Abstract
A biphasic developmental cycle whereby highly resistant small-cell variants (SCVs) are generated from large-cell variants (LCVs) is considered fundamental to the virulence of Coxiella burnetii, the causative agent of human Q fever. In this study a proteome analysis of C. burnetii developmental forms was conducted to provide insight into their unique biological and immunological properties. Silver-stained gels of SCV and LCV lysates separated by two-dimensional (2-D) gel electrophoresis resolved over 675 proteins in both developmental forms. Forty-eight proteins were greater than twofold more abundant in LCVs than in SCVs, with six proteins greater than twofold more abundant in SCVs than in LCVs. Four and 15 upregulated proteins of SCVs and LCVs, respectively, were identified by mass spectrometry, and their predicted functional roles are consistent with a metabolically active LCV and a structurally resistant SCV. One-dimensional and 2-D immunoblots of cell form lysates probed with sera from infected/vaccinated guinea pigs and convalescent-phase serum from human patients who had recovered from acute Q fever, respectively, revealed both unique SCV/LCV antigens and common SCV/LCV antigens that were often differentially synthesized. Antigens recognized during human infection were identified by mass spectroscopy and included both previously described immunodominant proteins of C. burnetii and novel immunogenic proteins that may be important in the pathophysiology of clinical Q fever and/or the induction of protective immunity.
Coxiella burnetii is the etiologic agent of an acute, disabling influenza-like illness in humans termed Q fever. In rare instances chronic disease can occur, usually presenting as hepatitis or endocarditis. C. burnetii infects a wide range of vertebrate and invertebrate hosts and is found worldwide with the exception of New Zealand. Most human Q fever cases are acquired through inhalation of contaminated aerosols originating from products shed from chronically infected domestic livestock. Indeed, massive numbers of coxiellae (>109/g) are present in the products of parturition (reviewed in reference 19).
C. burnetii has an obligate requirement for replication in a eukaryotic host cell vacuole with lysosomal characteristics (15). Resistance to this harsh intracellular environment correlates with remarkable environmental stability, a trait that distinguishes C. burnetii from other obligate intracellular bacteria. The pathogen is highly resistant to osmotic shock, elevated temperature, desiccation, UV light, and various chemical disinfectants (21, 36). Environmental stability, along with aerosol transmission and an infectious dose approaching one organism, has resulted in classification of C. burnetii as a CDC category B biothreat. The spore-like characteristics of C. burnetii are attributed to a stable small-cell variant (SCV) that arises as part of a C. burnetii biphasic developmental cycle (22). SCVs are rod shaped and 0.2 to 0.5 μm in length and have an electron-dense condensed chromatin with an array of intracytoplasmic membranes. Large-cell variants (LCVs) can exceed 1 μm in length and are pleomorphic with a dispersed chromatin (12, 22). C. burnetii preparations enriched in LCVs are more metabolically active in vitro than those enriched in SCVs (21).
By infecting Vero cells with SCVs, we have recently established the kinetics of C. burnetii morphological development and defined the LCV as the replicative form of the pathogen (5). The C. burnetii growth cycle has defined lag, log, and stationary phases (5). SCV-to-LCV morphogenesis occurs during a lag phase lasting approximately 2 days. Exponential replication of LCVs ensues over the next 4 days with LCVs condensing to SCVs in significant numbers during a stationary phase that starts approximately 6 days postinfection (p.i.).
Elucidation of the molecular events that regulate C. burnetii development has been hampered by an inability to genetically manipulate the organism; however, some progress has been made in identifying developmental-form-specific proteins. McCaul et al. (20) identified a 29.5-kDa immunogenic protein termed P1 that is differentially synthesized by the LCV. The encoding gene was later cloned by Varghees et al. (33), who described P1 as having porin activity. Additional proteins that are dramatically enriched in the LCV include EF-Tu and EF-Ts (26) and RpoS (28). Two DNA-binding proteins, ScvA and Hq1, are preferentially expressed by the SCV (13, 14).
Intracellular morphological differentiation to generate environmentally stable SCV and replicatively active LCV developmental forms is considered central to Coxiella virulence. However, the unique or upregulated proteins that confer the distinctive ultrastructural and biological characteristics of SCVs and LCVs are still largely unknown, as are the immunogenic constituents of developmental forms that are recognized during infection. The recent elucidation of the genome sequence of the Nine Mile isolate (RSA493) of C. burnetii (27) now allows a proteomic approach, i.e., two-dimensional (2-D) gel electrophoresis combined with peptide mass mapping and fingerprinting to address these problems. Using these approaches, we conducted an analysis of C. burnetii SCV and LCV protein composition and the developmental form antigens recognized during infection.
MATERIALS AND METHODS
Cultivation and purification of C. burnetii.
C. burnetii Nine Mile Crazy (RSA514) was grown in African green monkey kidney (Vero) fibroblasts (CCL-81; American Type Culture Collection) cultivated in RPMI (Invitrogen) supplemented with 10% fetal bovine serum. This strain was isolated from the placenta of a guinea pig that had been chronically infected for 343 days with the reference Nine Mile phase I strain (RSA439) (9). Bacteria were purified from infected cells at 7 days p.i. by Renografin gradient centrifugation (37). SCVs and LCVs were separated by equilibrium density centrifugation in 32% cesium chloride (CsCl2) as previously described (14). Cell variants were resuspended in K-36 (0.1 M KCl, 0.015 M NaCl, 0.05 M potassium phosphate, pH 7.0) and stored at −80°C.
Immune sera.
