Abstract
Brucella spp., like other pathogens, must cope with the environment of diverse host niches during the infection process. In doing this, pathogens evolved different type of transport systems to help them survive and disseminate within the host. Members of the TolC family have been shown to be involved in the export of chemically diverse molecules ranging from large protein toxins to small toxic compounds. The role of proteins from the TolC family in Brucella and other α-2-proteobacteria has been explored little. The gene encoding the unique member of the TolC family from Brucella suis (BepC) was cloned and expressed in an Escherichia coli mutant disrupted in the gene encoding TolC, which has the peculiarity of being involved in diverse transport functions. BepC fully complemented the resistance to drugs such as chloramphenicol and acriflavine but was incapable of restoring hemolysin secretion in the tolC mutant of E. coli. An insertional mutation in the bepC gene strongly affected the resistance phenotype of B. suis to bile salts and toxic chemicals such as ethidium bromide and rhodamine and significantly decreased the resistance to antibiotics such as erythromycin, ampicillin, tetracycline, and norfloxacin. Moreover, the B. suis bepC mutant was attenuated in the mouse model of infection. Taken together, these results suggest that BepC-dependent efflux processes of toxic compounds contribute to B. suis survival inside the host.
Brucella spp. are responsible for a zoonosis that causes a serious economical impact worldwide and a human disease that is difficult to treat (8, 16, 22).Brucella enters the host via the nasal, oral, and pharyngeal cavities, and after penetrating the mucosal epithelium, the organism is transported to the lymph nodes. During early infection, innate immunity mechanisms from the host contribute to reduce the initial number of infecting Brucella cells (38). Brucella has the ability to invade and survive within macrophages and nonphagocytic cells. After entering the host cell, Brucella is found in a membrane-associated vacuole (phagosome). Brucella subverts the intracellular endocytic pathway, bypassing the classical lysosomal pathway; the bacteria multiplicate and survive in a compartment associated with rough endoplasmic reticulum (for recent reviews, see references 33 and 69). By using this strategy, the bacterium escapes some bactericidal mechanisms (10).
The outer membrane of Brucella is considerably hydrophobic compared to those of other α-2-proteobacteria and therefore more permeable to lipophilic compounds (54). Conversely, the hydrophobic character of the outer membrane makes Brucella relatively resistant to polycationic peptides. In other species, low permeability to hydrophobic compounds goes together with efflux systems to increase protection against toxic molecules (47). Therefore, this unusual characteristic of the outer membrane raises interesting questions about the physiology of Brucella spp. Some outer membrane proteins (76) and the O side chain component of the lipopolysaccharide (4, 32, 52) have been shown to influence intracellular survival, probably altering the outer membrane properties.
In addition to the characteristics of outer membrane permeability, transport processes are crucial for protecting pathogens against toxic compounds. In fact, several gram-negative bacteria expel a broad range of antimicrobial compounds through the expression of different efflux systems (57, 83). Most of these systems are tripartite efflux pumps, in which an outer membrane protein channel from the TolC family works in association with inner membrane complexes to extrude different types of toxic compounds. The inner membrane complexes are formed by an inner membrane protein belonging to the RND (resistance nodulation division) or MFS (major facilitator superfamily) family and a protein from the MFP (membrane fusion protein) family that expands the periplasmic space (27, 60, 83). The paradigm of members from the outer membrane channel family is the multifunctional TolC of Escherichia coli. High-resolution crystal structure revealed that the TolC homotrimer has a 140-Å-long cylinder which is made up of a 100-Å-long α-helical barrel (the tunnel domain) extending the periplasmic space and a 40-Å-long β-barrel channel in the outer membrane (7). TolC together with an ABC-MFP complex is responsible for α-hemolysin (HlyA) translocation across both inner and outer membranes in a mechanism bypassing the periplasmic space (49, 79). From biochemical studies, it was shown that protein export was achieved by the recruitment of a trimeric TolC by the inner membrane translocase after it binds to its substrate (75). TolC works also in association with RND-MFP or MFS-MFP complexes to pump antimicrobial drugs outside the cell (40). It was proposed that substrate binding induces an open state by untwisting the tunnel α-helices of TolC; this conformation change allows the direct passage of proteins and drugs from the cytosol out of the cell (7, 41).
So far, only one efflux system belonging to the MATE (multidrug and toxic compound extrusion) family has been characterized in Brucella spp. This system was shown to be efficient in the elimination of drugs such as norfloxacin, ciprofloxacin, gentamicin, and acriflavine (18). In α-2-proteobacteria, the role of tripartite transport systems, particularly of TolC homologues, has been explored little. In this work, we investigated the role of the unique member of the TolC family identified in the Brucella suis genome. Heterologous and mutational approaches showed that the TolC homologue from B. suis is involved in the efflux of toxic and relatively hydrophobic compounds, influencing the survival of B. suis inside the host.
MATERIALS AND METHODS
Growth conditions, bacterial strains, and reagents.
The Brucella strains used in this study (B. suis 1330 [ATCC 23444] and derived mutants) were all grown in tryptic soy (TS; Bacto) medium in combination with the appropriate antibiotics (spectinomycin, 50 μg ml−1; chloramphenicol, 6 μg ml−1). E. coli strain DH5α was used as the recipient strain for cloning and was routinely grown in Luria-Bertani (LB) medium. The appropriate antibiotics (ampicillin, 50 μg ml−1; spectinomycin, 200 μg ml−1; chloramphenicol, 50 μg ml−1; tetracycline, 5 μg ml−1; and kanamycin, 50 μg ml−1) were added when needed. E. coli strain C600 and the tolC mutant (C600 tolC::Tn5) used in heterologous functional complementation assays were kindly provided by Philippe Delepelaire. The standard growth temperature for all bacterial strains was 37°C. Mueller-Hinton broth was obtained from Britania. The following drugs were used in sensitivity tests, with the compound class in parentheses: acriflavine (intercalator), ethidium bromide (intercalator), sodium deoxycholate (detergent, bile salt), erythromycin (macrolide), berberine hemisulfate (plant alkaloid), nalidixic acid (quinolone precursor), norfloxacin (fluoroquinolone), amikacin disulfate (aminoglucoside), cetyltrimethylammonium bromide (detergent), and crystal violet (dye, intercalator) were obtained from Sigma; carbenicillin (β-lactam) from Fisher Biotech; ampicillin (β-lactam) from Bago; sodium dodecyl sulfate (SDS) (detergent) from Promega; bile salts from Britania; chloramphenicol and rifampin from Parafarm; streptomycin (aminoglucoside) from Lab Richet; rhodamine 6G (dye) from Allied Chemical; and tetracycline from Amersham.
Cloning and disruption of bepC from B. suis.
