Abstract
The plant toxin ricin is one of the most potent and lethal substances known. Ricin inhibits protein synthesis by removing a specific adenine from the highly conserved α-sarcin/ricin loop in the large rRNA. Very little is known about how ricin interacts with ribosomes and the molecular mechanism by which it kills cells. To gain insight to the mechanism of ricin-induced cell death, we set up yeast (Saccharomyces cerevisiae) as a simple and genetically tractable system to isolate mutants defective in cytotoxicity. Ribosomes were depurinated in yeast cells expressing the precursor form of the A chain of ricin (pre-RTA), and these cells displayed apoptotic markers such as nuclear fragmentation, chromatin condensation, and accumulation of reactive oxygen species. We conducted a large-scale mutagenesis of pre-RTA and isolated a panel of nontoxic RTA mutants based on their inability to kill yeast cells. Several nontoxic RTA mutants depurinated ribosomes and inhibited translation to the same extent as wild-type RTA in vivo. The mutant proteins isolated from yeast depurinated ribosomes in vitro, indicating that they were catalytically active. However, cells expressing these mutants did not display hallmarks of apoptosis. These results provide the first evidence that the ability to depurinate ribosomes and inhibit translation does not always correlate with ricin-mediated cell death, indicating that ribosome depurination and translation inhibition do not account entirely for the cytotoxicity of ricin.
The plant toxins ricin and abrin and the bacterial toxins Shiga and Shiga-like toxins are type II ribosome-inactivating proteins (RIPs) that inhibit protein synthesis by removing a highly conserved adenine from the α-sarcin/ricin loop (SRL) of the large rRNA (7, 8, 41). They consist of a catalytic A chain covalently joined by a disulfide bond to a cell binding B chain and are highly toxic to eukaryotic cells (13, 34, 41). Ricin naturally exists in the seeds of Ricinus communis (castor bean), a plant native to Asia, the Middle East, and southern Europe (13, 34). The B chain of ricin (RTB) is a lectin that binds galactose or N-acetylgalactosamine receptors on the surface of target cells and promotes subsequent endocytosis of the A chain (RTA) (13, 34). RTA is an N-glycosidase that depurinates ribosomes in the cytosol by removing a specific adenine (A4324 in rat 28S rRNA) from the highly conserved SRL in the large rRNA (7, 8). The depurination of the SRL has been reported to interfere with the elongation factor 1-dependent binding of amino acyl-tRNA to the ribosome as well as the GTP-dependent binding of elongation factor 2 and to inhibit protein synthesis at the translocation step (27, 35). Since ricin and many other AB toxins are quite stable, one or a few molecules are sufficient to kill cells (13). RTA has been widely used in cancer therapy as the active moiety of immunotoxins selectively targeted to cancer cells (5).
Very little is known about the relationship between ribosome depurination and the cytotoxicity of ricin. There is evidence that ricin induces apoptosis in a wide variety of animal cells by mechanisms other than protein synthesis inhibition (32). Ricin-induced apoptosis in HeLa cells was associated with oxidative stress, glutathione depletion, and activation of the caspase 3 cascade, followed by downstream events leading to apoptotic cell death (32, 39).
In the castor bean, ricin A and B chains are encoded by a single gene, which is translated into a preproprotein of 576 amino acids. The ricin precursor consists of a 35-residue N-terminal extension which contains the signal sequence (13). The mature RTA, which consists of 267 residues, is joined to the 262-residue mature RTB by a 12-residue linker peptide (13). The signal peptide directs the protein into the endoplasmic reticulum (ER), where proricin is core glycosylated and disulfide bonds are formed within the protein (13). Four disulfide bonds form within the RTB sequence, and the fifth one joins RTA with RTB in the ricin holotoxin. After RTB binds to its receptor on the surface of animal cells, a portion of the endocytosed RTA reaches the Golgi complex. RTA undergoes retrograde transport from the Golgi to the ER and is thought to enter the cytosol from the ER (23).