A guinea pig was aerosol infected with 106 C. burnetii Nine Mile bacteria in phase I (RSA493) and bled for immune sera at 7, 14, and 28 days p.i. Guinea pig immune serum was also generated by vaccination and subsequent challenge with live organisms. Specifically, a guinea pig was vaccinated with 40 μg of formalin-fixed Nine Mile phase I bacteria mixed 1:1 in incomplete Freund's adjuvant and boosted 14 days later. After 2 weeks vaccinated animals were aerosol challenged with 106 live Nine Mile phase I bacteria, and immune serum was harvested at 28 days p.i. (Protocols for production of guinea pig immune sera were approved by the Texas A&M University Institutional Animal Care and Use Committee.) Human convalescent-phase sera from two patients who had recovered from acute Q fever were obtained from an Australian serum collection recently acquired by J.E.S. Generation of anti-ScvA serum has been previously described (14).
One-dimensional gel electrophoresis and immunoblotting.
Proteins of CsCl2-purified SCVs and LCVs were separated by polyacrylamide gel electrophoresis as previously described (14) using NextGel polyacrylamide solution (Amresco, Solon, OH). For both one-dimensional and 2-D gels (below), equal amounts of SCV and LCV protein were separated as determined by the RC DC protein assay (Bio-Rad). Gels were stained with a SilverQuest silver staining Kit (Invitrogen). Proteinase K digestion of C. burnetii proteins was conducted by first solubilizing organisms in lysis buffer (2% sodium dodecyl sulfate, 4% 2-mercaptoethanol, 10% glycerol, 1 M Tris [pH 8.8], and 0.2% bromophenol blue) at 100°C for 5 min. Samples were then cooled and treated with 20 μg of recombinant proteinase K (Roche Applied Science) for 4 h at 60°C while being mixed at 700 rpm in a Thermomixer (Brinkmann Instruments, Inc.). For immunoblotting, proteins were electrophoretically transferred to Immobilon P (Millipore) and blocked in phosphate-buffered saline (10 mM sodium phosphate, 150 mM sodium chloride, pH 7.4) plus 5% nonfat dry milk and 0.1% Tween 20. Guinea pig immune sera and human convalescent-phase sera were used at a 1:500 dilution, and rabbit anti-ScvA was used at a 1:4,000 dilution. Peroxidase-conjugated anti-guinea pig (Sigma), anti-human, or anti-rabbit (ICN) immunoglobulin G secondary antibodies were used at a 1:50,000 dilution. Bound antibodies were detected by chemiluminescence using Supersignal West Pico chemiluminescent substrate (Pierce) and Hyperfilm ECL (Amersham).
2-D electrophoresis.
Purified SCVs and LCVs were solubilized and processed using the Bio-Rad ReadyPrep system. Proteins were focused using pH 4 to 7, pH 7 to 10, or pH 3 to 10 nonlinear ReadyStrips (Bio-Rad). Focused proteins were separated by polyacrylamide gel electrophoresis and stained with silver as described above. Wet gels were scanned using a Hewlett-Packard ScanJet 5300C, and digitized images were analyzed using ImageJ (written by W. S. Rasband at the U.S. National Institutes of Health, Bethesda, MD, and available from http://rsb.info.nih.gov/ij/) and Phoretix 2-D Evolution (v2005) software (Nonlinear Dynamics, Newcastle-upon-Tyne, United Kingdom). Scanned images were converted to grayscale and inverted, and the integrated pixel density of identical areas of corresponding SCV and LCV spots was calculated. (The background staining of individual gels, as determined by averaging the minimum intensity from three different gel areas, was subtracted from each calculation.) Proteins were considered uniquely or differentially expressed between developmental forms if a greater-than-twofold difference in pixel density was observed.
Mass spectrometry.
Gel slices were destained, buffer equilibrated, dehydrated, and then rehydrated with 25 μl of porcine trypsin (Promega) at 20 μg/ml in 25 mM ammonium bicarbonate (pH 8.0). After an overnight incubation at 37°C, peptides were extracted and dried, and lysines were converted to homoarginine by adding 10 μl of 1 M O-methylisourea sulfate (pH 11) (Sigma) and incubating the mixture at 37°C for 2 h. Five microliters of 5% trifluoroacetic acid was added to the modified peptides, and they were subsequently purified using C18 Zip-Tips (Millipore). Purified peptides were dried and then rehydrated with 1.5 μl of matrix solution consisting of 2.5 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma) in a solution of 40% acetonitrile, 0.1% trifluoroacetic acid. An 0.5-μl volume of the peptide-matrix solution was spotted on the sample plate and allowed to air dry. Matrix-assisted laser desorption ionization-time of flight (TOF) mass spectra and peptide mass fingerprints were collected by using an Applied Biosystems/MDS Sciex 4800 matrix-assisted laser desorption ionization TOF/TOF mass spectrometry workstation (Applied Biosystems, Framington, MA). The resulting peptide peak list and peptide fragment list were submitted for a search of the CDS combined database (Celera Discovery System) by using the Mascot (Matrix Science) search software.
Transmission electron microscopy.
Purified SCVs and LCVs were fixed overnight at 4°C with 2.5% glutaraldehyde-4% paraformaldehyde in 100 mM sodium cacodylate buffer (pH 7.2). Cells were postfixed with 0.5% osmium tetroxide-0.8% potassium ferricyanide in 100 mM sodium cacodylate buffer followed by 1% tannic acid in distilled water. Samples were stained overnight with 1% uranyl acetate, washed with distilled water, dehydrated with a graded ethanol series, and embedded in Spurr's resin. Thin sections were stained with 1% uranyl acetate and Reynold's lead citrate. Sections were viewed at 80 kV on a Philips CM-10 transmission electron microscope (FEI, Hillsboro, OR). Digital images were acquired with an AMT digital camera (AMT, Chazy, NY) and processed with Adobe Photoshop (version 7.0; Adobe Systems, Mountain View, CA).
RESULTS
Purification of SCVs and LCVs.