A 1,932-bp DNA region containing the tolC homologue gene (bepC), corresponding to BR0945 of the annotated B. suis genome(http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?database=gbr), was amplified from B. suis 1330 genomic DNA by PCR, using the specific primer pair 5′-GAACGGGATGACGGGAAand 5′-GGCGTACCGTTTTCAATGCA, and cloned into pGEM-T Easy vector (Promega). The fidelity of the amplification reaction was confirmed by sequencing. The amplicon was subcloned as a SalI-SphI fragment in pBBAD18T vector (tetracycline resistance) (72) under the araC-inducible promoter. This construction (pFC25) was used for heterologous functional complementation assays. To generate a knockout mutant in the bepC gene, the 2-kb SmaI fragment containing the spectinomycin resistance cassette (Ω) (63) was ligated into the unique StuI site of bepC (712 bp downstream from the ATG start codon) cloned in pGEM-T Easy. The recombinant plasmid was electroporated into B. suis M1330 cells, and since the p-GEM-T-bepC::Ω plasmid cannot replicate in Brucella, the bepC insertional mutant (Br1) was obtained by selecting clones that were spectinomycin resistant and ampicillin sensitive. The Ω insertion was confirmed by Southern blot analysis. Briefly, chromosomal DNA was completely digested with EcoRI and ClaI, electrophoresed in 0.6% agarose, and capillary transferred to positively charged nylon membranes (Hybond-N). Membranes were hybridized with a 1.9-kb complete gene fragment labeled with radioactive phosphor using Prime-a-Gene (Promega) and washed with 2× SSC-0.1% SDS solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and once more with 0.1× SSC-0.1% SDS at 65°C. The washed blots were exposed on storage phosphor screen autoradiography and screened in a Storm 820 optical scanner (Amersham Pharmacia Biotech). For genetic complementation studies with Brucella, the SalI-SphI fragment containing the bepC gene (see above) was cloned into the broad-host-range pBBR1MCS vector, which confers resistance to chloramphenicol (42). The resulting plasmid (pFC115) was electroporated into the bepC Br1 mutant strain, and transformants were selected on TS agar supplemented with chloramphenicol and spectinomicin.
α-Hemolysin secretion and colicin E1 uptake.
Colicin E1 sensitivity was determined by spotting twofold serial dilutions of the colicin stock solution (Sigma) on bacterial lawns. Killing zones were recorded after 8 h of incubation at 37°C. α-Hemolysin (HlyA) secretion was analyzed using sheep blood agar plates (5% defibrinated blood); hemolysis zones around the colonies were observed after 10 h of incubation at 37°C. The presence of the 107-kDa HlyA polypeptide in the supernatant of assayed strains was also examined. Culture supernatant proteins were concentrated by precipitation with 10% trichloroacetic acid as described previously (70). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with 12% acrylamide and visualized by staining with Coomassie brilliant blue R-250.
Microdilution assay for drug susceptibility.
The MICs of drugs for E. coli strains were determined by a broth microdilution assay performed in Mueller-Hinton broth (Britania) supplemented with 0.2% l-arabinose. MIC was defined as the lowest concentration of a drug that completely inhibited growth. All tests were done by triplicate in accordance with the procedures established by the CLSI (formerly NCCLS). Briefly, the MIC was determined in microtiter plates with 96 flat-bottom wells in a final volume of 0.2 ml. Except for the growth controls in the absence of the drug, 100 μl of a twofold dilution of the drug was added to the wells. Next, except for the sterility (uninoculated) controls, 100 μl of a bacterial suspension (105 CFU/ml of E. coli cells grown in Mueller-Hinton broth) was added to the wells. The microtiter plates were shaken at 200 rpm during incubation, and bacterial growth was examined by measuring the optical density at 600 nm (OD600) with a microplate reader after 16 h of incubation at 37°C. The growth index was calculated by dividing the OD of the culture in the presence of drug by the OD in the absence of drug.
Disk diffusion tests.
Disk diffusion tests for E. coli were performed as outlined in the CLSI standard M2-A9, using Mueller-Hinton agar supplemented with 0.2% l-arabinose. Bacterial lawns of B. suis were grown on TS agar. Sterile paper disks (Whatman filters) 6 mm in diameter were placed on bacterial lawns, and 5 μl of each drug solution was pipetted onto separate disks on bacterial lawns. The plates were incubated at 37°C for 16 h for E. coli or 24 h for B. suis, and the diameters (in millimeters) of the inhibition zones were measured. Experiments were repeated at least twice, and all tests were performed in triplicate.
Infection in BALB/c mice.
Eight-week-old female BALB/c mice (5 mice per group) were inoculated intraperitoneally with 0.2 ml of a phosphate-buffered saline (PBS) suspension containing 5 × 105 CFU of wild-type B. suis 1330, the bepC mutant Br1, or Br1 complemented with bepC cloned in pFC115. At 2, 3, 5, and 7 weeks after infection, groups of five mice were sacrificed for spleen collection. The spleens were homogenized in 5 ml of PBS, and serial dilutions of the homogenates were plated on TS agar with the corresponding antibiotics to determine bacterial counts.
Cell infection assays.
Murine J774 macrophages seeded in 24-well plates (105 cells per well) were inoculated with 2 × 106 CFU (multiplicity of infection, 20:1) of wild-type B. suis 1330, the bepC mutant Br1, or Br1 complemented with bepC cloned in pFC115 in 0.5 ml of minimal essential medium (GIBCO, Paisley, Scotland) supplemented with 5% fetal calf serum and 2 mM glutamine (cell culture medium) without antibiotics. A similar procedure was followed for infecting HeLa cells, except that the inoculum size was 107 CFU (multiplicity of infection, 100:1). In order to ensure close contact between cells and bacteria, multiwell plates were centrifuged for 10 min at 141 × g at room temperature and placed in a 5% CO2 atmosphere at 37°C. After 1 h, the wells were washed three times with sterile PBS (pH 7.4) and further incubated with cell culture medium containing 50 mg of gentamicin per ml and 50 mg of streptomycin per ml to eliminate the remaining extracellular brucellae. At different times, the number of intracellular viable B. suis bacteria was determined as follows: cells were washed three times with PBS and treated for 10 min with 0.5 ml of 0.1% Triton X-100 in deionized sterile water, and lysates were serially diluted in PBS and plated on TS agar with the appropriate antibiotic to determine the number of CFU.
Phylogenetic analysis.
Molecular evolutionary relationships between 20 protein members of the TolC family were examined by the neighbor-joining method of tree construction. Alignment of the proteins was performed with ClustalX (1.81). Phylogenetic trees, bootstrap, and jackknife analysis to determine the statistical stability of each node were done using Paup 4.0b10. Trees were displayed by TreeView 1.6.6.
RESULTS
Phylogenetic analysis of BepC.
Proteins from the TolC family have been shown to be involved together with an inner membrane translocase in the export of different and diverse type of substrates, such as proteins, hydrophobic compounds, and cations. Available Brucella sp. genomes contain a unique protein from the TolC family, displaying 99% sequence identity with one another (not shown). The closest relatives to the TolC homologue of Brucella spp. detected by a BLAST search against the Swiss-Prot data bank were the multifunctional TolC from E. coli (25% identity), AprF from Pseudomonas aeruginosa involved in protease secretion (25% identity) (25), and TolC from Vibrio cholerae involved in the efflux of antimicrobial agents and cytotoxin secretion (24% identity) (14). The identity of the Brucella TolC homologue with TolC from E. coli was relatively low compared with other members of this family. In fact, TolC homologues from V. cholerae, Erwinia chrysanthemi, and Salmonella enterica serovar Typhimurium share more than 70% similarity with TolC of E. coli (9, 14, 71). Accordingly, these proteins were named “TolC.” The TolC homologue component from B. suis was named BepC for Brucella efflux protein (see below).
