toxicol. Lett. 91:121-127. [DOI] [PubMed] [Google Scholar]
- 37.Ye, D., D. C. Willhite, and S. R. Blanke. 1999. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J. Biol. Chem. 274:9277-9282. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama, R., Y. Fujii, Y. Noguchi, T. Nomura, M. Akita, K. Setsu, S. Yamamoto, and K. Okamoto. 2002. Physicochemical and biological properties of an extracellular serine protease of Aeromonas sobria. Microbiol. Immunol. 46:383-390. [DOI] [PubMed] [Google Scholar]