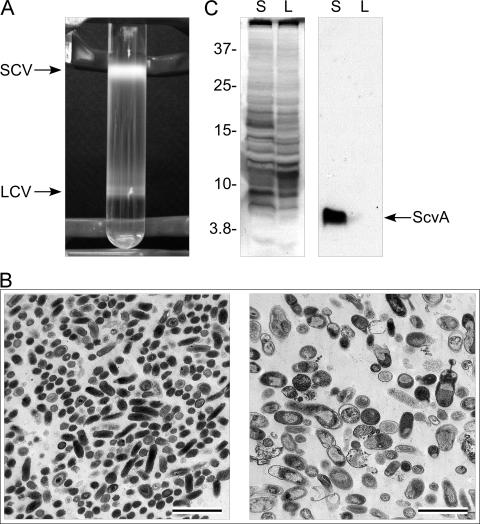
Earlier electrophoresis studies revealed distinct protein heterogeneity between SCVs and LCVs (12, 14, 20). To more completely assess the degree of differential protein synthesis by SCVs and LCVs, we conducted a proteomic analysis using 2-D electrophoresis and silver staining. Highly pure preparations of sample are required for reproducible proteome surveys. In a previous study, we fractionated SCVs and LCVs of the Nine Mile phase I strain by exploiting their different buoyant densities in CsCl2 equilibrium gradients (35). While nearly homogeneous populations of developmental forms were obtained, yields were low due to the poor infectivity of phase I C. burnetii for tissue culture cells, a characteristic partly attributed to its hydrophilic, full-length lipopolysaccharide (LPS) (23, 37). The Nine Mile phase II variant is approximately 500 times more infectious for cultured cells (23); however, a severely truncated LPS makes this isolate highly hydrophobic, which causes it to aggregate in CsCl2 (12). We therefore investigated whether the Nine Mile Crazy variant, which produces an intermediate-length LPS and is roughly 20 times more infectious for cell culture than the Nine Mile phase I strain (23), could be separated by CsCl2 equilibrium density centrifugation. Clear separation of Nine Mile Crazy was achieved with LCVs banding at a higher density than SCVs due to their presumed permeability to CsCl2 (35) (Fig. 1A). Although C. burnetii was harvested at 7 days p.i., when the intracellular numbers of SCVs and LCVs are roughly equal (5), the yield of purified SCVs always greatly exceeded that of LCVs, a result attributed to the fragile nature of the LCV. Ultrastructural analysis by transmission electron microscopy showed that nearly homogeneous SCVs and LCVs were isolated using this procedure (Fig. 1B). Prototypic SCVs were observed that were 0.2 to 0.5 μm in length and rod shaped and with an electron-dense condensed chromatin (Fig. 1B, left panel). The LCV ultrastructure was also typical with cell forms approximately 1 μm in length, pleomorphic, and displaying a granular appearance due to a dispersed chromatin (Fig. 1B, right panel). The homogeneity of SCV and LCV fractions was further confirmed by immunoblotting for ScvA, an SCV-specific protein (5, 14, 28) (Fig. 1C). ScvA was detected only in lysates of fractionated SCV.
FIG. 1.
Purification and characterization of the C. burnetii SCV and LCV. (A) The SCV and LCV of the C. burnetii Nine Mile Crazy variant were separated by equilibrium density centrifugation in 32% cesium chloride (CsCl2). LCVs (lower band) are denser than SCVs due to their permeability to CsCl2 (12). (B) Transmission electron micrographs of fractionated SCVs (left panel) and LCVs (right panel) showing the characteristic size and ultrastructure of developmental forms. Bars, 2.0 μm. (C) Confirmation of SCV and LCV fractionation by immunoblotting for ScvA. Shown are a silver-stained polyacrylamide gel of separated SCV (S) and LCV (L) lysates (left panel) and a parallel immunoblot (right panel) probed with polyclonal antiserum to ScvA, an SCV-specific protein (14). Molecular mass markers at the left are expressed in kDa.
Comparison of SCV and LCV proteomes.
Lysates of purified cell forms were separated in the first dimension using immobilized pH gradient strips having a pH range of 4 to 7 or 7 to 10. This protocol was employed to better resolve the C. burnetii proteome and to facilitate detection of low-abundance proteins. In both pH ranges combined, over 675 protein spots were detected in 2-D gels of both SCV and LCV lysates (Fig. 2).
FIG. 2.
Silver-stained 2-D gel profiles of SCV and LCV proteins. Equal amounts of SCV and LCV lysates were subjected to 2-D gel electrophoresis using pH 4 to 7 and pH 7 to 10 isoelectric focusing strips. Black circles denote proteins that are greater than twofold more abundant in LCVs than in SCVs as determined by scanning densitometry. (Black circles are placed at the same coordinates on SCV 2-D gels to facilitate protein level comparisons.) White circles denote proteins that are greater than twofold more abundant in SCVs than in LCVs as determined by scanning densitometry. (White circles are placed at the same coordinates on LCV 2-D gels to facilitate protein level comparisons.) Numbered circles denote proteins that were identified by mass spectrometry (Table 1). Molecular mass markers at left are expressed in kDa.
Based on densitometry, 48 proteins were greater than twofold more abundant in LCVs than in SCVs, with 32 and 16 of these proteins having isoelectric points between pH 4 and 7 and pH 7 and 10, respectively. Fifteen upregulated LCV proteins were identified by mass spectrometry, two of which were proteins of unknown function (Table 1). In general, identified proteins had predicted functions in general bacterial physiology including transcription (N utilization protein A [NusA]), translation (ribosomal protein S1 [RpsA], ribosomal protein L9 [RplI], and elongation factor EF-Tu), cell division (cell division protein FtsZ), chromosome partitioning (segregation and condensation protein B [ScpB] and chromosome partitioning protein ParB), riboflavin biosynthesis (6,7,-dimethyl-8-ribityllumazine synthase [RibH]), and protein folding (60-kDa chaperone [GroEL] and chaperone protein HtpG), and their upregulation is consistent with the LCV being more metabolically and replicatively active than the SCV (5, 21). Upregulation of stringent starvation protein (SspA) (10), universal stress protein A (UspA) (24), and thioredoxin peroxidase (Bcp) (38) may reflect a stress response to the lysosomal environment by the LCV.