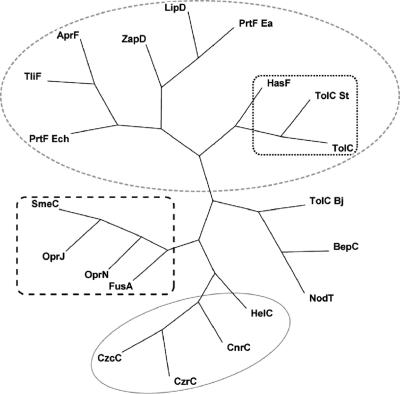
To investigate evolutionary relationships from which we can infer the possible substrates of BepC, a phylogenetic analysis of representative and characterized members of the TolC family was performed. We also included NodT and a TolC homologue from the close relatives Rhizobium leguminosarum and Bradyrhizobium japonicum, respectively, whose substrates have not been identified. In fact, although a role in Nod factor secretion by NodT was suggested, this has not been experimentally proved (67). BepC did not clearly cluster with any of the characterized TolC homologues (Fig. 1) but clustered in another group together with NodT and the TolC homologue from B. japonicum. As expected, members from the TolC family that participate in protein export clustered together and the same observation was made with proteins involved in drug efflux or cation export (Fig. 1). TolC from E. coli has the peculiarity to interact with many different inner membrane complexes to export either proteins or hydrophobic compounds (40) and to participate in the uptake of colicins (44). Interestingly, TolC formed a subcluster with HasF from Serratia marcescens, involved in both lipase secretion and drug efflux, and with the Salmonella enterica serovar Typhimurium TolCSt, which was capable of pumping hydrophobic compounds and secreting a heterologous protein (Fig. 1). Therefore, phylogenetic analysis did not allow the prediction of a possible substrate; BepC could be involved in one or several of the functions so far described for this family of proteins. To investigate potential roles for BepC, functional complementation studies with E. coli were performed.
FIG. 1.
Phylogenetic tree of the TolC family. Twenty protein members from the TolC family for which function is known, or strongly implicated by the location of their genes, are sorted by TreeView based on sequence alignment using ClustalX, and the tree is arranged using Paup 4.0. Sequence similarity correlates with substrate specificity; indeed, proteins can be grouped into three subfamilies corresponding to their roles.  , group 1, protein secretion;
, group 1, protein secretion;  , subgroup of multifunctional proteins;
, subgroup of multifunctional proteins;  , group 2, cation efflux;
, group 2, cation efflux;  , group 3, drug efflux. The substrate(s) and organism of the TolC homologues from group 1 are as follows: TliF, lipase, Pseudomonas fluorescens (1); AprF, alkaline protease, Pseudomonas aeruginosa (25); PrtF, protease, Erwinia chrysanthemi (45); LipD, lipase, Serratia marcescens (3); PrtF, protease, Erwinia amylovora (84); ZapD, metalloprotease, Proteus mirabilis (80); TolC, α-hemolysin, multiple drugs, multifunctional, Escherichia coli (40, 79); HasF, multifunctional, Serratia marcescens (2, 15, 43); and TolCSt, multifunctional, Salmonella enterica serovar Typhimurium (11), (12). The substrate(s) and organism of the TolC homologues from group 2 are as follows: CzrC, cadmium and zinc, Pseudomonas aeruginosa (36); CzcC, divalent cations, Ralstonia metallidurans (55); CnrC, cobalt and nickel efflux, Cupriavidus metallidurans (34); and HelC, heme, Legionella pneumophila (19). The substrate(s) and organism of the TolC homologues from group 3 are as follows: OprN, multiple drugs, Pseudomonas aeruginosa (51); OprJ, multiple drugs, Pseudomonas aeruginosa (62); SmeC, multiple drugs, Xanthomonas maltophilia (Stenotrophomonas maltophilia) (46); and FusA, fusaric acid, Burkholderia cepacia (78). BepC corresponds to the TolC homologue from B. suis M1330. This protein does not clearly belong to any of those groups and forms a new group with NodT from Rhizobium leguminosarum (unknown substrate) (73) and TolC from Bradyrhizobium japonicum (unknown substrate) (37).
, group 3, drug efflux. The substrate(s) and organism of the TolC homologues from group 1 are as follows: TliF, lipase, Pseudomonas fluorescens (1); AprF, alkaline protease, Pseudomonas aeruginosa (25); PrtF, protease, Erwinia chrysanthemi (45); LipD, lipase, Serratia marcescens (3); PrtF, protease, Erwinia amylovora (84); ZapD, metalloprotease, Proteus mirabilis (80); TolC, α-hemolysin, multiple drugs, multifunctional, Escherichia coli (40, 79); HasF, multifunctional, Serratia marcescens (2, 15, 43); and TolCSt, multifunctional, Salmonella enterica serovar Typhimurium (11), (12). The substrate(s) and organism of the TolC homologues from group 2 are as follows: CzrC, cadmium and zinc, Pseudomonas aeruginosa (36); CzcC, divalent cations, Ralstonia metallidurans (55); CnrC, cobalt and nickel efflux, Cupriavidus metallidurans (34); and HelC, heme, Legionella pneumophila (19). The substrate(s) and organism of the TolC homologues from group 3 are as follows: OprN, multiple drugs, Pseudomonas aeruginosa (51); OprJ, multiple drugs, Pseudomonas aeruginosa (62); SmeC, multiple drugs, Xanthomonas maltophilia (Stenotrophomonas maltophilia) (46); and FusA, fusaric acid, Burkholderia cepacia (78). BepC corresponds to the TolC homologue from B. suis M1330. This protein does not clearly belong to any of those groups and forms a new group with NodT from Rhizobium leguminosarum (unknown substrate) (73) and TolC from Bradyrhizobium japonicum (unknown substrate) (37).
B. suis BepC restores drug efflux in tolC-deficient E. coli.
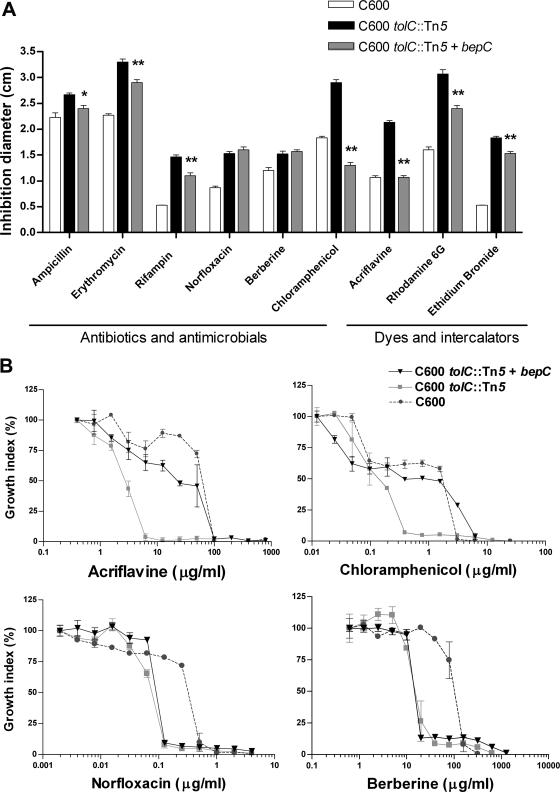
Since TolC of E. coli has the distinctive feature of participating in many transport processes, the potential role of BepC was initially analyzed by heterologous complementation of a tolC mutant. The bepC gene of B. suis (BR0945) (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?database=gbr) was amplified by PCR and cloned into pBBad-18T, under the control of the inducible araC promoter. The resulting plasmid (pFC25) was electroporated into the hypersensitive tolC::Tn5 mutant of E. coli (40), and drug sensitivity was assayed by using an agar diffusion plate assay. The bepC gene cloned in pFC25 restored the wild-type levels of resistance to drugs such as chloramphenicol and acriflavine (Fig. 2A). The presence of the bepC gene also resulted in a considerable reduction of susceptibility to other hydrophobic compounds such as ampicillin, erythromycin, rifampin, rhodamine 6G, and ethidium bromide (Fig. 2A). No effect of bepC expression on the hypersensitivity phenotypes of the tolC mutant to other compounds such as norfloxacin and berberine (Fig. 2A) as well as to sodium deoxycholate, nalidixic acid, crystal violet, SDS, streptomycin, gentamicin, and carbenicillin (data not shown) was observed. These results were confirmed using drug microdilution assays in a multiwell plate. We studied the effect of bepC expression on the growth of the tolC mutant in the presence of twofold serial dilutions of both chloramphenicol and acriflavine, using berberine and norfloxacin, two drugs that were equally toxic in the presence and absence of bepC, as controls (Fig. 2A). Expression of bepC resulted in an increase of the MIC of chloramphenicol to 3.12 μg/ml, a value higher than that for the wild type (1.56 μg/ml) (Fig. 2B). Likewise, bepC was also able to complement the susceptibility phenotype to acriflavine, resulting in an MIC very similar to that of the wild type (50 μg/ml) (Fig. 2B). As expected, bepC expression in the tolC mutant had no effect on the MICs of berberine and norfloxacin (Fig. 2B). These results indicate that BepC reverts the tolC-deficient E. coli hypersusceptibility to several diverse hydrophobic compounds.
FIG. 2.
Heterologous complementation of the resistance phenotype of the E. coli tolC mutant by bepC from B. suis. (A) The sensitivities of the E. coli wild type, the tolC mutant, and the tolC-plus-bepC strain were evaluated by the disk diffusion test in triplicate. The inoculated plates were incubated at 37°C for 16 h, and inhibition zones were compared. While partial complementation of the sensitive phenotype was observed for several drugs, sensitivity to acriflavine and chloramphenicol was fully complemented. A representative experiment is shown. For each drug, the values corresponding to the tolC mutant harboring the bepC gene marked with asterisks were significantly different from that of the tolC mutant (**, P < 0.001; *, P < 0.05). The statistical analysis was done by one-way analysis of variance and Bonferroni's multiple-comparison test. (B) MICs were determined for acriflavine and chloramphenicol as well as for two negative controls (norfloxacin and berberine). Microdilution assays were performed in Mueller-Hinton broth by broth microdilution tests in the presence of 0.2% l-arabinose. The growth index was calculated by dividing the OD600 of the culture in the presence of drug by the OD600 of the culture in the absence of drug. MIC was defined as the lowest concentration that completely inhibited growth. The experiments were performed at least twice in triplicate, and the values are means ± standard deviations.
BepC fully complemented colicin E1 uptake in the tolC mutant of E. coli.
Colicins are citotoxic peptides against susceptible relatives produced by bacteria of the Enterobacteriaceae family (31). TolC is involved in the uptake of colicins E1 and 10 by a specific interaction with the colicin amino-terminal translocator domain (56, 82). Consequently, mutagenesis of tolC results in a colicin-tolerant phenotype. In order to assess whether BepC complements TolC absence, different colicin E1 quantities were spotted on agar plates seeded with the E. coli wild type (C600), the tolC mutant, or the tolC mutant harboring the bepC gene on pFC25. As shown in Fig. 3 wild-type E. coli as well as the tolC mutant carrying bepC showed similar levels of colicin E1 susceptibility. This was an interesting result because HasF, a TolC homologue from Serratia marcescens that shares higher identity with TolC from E. coli (74%) than with BepC, was unable to complement colicin E1 sensitivity in the tolC mutant (15). This result suggests that although TolC and BepC share a low global degree of amino acid sequence similarity, some local sequence similarity or structural features within the putative colicin E1 receptor (and probably translocator) domain are conserved in both proteins. To assess a possible biological implication of such similarity, we examined the sensitivity of B. suis to colicin E1. Wild-type brucellae spread on a TS agar plate were not susceptible to colicin E1 over a concentration range similar to that used in experiments with E. coli (data not shown). This observation indicates that the receptor and uptake activities conferred by BepC are not sufficient to mediate colicin susceptibility and that other bacterial components required for colicin sensitivity (31) are probably not conserved in B. suis. In addition, the possibility that BepC of B. suis may be recognized as a specific receptor by other cytotoxic peptides cannot be rule out.
FIG. 3.
Heterologous complementation of colicin E1 susceptibility. Colicin E1 sensitivity in wild-type E. coli (C600) and the lack of sensitivity in the tolC mutant (C600 tolC::Tn5) are shown. Cloned bepC is capable of restoring sensitivity to colicin E1. The concentration of colicin E1 decreases clockwise.
Hemolysin secretion in the E. coli tolC mutant was not restored by BepC.
Most gram-negative bacteria, including members of the Rhizobiaceae family, export proteins to the extracellular medium through tripartite (ABC-MFP-TolC homologue) type I secretion systems. In R. leguminosarum, a close relative of Brucella spp., the inner membrane PrsD-PrsE translocase is responsible for the secretion of several proteins involved in biofilm formation and nodulation (28, 29, 70). However, the TolC homologue that works in association with PrsD-PrsE has not been identified. In E. coli, TolC together with the inner membrane translocase HlyB-HlyD is responsible for alpha-hemolysin (HlyA) secretion (79). Since BepC is the unique member of the TolC family that came out from the B. suis genome analysis, we explored the possibility that BepC was involved in protein secretion by testing whether BepC can restore hemolysin secretion in the tolC mutant expressing the hlyCABD genes (48, 79). The locus encoding HlyC, HlyA, HlyB, and HlyD cloned in pSF4000 was transferred by electroporation into the tolC mutant of E. coli. Hemolysin secretion was analyzed by a blood plate assay and by SDS-PAGE analysis of proteins secreted in the presence or the absence of bepC cloned in the pFC25 plasmid, which is compatible with pSF4000. A low level of hemolysis around the colony of the tolC mutant harboring both pSF4000 and pFC25 was observed, while no halo was detected around the E. coli tolC mutant containing only pSF4000 (data no shown). However, analysis of the trichloroacetic acid-precipitated extracellular proteins by SDS-PAGE clearly showed that secretion of HlyA was not restored by pFC25 in the tolC mutant (data not shown). The faint halo of hemolysis observed around the colonies might be due to a deleterious effect on cell membrane integrity by the expression of several genes encoding membrane proteins. Therefore, BepC was not capable of replacing TolC for HlyA secretion. This result suggests that TolC domains responsible for the interaction with the HlyB-HlyD translocase are not conserved in BepC or, alternatively, that some other intrinsic structural features of BepC do not allow for the secretion of such large and hydrophilic molecules.