TABLE 1.
Identification of differentially expressed SCV and LCV proteins
| No. | SCV/LCVa | Gene | Description | Predicted mass (kDa) | Predicted pI |
|---|---|---|---|---|---|
| 1 | LCV | CBU0528 | Ribosomal protein S1 (RpsA) | 62.1 | 5.0 |
| 2 | LCV | CBU0867 | Ribosomal protein L9 (RplI) | 16.6 | 6.6 |
| 3 | LCV | CBU1433 | N utilization substance protein A (NusA) | 56.2 | 4.3 |
| 4 | LCV | CBU0236 | Elongation factor EF-Tu | 43.5 | 5.2 |
| 5 | LCV | CBU1718 | 60-kDa chaperone (GroEL) | 58.3 | 4.9 |
| 6 | LCV | CBU0309 | Chaperone protein HtpG (HtpG) | 72.8 | 5.0 |
| 7 | LCV | CBU1747 | Stringent starvation protein A (SspA) | 24.4 | 5.6 |
| 8 | LCV | CBU0141 | Cell division protein FtsZ (FtsZ) | 40.7 | 4.3 |
| 9 | LCV | CBU1060 | Segregation and condensation protein (ScpB) | 23.7 | 4.4 |
| 10 | LCV | CBU0658 | Hypothetical protein | 16.0 | 4.5 |
| 11 | LCV | CBU1754 | Hypothetical protein | 22.3 | 4.7 |
| 12 | LCV | CBU1927 | Chromosome partitioning protein ParB (ParB) | 32.0 | 9.4 |
| 13 | LCV | CBU0963 | Thioredoxin peroxidase (Bcp) | 16.9 | 8.0 |
| 14 | LCV | CBU0648 | DMRLb synthase (RibH) | 16.6 | 7.3 |
| 15 | LCV | CBU1916 | Universal stress protein A (UspA) | 15.9 | 7.2 |
| 16 | SCV | CBU0090 | TolB protein (TolB) | 47.8 | 8.9 |
| 17 | SCV | CBU2025 | Cystathionine beta-lyase (MetC) | 42.7 | 8.3 |
| 18 | SCV | CBU2079 | Hypothetical | 14.2 | 8.6 |
| 19 | SCV | CBU1502 | GTP binding protein Era homolog (Era) | 33.9 | 9.4 |
Unique to or upregulated greater than twofold in the LCV or the SCV.
DMRL, 6,7,-dimethyl-8-ribityllumazine.
Six proteins were detected that were greater than twofold more abundant in SCVs than in LCVs, all having isoelectric points between 7 and 10 (Fig. 2). Four of these were identified by mass spectrometry as TolB, a protein involved in outer membrane stability (17); GTP binding protein Era homolog (Era) (3); cystathionine beta-lyase (MetC); and a hypothetical protein (Table 1). C. burnetii genes encode an unusually high number of basic proteins with ∼24% having a predicted isoelectric point greater than 10 (27). Obvious stacking of unresolved high-isoelectric-point proteins was observed at the basic end of pH 7 to 10 2-D gels, particularly with SCV lysates (Fig. 2).
Immunodominant SCV and LCV antigens recognized by guinea pigs.
Differentially expressed components of SCVs and LCVs might include immunogenic determinants that play important roles in the adaptive immune response to C. burnetii infection. To date, just one developmentally regulated antigen has been identified: P1, a porin protein preferentially expressed by LCVs (33). To determine the extent of differential antigen synthesis by SCVs and LCVs, immunoblots of developmental form lysates were probed with sera from infected and vaccinated/challenged guinea pigs and with sera from human patients who had recovered from acute Q fever.
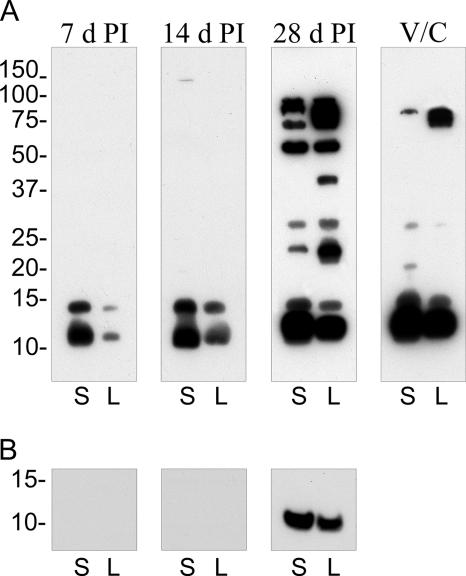
An increasingly complex array of antigens was recognized by serum derived from a single infected guinea pig at 7, 14, and 28 days p.i. (Fig. 3A). Two predominant low-molecular-mass antigens of 12.5 and 15 kDa were recognized throughout the course of infection in both SCV and LCV lysates. LPS of the Nine Mile Crazy variant migrates on a polyacrylamide gel with a molecular mass of ∼12 to 14 kDa (2). To determine if serum reactivity to LPS was associated with either low-molecular-mass antigen, lysates were digested with proteinase K before immunoblotting. A proteinase-resistant antigen of 12.5 kDa was detected in SCV and LCV lysates with 28-day-p.i. serum, but not 7- or 14-day-p.i. serum (Fig. 3b), that comigrated with purified C. burnetii Crazy LPS (data not shown). Fourteen-day-postinfection serum recognized an additional 150-kDa antigen in SCVs. Serum obtained at 28 days p.i. recognized 12.5-, 15-, 24-, 30-, 55-, 75-, 92-, and 94-kDa antigens in both SCV and LCV fractions. This serum also uniquely recognized two LCV antigens with approximate molecular masses of 42 and 84 kDa. (The four bands comprising the 75- to 94-kDa LCV reactivity are evident with shorter exposures.) Guinea pig immune serum generated by vaccination with formalin-fixed C. burnetii and subsequent challenge with viable organisms recognized fewer antigens in both SCVs and LCVs than did 28-day-p.i. serum. Serum reactivity was observed with the 12.5-, 15-, 30-, and 92-kDa common LCV/SCV antigens, and the 84-kDa LCV-specific antigen, that was recognized with 28-day-p.i. serum. Also detected was a novel 22-kDa antigen specific to the SCV. No SCV or LCV antigens were recognized by sera from naïve guinea pigs (data not shown). Collectively, these results indicate that SCVs and LCVs express unique antigens. Moreover, based on immunoblot signal intensities, some shared antigens are associated with developmental forms in different amounts.