Sensitivity of the bepC::Ω mutant to different hydrophobic compounds.
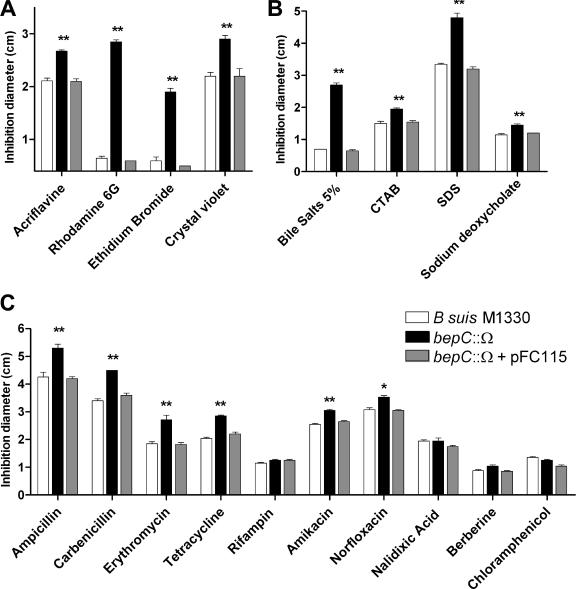
The heterologous complementation studies suggest that BepC is more likely to have a role in the transport of small and relatively hydrophobic compounds. A mutational approach was used to further investigate the role of BepC of B. suis in the efflux of toxic compounds. The protein secretion phenotype was not investigated since we and others have observed that the amount of protein exported by Brucella spp. to the extracellular medium during in vitro cultivation is very small (24; D. J. Comerci and R. A. Ugalde, personal communication). The spectinomycin cassette (Ω) was inserted in the StuI site of bepC, and a knockout mutant was generated by double recombination of the bepC::Ω allele into the B. suis genome. Mutation of bepC did not affect the growth rate of B. suis in tryptic soy broth-rich medium (data not shown). The sensitivity phenotype of the bepC::Ω mutant to several chemicals was analyzed in tryptic soy broth by the disk diffusion plate assay. A marked growth inhibition phenotype of the bepC::Ω mutant by the rhodamine 6G dye and the ethidium bromide intercalator was observed (Fig. 4A). Crystal violet (dye) and acriflavine (intercalator) also induced halos of growth inhibition greater than those seen for the wild-type strain (Fig. 4A). The sensitive phenotypes to all of these drugs were complemented by pFC115 carrying the bepC gene (Fig. 4A). Interestingly, the bepC::Ω mutant showed a hypersensitive phenotype towards a mixture of bile salts (Fig. 4B). A significant increase in sensitivity of the bepC mutant was also observed in the presence of the pure steroid sodium deoxycholate and detergents such as SDS and cetyltrimethylammonium bromide (Fig. 4B). In addition, the bepC::Ω mutant was significantly more sensitive than the wild-type strain to β-lactam antibiotics such as carbenicillin and ampicillin, a macrolide (erythromycin), tetracycline, an aminoglycoside (amikacin), and a quinolone (norfloxacin) (Fig. 4C). The bepC gene cloned in pFC115 restored the resistance level to bile salts, the detergents, and all of the antibiotics (Fig. 4B and C). Mutation in bepC did not alter the susceptibility phenotypes of B. suis to berberine (plant antimicrobial), rifampin, chloramphenicol, and nalidixic acid (quinolone precursor) (Fig. 4C). These results confirm that the BepC outer membrane protein of B. suis plays a role in the efflux of toxic compounds and suggest that BepC is more efficient in extruding relatively hydrophobic or amphipathic molecules.
FIG. 4.
Susceptibility phenotypes of the bepC mutant of B. suis. The drug sensitivity of the bepC::Ω insertional mutant was evaluated in agar plate diffusion tests. The mutation in the bepC gene significantly affected the resistance phenotype of B. suis to different compounds such as intercalators and dyes (A), detergents and bile salts (B), and antibiotics and antimicrobials (C). The experiments were repeated at least twice in triplicate. The data shown are expressed as mean values ± standard errors; asterisks above the values corresponding to the bepC mutant indicate that they were significantly different from the values for the wild-type strain 1330 (*, P < 0.01; **, P < 0.001). The statistical analysis was done by one-way analysis of variance and Bonferroni's multiple-comparison test. CTAB, cetyltrimethylammonium bromide.
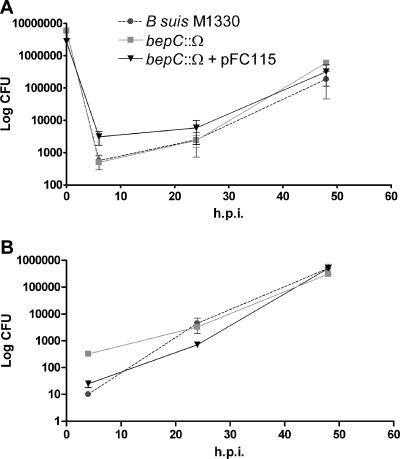
Survival of the bepC::Ω mutant in cultured cells.
To analyze whether mutation in bepC affects intracellular survival, murine J774 macrophages or HeLa cells were infected with wild-type B. suis 1330, the bepC::Ω mutant, or the bepC::Ω mutant harboring the pFC115 complementing plasmid. Figure 5A shows similar biphasic curves of viable brucellae recovered from macrophages infected with the wild type, the bepC mutant, and the complemented mutant over a 48-h experiment. Similarly, no significant differences in the numbers of brucellae recovered from HeLa cells infected with wild-type B. suis 1330, the bepC mutant, or the complemented strain were observed (Fig. 5B). This indicates that the BepC protein is not crucial for in vitro intracellular survival.
FIG. 5.
Intracellular replication of B. suis M1330, the bepC::Ω mutant, and the complemented strain (bepC::Ω + pFC115) in murine J774 macrophages or HeLa cells. The number of intracellular viable bacteria was determined at 6, 24, and 48 h postinfection in J774 macrophages (A) or at 4, 24, and 48 h postinfection in HeLa cells (B). No significant differences in the uptake and intracellular growth in both cell types between the bepC mutant and the parent strain were seen. The data presented are the results of a representative experiment and are means ± standard deviations (error bars) of plate counts.
Virulence of bepC::Ω in the mouse model.
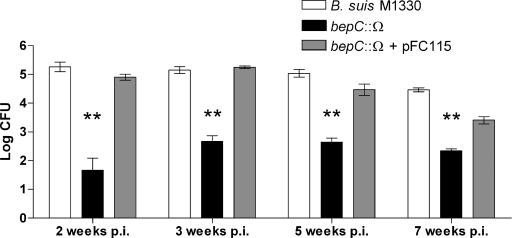
In the mouse model of infection, Brucella is able to establish a chronic infection characterized by a large accumulation of brucellae in the spleen. To study the role of the BepC outer membrane protein in B. suis survival in vivo, groups of five mice were infected intraperitoneally with 5 × 105 CFU of the wild-type strain, the bepC::Ω mutant, or the complemented strain. The number of CFU recovered from spleens was determined at 2, 3, 5, and 7 weeks postinfection (p.i.). Recovery of the bepC::Ω mutant from the spleen was reduced 3 logs relative to that of the wild type at 2 weeks postinoculation and was reduced 2 to 2.5 logs at 3, 5, and 7 weeks postinoculation. The bepC gene cloned in pFC115 fully complemented the virulence phenotype at 2, 3, and 5 weeks p.i. and partially complemented the virulence phenotype at 7 weeks p.i., probably due to some plasmid instability (Fig. 6). These observations show that the BepC outer membrane channel might be required for in vivo B. suis survival.