FIG. 3.
Temporal analysis of antibody development against SCV and LCV antigens. (A) Immunoblots containing an equal amount of SCV (S) and LCV (L) lysates were probed with sera derived from a C. burnetii-infected guinea pig at 7, 14, and 28 days p.i. or with serum derived from a guinea pig vaccinated with formalin-fixed C. burnetii and then challenged with live organisms (V/C) (see Materials and Methods). (B) Immunoblot of proteinase K-digested lysates probed with infected guinea pig sera. Depicted is the region of the immunoblot showing a digestion-resistant product that comigrates with Nine Mile Crazy LPS. Film exposures for all immunoblots were conducted for 30 s. Molecular mass markers at left are expressed in kDa.
SCV and LCV antigens recognized by convalescent-phase sera from patients who recovered from acute Q fever.
Convalescent-phase immune sera from two patients who had recovered from acute Q fever (2036 and 3004) were analyzed for immunoreactivity against SCV and LCV antigens separated by 2-D gel electrophoresis (Fig. 4). Serum 2036 recognized 20 SCV and 26 LCV antigens. Fifteen antigens were common to both cell variants, five were unique to SCV, and 11 were unique to LCV. By overlaying films of immunoblots with parallel silver-stained gels, we identified by mass spectrometry 10 strongly immunogenic antigens that were generally shared by SCVs and LCVs. Identified common cell variant antigens recognized by patient serum 2036 were GroEL, ribosomal protein L7/L12 (RplJ), acute disease antigen A (AdaA), trigger factor (Tig), and 3-oxoacyl-acyl carrier protein synthase II (FabF) (Table 2). Identified LCV-specific antigens included Bcp and repressor protein C2. Serum 3004 recognized 24 SCV and 27 LCV antigens. Thirteen antigens were common to both developmental forms with 11 and 14 antigens unique to SCV and LCV, respectively. Identified antigens common to both cell variants included GroEL, elongation factor EF-Tu, RplJ, ATP synthase F-1 alpha subunit (AtpA), FabF, and isocitrate dehydrogenase (Icd) (Table 2). Tig was recognized only in LCV lysates. We were unable to identify SCV-specific antigens recognized by either serum.
FIG. 4.
Two-dimensional immunoblots of SCV and LCV lysates showing antigens recognized by convalescent-phase sera from two human patients who had recovered from Q fever. Isoelectric focusing of proteins was conducted using pH 3 to 10 nonlinear (NL) strips. Immunoreactive spots were cross-referenced with parallel silver-stained gels. Stained proteins that clearly correlated with immunoreactive spots were processed for mass spectrometry. Identified antigens are denoted with circles and numbers. Spots on other gels that have coordinates identical to those of mass spectrometry-identified antigens are denoted with circles and asterisked numbers. Identified antigens are listed in Table 2. (Some immunoreactive protein spots are difficult to visualize in the figure but are clearly visible in the original films.) Molecular mass markers at left are expressed in kDa.
TABLE 2.
Identification of immunoreactive SCV and LCV proteins
| No. | Gene | Description | Predicted mass (kDa) | Predicted pI | Serum 2036
|
Serum 3004
|
||
|---|---|---|---|---|---|---|---|---|
| Cell variant MS/MS identificationa | Spot in other cell variant lysateb | Cell variant MS/MS identificationa | Spot in other cell variant lysateb | |||||
| 1 | CBU1718 | 60-kDa chaperone (GroEL) | 58.3 | 4.9 | S | + | S+L | NAc |
| 2 | CBU0236 | Elongation factor EF-Tu | 43.5 | 5.2 | NA | S+L | NA | |
| 3 | CBU0229 | Ribosomal protein L7/L12 (RplJ) | 13.2 | 4.4 | S | + | S+L | NA |
| 4 | CBU1943 | ATP synthase F-1 alpha subunit (AtpA) | 58.8 | 6.0 | NA | L | + | |
| 5 | CBU1416 | Repressor protein C2 | 24.3 | 7.6 | L | NA | ||
| 6 | CBU0952 | Acute disease antigen A (AdaA) | 26.0 | 9.3 | L | + | NA | |
| 7 | CBU0963 | Thioredoxin peroxidase (Bcp) | 16.9 | 8.0 | L | NA | ||
| 8 | CBU0737 | Trigger factor (Tig) | 50.2 | 5.1 | +d | L | ||
| 9 | CBU0497 | 3-Oxoacyl-acyl carrier protein synthase II (FabF) | 44.1 | 5.5 | +d | L | + | |
| 10 | CBU1200 | Isocitrate dehydrogenase (Icd) | 46.6 | 6.4 | NA | L | + | |
Protein identified by mass spectrometry in lysates of the SCV (S), the LCV (L), or both (S+L).
Corresponding spot present in immunoblot of lysates derived from other cell variants is indicated with a +.
NA, not applicable.
Immunoreactive spot at the same coordinates in both SCV and LCV 2-D immunoblots as those of the protein identified by serum 3004 in the LCV.