FIG. 6.
Virulence of B. suis M1330, the bepC mutant, and the complemented strain in BALB/c mice. Mice were infected by intraperitoneal injection with 105 brucellae. At 2, 3, 5, and 7 weeks after infection, groups of five mice were sacrificed for spleen collection and bacterial counts were determined. Values are means (log number of CFU per spleen) ± standard deviations (error bars) (n = 5). The mean numbers of bacteria in the spleens of bepC-infected mice were always significantly lower (**, P < 0.001) than the mean numbers of bacteria in the spleens of wild-type strain 1330- or complemented strain-infected mice. These experiments also showed a clear 2- to 3-log reduction of CFU by the bepC mutant.
DISCUSSION
In this work, we have analyzed the possible roles of the unique member of the TolC family (BepC) encoded by the B. suis genome. Members of the TolC family are recruited by different types of inner membrane translocases to allow a direct passage of diverse substrates from the cytoplasm to the external medium. Phylogenetic analysis of BepC did not clearly show a possible substrate for BepC. However, functional complementation studies with the hypersensitive and protein secretion-defective tolC mutant from E. coli support a role for BepC in the efflux of small and hydrophobic molecules. Mutation in the bepC gene of B. suis strongly increased the sensitivity of B. suis to bile salts, dyes such as rhodamine 6G, and ethidium bromide and significantly affected its resistance to antimicrobials such as erythromycin, tetracycline, and norfloxacin. These results indicate that BepC is capable of participating in the efflux of small and relatively hydrophobic compounds.
One obvious question is the following: which translocases are the partners of BepC? Or, more precisely, which are the putative inner membrane transporters encoded by the B. suis genome that work in association with BepC to pump toxic compounds? Our genomic analysis and the annotation of the B. suis genome (58) indicate that there are possible ABC-MFP and several putative RND-MFP “translocases” encoded by the B. suis genome. In theory, all of these candidates could work with BepC. In fact, our preliminary data suggest that at least one ABC-MFP complex and one of the RND-MFP complexes are involved in the extrusion of toxic molecules (F. A. Martín et al., unpublished data). Therefore, these inner membrane translocases probably form transenvelope tripartite complexes with BepC, allowing for the efflux of toxic compounds.
Detoxifying mechanisms in bacteria are of increasing interest because augmented resistance of several human pathogens to diverse antibiotics often involves the overexpression of efflux systems, including tripartite pumps (50, 61, 81). However, it is clear that this bacterial strategy emerged after the extensive use of antibiotics. Therefore, another question commonly raised about efflux systems concerns the in vivo physiological substrates. BepC-dependent efflux processes might contribute to resistance to compounds naturally present in the mammalian host. We have found that while the wild-type strain of B. suis was considerably resistant to 5% of a mixture of bile salts, the growth of the bepC knockout mutant was strongly inhibited by 5% of the bile salts. This observation suggests that BepC together with an inner membrane translocase may efficiently extrude bile salts produced by the host. Bile salts are very abundant in the mammalian intestine, which is not a preferred host niche for Brucella replication. Nevertheless, the efflux of bile salts may be important for Brucella survival during the intestinal transit of the bacterium in orally acquired infections. Another possibility is that BepC participates in the efflux of other steroid-like molecules encountered within the host during infection (26). In any case, these abilities may be particularly relevant for survival inside the host since the outer membrane of Brucella spp. was found to be particularly permeable to hydrophobic molecules (54). A similar feature was also reported for the outer membrane of Vibrio cholerae, an enteropathogen that circumvents the bactericidal activity of bile salts in the intestine through efflux pumps (21).
Another alternative role has been proposed for tripartite systems; these systems might be responsible of exporting hydrophobic quorum sensing signals, such as long-chain N-acylhomoserine lactones and quinolones (39, 59). Indeed, analysis by high-performance liquid chromatography and mass spectrometry of a dichloromethane extract of a spent culture supernatant from B. melitensis identified an N-dodecanoylhomoserine lactone (74). The transport of this putative signal to the extracellular milieu by Brucella spp. may be BepC dependent.
BepC did not complement the protein secretion phenotype of the tolC mutant, suggesting that BepC may not be involved in protein secretion. A protein secretion phenotype of the bepC mutant was not analyzed because it is well known that cultured Brucella does not secret detectable amounts of protein (24; Comerci and Ugalde, personal communication). Besides, our own analysis and description of predicted proteins by the published Brucella genomes (20, 23, 24, 35, 58) support the idea that there are no ABC-MFP candidates that could recruit BepC for protein secretion.
Our observations indicate that the BepC outer membrane protein significantly contributes to the intrinsic resistance/susceptibility phenotype of B. suis to several antibiotics. Human brucellosis is normally treated with doxycycline (a tetracycline), streptomycin, rifampin, fluoroquinolones, or combinations of these antibiotics. Emerged resistance to these or other antimicrobials in Brucella spp. is not a serious cause for concern. However, the presence of BepC-dependent efflux pumps in B. suis that are able to extrude antibiotics make this subject deserving of special attention. In fact, some strains nonsusceptible to rifampin have been isolated in Turkey and South Arabia (13, 53). In addition, a lack of effective bactericidal activity of fluoroquinolones against Brucella spp. has often been reported (5, 6, 30, 64, 68). In this regard, it should be noted that TolC-dependent efflux processes (Fig. 1) are responsible for the multiresistant phenotype of the Salmonella enterica serovar Typhimurium DT104 strain (12). In addition, it has been shown that ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC (66).
The observation that BepC is able to replace TolC for the sensitivity phenotype of E. coli towards colicin is intriguing. The biological significance of this is not known since B. suis was not found to be affected by colicin E1. To enter cells, colicin parasitizes multiprotein systems used by sensitive cells for important biological functions (17, 44). The fact that BepC was functional for this role might be fortuitous or may imply other unknown receptor and/or uptake functions.
Significantly fewer brucellae were recovered at 2, 3, 5, and 7 weeks p.i. from spleens of mice infected with the bepC mutant than from spleens of mice infected with the wild-type strain 1330. This phenotype was fully complemented by the cloned bepC mutant, indicating a direct association between mutation in the bepC gene and attenuation. However, no difference in the abilities to invade and replicate intracellularly in murine macrophages and HeLa cells between the bepC mutant and the parental strain was observed. This discrepancy between the in vitro and in vivo virulence phenotypes could be explained by different hypotheses. First, different surface molecules were proposed to control the initial number of infecting bacteria (38). In fact, a knockout mutant in the gene encoding the outer membrane lipoprotein Omp10 of Brucella abortus showed a phenotype similar to that of the bepC mutant, i.e., it was significantly attenuated in vivo but not in vitro. Similarly, a mutant in another outer membrane lipoprotein (Omp19) of B. abortus showed an in vitro phenotype indistinguishable from that of the parental strain by using bovine macrophages but was significantly attenuated in vivo. It was suggested that the attenuation associated with the lack of these outer membrane proteins is due to an increased sensitivity to serum complement (76). Another class of mutants that was frequently attenuated in vivo but not in vitro is related to lipopolysaccharide-altered phenotypes (32, 77). The lipopolysaccharide mutants may be more sensitive to mechanisms of the immune response in the early stage of infection. In all of these cases, there might be a contribution in vivo of lysis mediated by complement, a lectin pathway, or other host factors that influence extracellular survival (38). These processes may have an impact on Brucella dissemination (65) and on the number of bacteria contacting host target cells, especially during the initial phase of infection. The BepC outer membrane protein may have a similar immunomodulatory effect.
A relatively larger increase in the CFU of mice infected with the bepC mutant from 2 to 5 weeks p.i. than in the CFU of mice infected with the wild-type or complemented strain was observed. The comparatively larger error in the first time point may explain to some extent this difference. In addition, this difference may be due to a shift of the intracellular growth curve of the bepC mutant relative to the curve of the wild-type strain. During Brucella infection in mice, an initial increase of spleen CFU is usually observed, which is followed by a slow but steady decrease beginning around weeks 2 to 4 p.i. (76). In every case, the splenic load seems to reach a maximum and to decline later. In our case, the number of spleen CFU of wild-type B. suis seemed to have reached a plateau between weeks 2 and 3 p.i. In contrast, the splenic load of the bepC mutant showed a slight increase between these time points. This difference may be due to a lower initial inoculum of splenic cells in mice infected with the bepC mutant than in mice infected with the wild-type strain (because of the immunomodulatory effect described above). The maximum capacity of spleen cells to sustain Brucella replication may have been reached at 2 weeks for the wild-type infection but not for the infection with the bepC mutant.
Another plausible hypothesis for a role for BepC in extracellular survival would be more directly related with its efflux role. Toxic compounds encountered in the host during infection or other endogenously generated compounds might be removed by BepC-dependent efflux mechanisms. This, in turn, could have a direct influence on survival and dissemination within the host (65). In addition, the evidence presented in this work does not completely rule out a role for BepC in intracellular survival. BepC may participate in the efflux of antimicrobial compounds produced by macrophages induced only in vivo upon stimulation by cytokines generated locally by other cells in response to infection.
Acknowledgments
We thank Marcelo E. Tolmasky for critical reading the manuscript. We also thank Philippe Delepelaire for kindly providing the E. coli tolC mutant and the pSF4000 plasmid. We thank Carlos A. Fossati, Diego A. Laplagne, and Adrián A. Vojnov for their support and advice and Alfonso Soler Bistué, Daniela M. Russo, and Lorena Haurigot for helpful discussions. We thank Marta Bravo and Jimena Ortega for DNA sequencing.
This work was supported by the University of Buenos Aires (UBACyT X-245) and Agencia de Promoción Científica y Tecnológica (PICT 8266). A.Z. is a member of CONICET and a professor of the University of Buenos Aires; D.M.P. was supported by a CONICET fellowship and F.A.M. by a University of Buenos Aires fellowship.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Ahn, J. H., J. G. Pan, and J. S. Rhee. 1999. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J. Bacteriol. 181:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akatsuka, H., R. Binet, E. Kawai, C. Wandersman, and K. Omori. 1997. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J. Bacteriol. 179:4754-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akatsuka, H., E. Kawai, K. Omori, and T. Shibatani. 1995. The three genes lipB, lipC, and lipD involved in the extracellular secretion of the Serratia marcescens lipase which lacks an N-terminal signal peptide. J. Bacteriol. 177:6381-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, C. A., L. G. Adams, and T. A. Ficht. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.al-Sibai, M. B., M. A. Halim, M. M. el-Shaker, B. A. Khan, and S. M. Qadri. 1992. Efficacy of ciprofloxacin for treatment of Brucella melitensis infections. Antimicrob. Agents Chemother. 36:150-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.al-Sibai, M. B., and S. M. Qadri. 1990. Development of ciprofloxacin resistance in Brucella melitensis. J. Antimicrob. Chemother. 25:302-303. [DOI] [PubMed] [Google Scholar]
- 7.Andersen, C., C. Hughes, and V. Koronakis. 2000. Chunnel vision. Export and efflux through bacterial channel-tunnels. EMBO Rep. 1:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldi, P. C., G. H. Giambartolomei, J. C. Wallach, C. A. Velikovsky, and C. A. Fossati. 2001. Limited diagnostic usefulness of antibodies to cytoplasmic proteins of Brucella in early-treated human brucellosis. Scand. J. Infect. Dis. 33:200-205. [DOI] [PubMed] [Google Scholar]
- 9.Barabote, R. D., O. L. Johnson, E. Zetina, S. K. San Francisco, J. A. Fralick, and M. J. San Francisco. 2003. Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J. Bacteriol. 185:5772-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batut, J., S. G. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933-945. [DOI] [PubMed] [Google Scholar]
- 11.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 12.Baucheron, S., S. Tyler, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baykam, N., H. Esener, O. Ergonul, S. Eren, A. K. Celikbas, and B. Dokuzoguz. 2004. In vitro antimicrobial susceptibility of Brucella species. Int. J. Antimicrob. Agents 23:405-407. [DOI] [PubMed] [Google Scholar]
- 14.Bina, J. E., and J. J. Mekalanos. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 69:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binet, R., and C. Wandersman. 1996. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analogue. Mol. Microbiol. 22:265-273. [DOI] [PubMed] [Google Scholar]
- 16.Boschiroli, M. L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 17.Bouveret, E., A. Rigal, C. Lazdunski, and H. Benedetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 18.Braibant, M., L. Guilloteau, and M. S. Zygmunt. 2002. Functional characterization of Brucella melitensis NorMI, an efflux pump belonging to the multidrug and toxic compound extrusion family. Antimicrob. Agents Chemother. 46:3050-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 20.Chain, P. S., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 73:8353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee, A., S. Chaudhuri, G. Saha, S. Gupta, and R. Chowdhury. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 186:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbel, M. J. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DelVecchio, V. G., V. Kapatral, P. Elzer, G. Patra, and C. V. Mujer. 2002. The genome of Brucella melitensis. Vet. Microbiol. 90:587-592. [DOI] [PubMed] [Google Scholar]
- 24.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duong, F., A. Lazdunski, B. Cami, and M. Murgier. 1992. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene 121:47-54. [DOI] [PubMed] [Google Scholar]
- 26.Elkins, C. A., and L. B. Mullis. 2006. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J. Bacteriol. 188:1191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eswaran, J., E. Koronakis, M. K. Higgins, C. Hughes, and V. Koronakis. 2004. Three's company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 14:741-747. [DOI] [PubMed] [Google Scholar]
- 28.Finnie, C., N. M. Hartley, K. C. Findlay, and J. A. Downie. 1997. The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol. Microbiol. 25:135-146. [DOI] [PubMed] [Google Scholar]
- 29.Finnie, C., A. Zorreguieta, N. M. Hartley, and J. A. Downie. 1998. Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J. Bacteriol. 180:1691-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Rodriguez, J. A., J. E. Garcia Sanchez, and I. Trujillano. 1991. Lack of effective bactericidal activity of new quinolones against Brucella spp. Antimicrob. Agents Chemother. 35:756-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillor, O., B. C. Kirkup, and M. A. Riley. 2004. Colicins and microcins: the next generation antimicrobials. Adv. Appl. Microbiol. 54:129-146. [DOI] [PubMed] [Google Scholar]