DISCUSSION
We have previously demonstrated by gel electrophoresis a few differences in protein composition between C. burnetii SCV and LCV developmental forms (12, 14); however, attempts to extensively identify developmental form-specific proteins were not undertaken due to low yields of purified cell variants coupled with the lack of sensitive methods for protein identification. Moreover, previous studies were hampered by poor solubilization of cell variants, with fewer than 200 SCV and LCV proteins resolved by 2-D gel electrophoresis and silver staining (12). In this study we exploited improved yields and solubilization of C. burnetii developmental forms along with a dual-pH-range 2-D electrophoresis procedure to develop a more complete proteome map of the SCV and the LCV by more than tripling the number of proteins resolved by silver staining over a previous study (12). Forty-eight proteins are greater than twofold more abundant in LCVs than in SCVs, with six proteins greater than twofold more abundant in SCVs than in LCVs. Thus, the morphological differences observed between C. burnetii developmental forms clearly correlate with differences in protein composition.
Using mass spectroscopy, we identified 15 of the 48 proteins differentially synthesized by the LCV. These proteins are generally involved in cell division, RNA synthesis, and protein synthesis/processing. As the more metabolically active cell form of C. burnetii (5, 21), the LCV would be expected to have increased ribosome content. Consistent with this idea, we identified upregulation by the LCV of N utilization substance protein A (NusA), a protein required for robust transcription of rRNA (34), and the ribosomal proteins S1 and L9. Further enhancing the translational machinery of the LCV is upregulation of EF-Tu, a finding in keeping with a previous study (26). We have recently confirmed that the LCV is the replicatively active cell form of the C. burnetii developmental cycle (5). As such, and in accordance with increased metabolic activity, the LCV upregulates FtsZ, required for bacterial binary fission, and two proteins involved in chromosome partitioning, ScpB and ParB (11, 30), relative to the SCV.
At first glance, upregulation of stringent starvation protein A (SspA) by the LCV seems counterintuitive as, among other effects, expression of this protein in Escherichia coli results in increased synthesis of the stationary-phase sigma factor RpoS (10). However, we (5) and others (28) have previously shown that RpoS is upregulated by the LCV during exponential growth, where it potentially induces genes involved in protecting C. burnetii against lysosomal stress. Indeed, RpoS induction of hydroperoxidase I in exponentially growing Vibrio vulnificus is critical for survival of this organism during oxidative stress (25). In addition to regulating RpoS levels, SspA is thought to be a global regulator of bacterial virulence genes (10). Along with SspA, elevated levels of Bcp, UspA, GroEL, and HtpG in the LCV also support the idea that this developmental form is responding to lysosomal stress. As a thioredoxin peroxidase, Bcp resides within the general family of thiol-specific antioxidant proteins termed peroxiredoxins. They are abundant proteins in bacteria and serve to detoxify various peroxide compounds (38). In E. coli, UspA is essential for survival of a variety of insults including exposure to hydrogen peroxide (24). While the chaperones GroEL and HtpG are abundant in bacteria growing under normal conditions, they are also upregulated in response to a variety of adverse conditions (6, 29). Moreover, GroEL and HtpG are implicated in protein secretion (16, 32), and we have previously speculated that the LCV is the secretion-competent form of C. burnetii (5).
The most abundant LCV-upregulated protein that we identified is annotated as a hypothetical protein (CBU0658). The Coxiella genome encodes a high proportion of hypothetical proteins (33.7% of coding sequences) (27). It is reasonable to suspect that these play critical roles in C. burnetii morphological development and pathogenesis; however, their functional assignments await future biochemical and genetic analysis. We did not exhaustively attempt to identify by mass spectrometry the remaining subset of LCV-upregulated proteins. However, it likely includes RpoS and EF-Ts, previously described as differentially synthesized by the LCV (35, 37), as LCV-upregulated proteins with molecular masses and pIs approximating those of these proteins are present on 2-D gels.
We identified four proteins differentially synthesized by the SCV with three, TolB, Era homolog, and MetC, having predicted functions. In E. coli, TolB interacts with other Tol proteins to stabilize the outer membrane (17). Elevated TolB levels in the SCV may partially explain this developmental form's noted resistance to physical disruption. The E. coli protein Era is a member of the small regulatory GTPase superfamily (3). Among the proposed functions of Era and its homologs are regulation of the cell cycle and cellular differentiation (3). For example, Era homologs of Bacillus subtilis are required for normal cell division and spore formation (1, 4). Thus, this protein may be a key regulator of SCV morphological differentiation. Upregulation of MetC, a biosynthetic enzyme, by the SCV is puzzling. This enzyme catalyzes the last intermediate of the methionine biosynthetic pathway, converting cystathionine to l-homocysteine. In E. coli, homocysteine globally affects transcription and inhibits growth by perturbing the activity of threonine deaminase, which results in depletion of branched-chain amino acids (8, 31). Consequently, homocysteine may serve a similar signaling role in the SCV to downregulate metabolism. Alternatively, SCV may require more methionine for production of S-adenosylmethionine. Among the many uses of S-adenosylmethionine is production of spermine and spermidine (7). These cationic polyamines bind to DNA which stabilizes the molecule and facilitates chromatin condensation (1), a hallmark of the SCV. Two very basic SCV-specific DNA-binding proteins, ScvA and Hq1, have been previously described (13, 14) that also presumably play roles in chromatin condensation. Interestingly, all SCV-upregulated proteins were found in pH 7 to 10 2-D gels. Moreover, there is an abundance of unresolved SCV proteins that “stack up” at the basic end of pH 7 to 10 2-D gels relative to the LCV. In addition to ScvA and Hq1, these likely include additional SCV-upregulated proteins involved in chromatin condensation or other processes that contribute to the resistance properties of SCV. Resolution and identification of these proteins will require an alternative method of protein fractionation. Similarly to the LCV, the most abundant SCV-upregulated protein based on densitometry is annotated as a hypothetical protein (CBU2079).
P1 is the only surface antigen known to be differentially synthesized by C. burnetii developmental forms (20, 33). Here we extensively examined SCV and LCV antigens recognized in the context of both infection and vaccination with killed organisms. An infected guinea pig developed antibodies to an increasingly complex array of SCV and LCV antigens over the time course of infection. Although different levels of expression are apparent, all SCV antigens recognized at 28 days p.i. are also present in the LCV. A small subset of these common antigens is also recognized by vaccinated animals that were challenged with live organisms. The less complex humoral response elicited by vaccination and subsequent infection is not unexpected and has been reported elsewhere for C. burnetii (40). At least two antigens with approximate molecular masses of 42 and 84 kDa appear unique to the LCV. Like P1, these antigens may be surface exposed and involved in nutrient uptake by the LCV. Moreover, antigenic variation in vivo between the SCV and the LCV may play a role in the immune evasion and persistence of C. burnetii that lead to chronic infections (39).
An infected guinea pig first generates antibodies to 12.5- and 15-kDa antigens with strong recognition of these proteins throughout the course of infection. Similar reactivities are observed with sera derived from vaccinated animals. Based on immunoblot signal intensities, these antigens appear more abundant in the SCV than in the LCV. Diagnosis of Q fever is usually by serological methods with the reference technique being microimmunofluorescence (19). This test employs inactivated whole-cell C. burnetii as antigen. A serodiagnostic test based on recombinant antigen would eliminate the need for cultivation and purification of C. burnetii and would likely be more specific. As the 12.5- and 15-kDa antigens are the predominant antigens recognized during the early stages of infection, they may have utility as serological diagnostic target antigens in the form of recombinant protein. At 28 days p.i., LPS comprises a portion of the 12.5-kDa-antigen reactivity. C. burnetii LPS is strongly immunogenic and also comprises the organism's phase I antigen (41). Recognition of LPS only late in infection is in keeping with the serological phenomenon of phase variation whereby only late antiserum (>20 days p.i.) of an infected animal binds to phase I antigen (i.e., LPS) (23). Consistent with a previous report (20), there appears to be more LPS associated with the SCV than with the LCV.
To develop a more complete understanding of the human humoral response to C. burnetii, we identified immunogenic proteins recognized in the context of human acute Q fever. Similar to the reactivity observed with infected guinea pig sera, human convalescent-phase sera recognize a greater number of LCV than SCV proteins. While a number of SCV- and LCV-specific proteins are recognized by convalescent-phase sera, in most cases these are weakly immunogenic. However, one human serum strongly recognized two LCV-specific proteins: Bcp and repressor protein C2. C2 is an S24 peptidase family autopeptidase which is commonly involved in activation of RecA (18). As discussed earlier, Bcp may protect C. burnetii against oxidative stress. Selective recognition by the human immune system of both proteins only in the LCV is consistent with an elevated stress response by this developmental form. The remaining identified antigens are present in both the SCV and the LCV with GroEL, RplJ, and FabF recognized by both human sera in both cell forms. GroEL and the ribosomal protein L9 were recently identified as immunogens by screening a C. burnetii expression library with serum from Coxiella-infected mice (40). FabF, an enzyme involved in fatty acid biosynthesis; Icd, a Krebs' cycle enzyme; and AtpA, a subunit of ATP synthase, are considered housekeeping enzymes. However, antigenic C. burnetii housekeeping proteins have been reported elsewhere (40), indicating that general metabolic enzymes of the organism are exposed to the host immune system. AdaA, EF-Tu, AtpA, and Icd were recognized in both SCV and LCV lysates by a single convalescent-phase serum. AdaA was recently identified as an immunodominant outer membrane protein synthesized only by isolates acquired from patients with acute disease (42), and mice generate an antibody response to EF-Tu when vaccinated with formalin-fixed C. burnetii (26). One human serum recognized Tig, an abundant ribosome-associated chaperone (6), in both SCV and LCV lysates while the other human serum recognized Tig only in LCV lysates.
In summary, this study provides an extensive proteome analysis of C. burnetii developmental forms. Identification of cell-form-specific and common antigens provides new information for development of rationally designed subunit vaccines and new diagnostic tests. This is particularly relevant with respect to surface antigens of the SCV, the environmentally stable cell form.
Acknowledgments
We thank Harlan Caldwell, Ted Hackstadt, and Shelly Robertson for critical review of the manuscript; Scott Grieshaber and Chris Burlak for assistance with 2-D gel quantitation; and Gary Hettrick and Anita Mora for graphics.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R.A.H.), and by Public Heath Service grant AI057156 from the National Institute of Allergy and Infectious Diseases (J.E.S.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Bachrach, U. 2005. Naturally occurring polyamines: interaction with macromolecules. Curr. Protein Pept. Sci. 6:559-566. [DOI] [PubMed] [Google Scholar]
- 2.Beare, P. A., J. E. Samuel, D. Howe, K. Virtaneva, S. F. Porcella, and R. A. Heinzen. 2006. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J. Bacteriol. 188:2309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135-139. [DOI] [PubMed] [Google Scholar]
- 4.Cladiere, L., K. Hamze, E. Madec, V. M. Levdikov, A. J. Wilkinson, I. B. Holland, and S. J. Seror. 2006. The GTPase, CpgA(YloQ), a putative translation factor, is implicated in morphogenesis in Bacillus subtilis. Mol. Genet. Genomics 275:409-420. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, S. A., E. R. Fischer, D. Howe, D. J. Mead, and R. A. Heinzen. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deuerling, E., and B. Bukau. 2004. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit. Rev. Biochem. Mol. Biol. 39:261-277. [DOI] [PubMed] [Google Scholar]
- 7.Fontecave, M., M. Atta, and E. Mulliez. 2004. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29:243-249. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, K. R., N. L. Tuite, A. Bhagwat, and C. P. O'Byrne. 2006. Global effects of homocysteine on transcription in Escherichia coli: induction of the gene for the major cold-shock protein, CspA. Microbiology 152:2221-2231. [DOI] [PubMed] [Google Scholar]
- 9.Hackstadt, T., M. G. Peacock, P. J. Hitchcock, and R. L. Cole. 1985. Lipopolysaccharide variation in Coxiella burnetii: intrastrain heterogeneity in structure and antigenicity. Infect. Immun. 48:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, A. M., Y. Qiu, N. Yeh, F. R. Blattner, T. Durfee, and D. J. Jin. 2005. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56:719-734. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, F., and D. Barilla. 2006. Assembling the bacterial segrosome. Trends Biochem. Sci. 31:247-250. [DOI] [PubMed] [Google Scholar]
- 12.Heinzen, R. A. 1997. Intracellular development of Coxiella burnetii, p. 99-129. In B. Anderson, M. Bendinelli, and H. Friedman (ed.), Rickettsial infection and immunity. Plenum Publishing Corp., New York, NY.