- 32.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorvel, J. P., and E. Moreno. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90:281-297. [DOI] [PubMed] [Google Scholar]
- 34.Grass, G., C. Grosse, and D. H. Nies. 2000. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J. Bacteriol. 182:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187:2715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan, M. T., D. van der Lelie, D. Springael, U. Romling, N. Ahmed, and M. Mergeay. 1999. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene. 238:417-425. [DOI] [PubMed] [Google Scholar]
- 37.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110 (supplement). DNA Res. 9:225-256. [DOI] [PubMed] [Google Scholar]
- 38.Ko, J., and G. A. Splitter. 2003. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin. Microbiol. Rev. 16:65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köhler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J.-C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koronakis, V. 2003. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 555:66-71. [DOI] [PubMed] [Google Scholar]
- 41.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 42.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 43.Kumar, A., and E. A. Worobec. 2005. HasF, a TolC-homolog of Serratia marcescens, is involved in energy-dependent efflux. Can. J. Microbiol. 51:497-500. [DOI] [PubMed] [Google Scholar]
- 44.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubes, and H. Benedetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letoffe, S., P. Delepelaire, and C. Wandersman. 1990. Protease secretion by Erwinia chrysanthemi: the specific secretion functions are analogous to those of Escherichia coli alpha-haemolysin. EMBO J. 9:1375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, X. Z., L. Zhang, and K. Poole. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 46:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 48.Mackman, N., J. M. Nicaud, L. Gray, and I. B. Holland. 1985. Genetical and functional organisation of the Escherichia coli haemolysin determinant 2001. Mol. Gen. Genet. 201:282-288. [DOI] [PubMed] [Google Scholar]
- 49.Mackman, N., J. M. Nicaud, L. Gray, and I. B. Holland. 1986. Secretion of haemolysin by Escherichia coli. Curr. Top. Microbiol. Immunol. 125:159-181. [DOI] [PubMed] [Google Scholar]
- 50.Maneewannakul, K., and S. B. Levy. 1996. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McQuiston, J. R., R. Vemulapalli, T. J. Inzana, G. G. Schurig, N. Sriranganathan, D. Fritzinger, T. L. Hadfield, R. A. Warren, L. E. Lindler, N. Snellings, D. Hoover, S. M. Halling, and S. M. Boyle. 1999. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect. Immun. 67:3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Memish, Z., M. W. Mah, S. Al Mahmoud, M. Al Shaalan, and M. Y. Khan. 2000. Brucella bacteraemia: clinical and laboratory observations in 160 patients. J. Infect. 40:59-63. [DOI] [PubMed] [Google Scholar]
- 54.Moreno, E., and I. Moriyon. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 86:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otsuji, N., T. Soejima, S. Maki, and H. Shinagawa. 1982. Cloning of colicin E1 tolerant tolC (mtcB) gene of Escherichia coli K12 and identification of its gene product. Mol. Gen. Genet. 187:30-36. [DOI] [PubMed] [Google Scholar]
- 57.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446-451. [DOI] [PubMed] [Google Scholar]
- 58.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poole, K. 2002. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 92(Suppl.):55S-64S. [PubMed] [Google Scholar]
- 61.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 62.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 63.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 64.Qadri, S. M., M. Akhtar, Y. Ueno, and M. B. al-Sibai. 1989. Susceptibility of Brucella melitensis to fluoroquinolones. Drugs Exp. Clin. Res. 15:483-485. [PubMed] [Google Scholar]
- 65.Rajashekara, G., D. A. Glover, M. Krepps, and G. A. Splitter. 2005. Temporal analysis of pathogenic events in virulent and avirulent Brucella melitensis infections. Cell. Microbiol. 7:1459-1473. [DOI] [PubMed] [Google Scholar]
- 66.Ricci, V., P. Tzakas, A. Buckley, and L. J. Piddock. 2006. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 50:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivilla, R., J. M. Sutton, and J. A. Downie. 1995. Rhizobium leguminosarum NodT is related to a family of outer-membrane transport proteins that includes TolC, PrtF, CyaE and AprF. Gene 161:27-31. [DOI] [PubMed] [Google Scholar]
- 68.Rolain, J. M., M. Maurin, and D. Raoult. 2000. Bactericidal effect of antibiotics on Bartonella and Brucella spp.: clinical implications. J. Antimicrob. Chemother. 46:811-814. [DOI] [PubMed] [Google Scholar]
- 69.Roop, R. M., II, B. H. Bellaire, M. W. Valderas, and J. A. Cardelli. 2004. Adaptation of the brucellae to their intracellular niche. Mol. Microbiol. 52:621-630. [DOI] [PubMed] [Google Scholar]
- 70.Russo, D. M., A. Williams, A. Edwards, D. M. Posadas, C. Finnie, M. Dankert, J. A. Downie, and A. Zorreguieta. 2006. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188:4474-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santiviago, C. A., J. A. Fuentes, S. M. Bueno, A. N. Trombert, A. A. Hildago, L. T. Socias, P. Youderian, and G. C. Mora. 2002. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46:687-698. [DOI] [PubMed] [Google Scholar]
- 72.Sukchawalit, R., P. Vattanaviboon, R. Sallabhan, and S. Mongkolsuk. 1999. Construction and characterization of regulated l-arabinose-inducible broad host range expression vectors in Xanthomonas. FEMS Microbiol. Lett. 181:217-223. [DOI] [PubMed] [Google Scholar]
- 73.Surin, B. P., J. M. Watson, W. D. Hamilton, A. Economou, and J. A. Downie. 1990. Molecular characterization of the nodulation gene, nodT, from two biovars of Rhizobium leguminosarum. Mol. Microbiol. 4:245-252. [DOI] [PubMed] [Google Scholar]
- 74.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tibor, A., V. Wansard, V. Bielartz, R. M. Delrue, I. Danese, P. Michel, K. Walravens, J. Godfroid, and J. J. Letesson. 2002. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70:5540-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ugalde, J. E., C. Czibener, M. F. Feldman, and R. A. Ugalde. 2000. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68:5716-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Utsumi, R., T. Yagi, S. Katayama, K. Katsuragi, K. Tachibana, H. Toyoda, S. Ouchi, K. Obata, Y. Shibano, and M. Noda. 1991. Molecular cloning and characterization of the fusaric acid-resistance gene from Pseudomonas cepacia. Agric. Biol. Chem. 55:1913-1918. [PubMed] [Google Scholar]
- 79.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wassif, C., D. Cheek, and R. Belas. 1995. Molecular analysis of a metalloprotease from Proteus mirabilis. J. Bacteriol. 177:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Webber, M. A., and L. J. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zakharov, S. D., V. Y. Eroukova, T. I. Rokitskaya, M. V. Zhalnina, O. Sharma, P. J. Loll, H. I. Zgurskaya, Y. N. Antonenko, and W. A. Cramer. 2004. Colicin occlusion of OmpF and TolC channels: outer membrane translocons for colicin import. Biophys. J. 87:3901-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, Y., D. D. Bak, H. Heid, and K. Geider. 1999. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Biol. 289:1239-1251. [DOI] [PubMed] [Google Scholar]