- 13.Heinzen, R. A., and T. Hackstadt. 1996. A developmental stage-specific histone H1 homolog of Coxiella burnetii. J. Bacteriol. 178:5049-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinzen, R. A., D. Howe, L. P. Mallavia, D. D. Rockey, and T. Hackstadt. 1996. Developmentally regulated synthesis of an unusually small, basic peptide by Coxiella burnetii. Mol. Microbiol. 22:9-19. [DOI] [PubMed] [Google Scholar]
- 15.Heinzen, R. A., M. A. Scidmore, D. D. Rockey, and T. Hackstadt. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusukawa, N., T. Yura, C. Ueguchi, Y. Akiyama, and K. Ito. 1989. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 8:3517-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Y., R. A. Pfuetzner, S. Mosimann, M. Paetzel, E. A. Frey, M. Cherney, B. Kim, J. W. Little, and N. C. Strynadka. 2001. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell 106:585-594. [DOI] [PubMed] [Google Scholar]
- 19.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCaul, T. F., N. Banerjee-Bhatnagar, and J. C. Williams. 1991. Antigenic differences between Coxiella burnetii cells revealed by postembedding immunoelectron microscopy and immunoblotting. Infect. Immun. 59:3243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaul, T. F., T. Hackstadt, and J. C. Williams. 1981. Ultrastructural and biological aspects of Coxiella burnetii under physical disruptions, p. 267-280. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY.
- 22.McCaul, T. F., and J. C. Williams. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moos, A., and T. Hackstadt. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect. Immun. 55:1144-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystrom, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537-544. [DOI] [PubMed] [Google Scholar]
- 25.Park, K. J., M. J. Kang, S. H. Kim, H. J. Lee, J. K. Lim, S. H. Choi, S. J. Park, and K. H. Lee. 2004. Isolation and characterization of rpoS from a pathogenic bacterium, Vibrio vulnificus: role of σS in survival of exponential-phase cells under oxidative stress. J. Bacteriol. 186:3304-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seshadri, R., L. R. Hendrix, and J. E. Samuel. 1999. Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect. Immun. 67:6026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshadri, R., and J. E. Samuel. 2001. Characterization of a stress-induced alternate sigma factor, RpoS, of Coxiella burnetii and its expression during the development cycle. Infect. Immun. 69:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stechmann, A., and T. Cavalier-Smith. 2004. Evolutionary origins of Hsp90 chaperones and a deep paralogy in their bacterial ancestors. J. Eukaryot. Microbiol. 51:364-373. [DOI] [PubMed] [Google Scholar]
- 30.Strunnikov, A. V. 2006. SMC complexes in bacterial chromosome condensation and segregation. Plasmid 55:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuite, N. L., K. R. Fraser, and P. C. O'Byrne. 2005. Homocysteine toxicity in Escherichia coli is caused by a perturbation of branched-chain amino acid biosynthesis. J. Bacteriol. 187:4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueguchi, C., and K. Ito. 1992. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J. Bacteriol. 174:1454-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghees, S., K. Kiss, G. Frans, O. Braha, and J. E. Samuel. 2002. Cloning and porin activity of the major outer membrane protein P1 from Coxiella burnetii. Infect. Immun. 70:6741-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel, U., and K. F. Jensen. 1997. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 272:12265-12271. [DOI] [PubMed] [Google Scholar]
- 35.Wiebe, M. E., P. R. Burton, and D. M. Shankel. 1972. Isolation and characterization of two cell types of Coxiella burnetii phase I. J. Bacteriol. 110:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, J. C. 1991. Infectivity, virulence, and pathogenicity of Coxiella burnetii for various hosts, p. 21-71. In J. C. Williams and H. A. Thompson (ed.), Q fever: the biology of Coxiella burnetii. CRC Press, Boca Raton, FL.
- 37.Williams, J. C., M. G. Peacock, and T. F. McCaul. 1981. Immunological and biological characterization of Coxiella burnetii, phases I and II, separated from host components. Infect. Immun. 32:840-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood, Z. A., L. B. Poole, and P. A. Karplus. 2003. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300:650-653. [DOI] [PubMed] [Google Scholar]
- 39.Young, D., T. Hussell, and G. Dougan. 2002. Chronic bacterial infections: living with unwanted guests. Nat. Immunol. 3:1026-1032. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, G., K. Kiss, R. Seshadri, L. R. Hendrix, and J. E. Samuel. 2004. Identification and cloning of immunodominant antigens of Coxiella burnetii. Infect. Immun. 72:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, G., and J. E. Samuel. 2004. Vaccines against Coxiella infection. Expert Rev. Vaccines 3:577-584. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, G., H. To, K. E. Russell, L. R. Hendrix, T. Yamaguchi, H. Fukushi, K. Hirai, and J. E. Samuel. 2005. Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease. Infect. Immun. 73:1561-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]