Due to its potent cytotoxicity and wide availability, ricin has been exploited as a biological weapon and an agent of bioterrorism (2, 19) and has been classified as a level B biothreat by the Centers for Disease Control and Prevention. Inhalation of small amounts of ricin aerosol can rapidly and irreversibly damage cells of the respiratory tract, leading to severe pulmonary incapacitation or death (3, 12). Currently, there is no approved antidote, vaccine, or other specific medical treatment option for ricin exposure. Since the wild-type RTA is highly toxic to humans, nontoxic recombinant vaccines need to be developed based on detailed understanding of the structure and molecular mechanism of action of ricin. Since several immunodominant human T- and B-cell epitopes map to the region surrounding the active site of RTA (6, 43), nontoxic forms which contain mutations outside the active site may lead to the development of safe and effective vaccines. A promising recombinant attenuated vaccine which contains two point mutations in RTA, one at a substrate binding site and another at a vascular leak syndrome-inducing site, is in clinical trials (46).
The search for residues critical for enzymatic activity has been carried out by expressing mutated RTA in Escherichia coli, followed by the subsequent analysis of in vitro enzymatic activity. By passaging the plasmid containing the mature RTA through an E. coli mutator strain, five different mutations were identified from residues at the active-site cleft, including Glu177, Trp211, Gly212, and Ser215, which are located on the same helix, and Ile252, which is located close to the C-terminal end(10). Recently, another large-scale mutagenesis screen was conducted using error-prone PCR of the mature RTA (1). While these studies identified several residues essential for enzymatic activity, the correlation between ribosome depurination and cytotoxicity has not been addressed. Here, we conducted a large-scale mutagenesis of the precursor form of RTA (pre-RTA) in the yeast Saccharomyces cerevisiae and isolated mutant forms of RTA based on their inability to kill yeast cells. The nontoxic RTA mutants were characterized with respect to their ability to depurinate ribosomes, inhibit translation, and cause cell death. To gain insight into the mechanism of ricin-induced cell death, we examined the hallmarks of apoptosis in cells expressing the wild-type form and the nontoxic forms of RTA. Apoptotic markers, such as chromatin condensation, nuclear fragmentation, and reactive oxygen species (ROS) production, were observed for yeast expressing the wild-type RTA but not for cells expressing the nontoxic mutants, even though they depurinated ribosomes and inhibited translation. These results provide evidence that ribosome depurination and translation inhibition are not sufficient for the cytotoxicity of ricin.
MATERIALS AND METHODS
Yeast expression vectors.
The pre-RTA cDNA was constructed in our laboratory by synthesizing the signal sequence (38) (Genewiz, North Brunswick, NJ) and ligating it to mature RTA in pRAIBI30 (30). The pre-RTA cDNA was then cloned into the yeast expression vector YEp351 downstream of the galactose-inducible GAL1 promoter (45). The pre-RTA plasmid was transformed into Saccharomyces cerevisiae strain W303 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 [from B. Thomas, Columbia University, New York, NY]), and transformants were selected on SD-Leu mediumcontaining 2% glucose.
Mutagenesis of pre-RTA.
Plasmid DNA mutagenesis was carried out as previously described (16). Briefly, the pre-RTA plasmid was incubated with 7% hydroxylamine for 20 h at 37°C and then precipitated and transformed into yeast. Yeast cells were plated onto SD-Leu supplemented with 2% glucose and replica plated onto SD-Leu containing 2% galactose. The pre-RTA plasmid was isolated from the colonies, which were able to grow on galactose and retransformed into yeast to confirm that the resistance was due to the plasmid. Plasmids isolated from colonies expressing RTA were characterized by sequence analysis.
Analysis of pre-RTA expression.
Yeast cells harboring the pre-RTA plasmid were grown on SD-Leu containing 2% glucose to an A600 of 0.3. Cells were pelleted at 2,000 × g for 5 min, resuspended in SD-Leu medium containing 2% galactose, and grown for 6 h to induce RTA expression. For immunoblot analysis, ER membrane fractions were isolated as previously described (36). The membrane fraction was dissolved in sodium dodecyl sulfate (SDS) buffer and heated at 37°C for 10 min before loading onto a 12% SDS-polyacrylamide gel. The blots were probed using polyclonal anti-RTA antibodies (1:3,000) produced in rabbits (Covance Research Products, Denver, PA). The blots were then stripped for 30 to 45 min with 8 M guanidine hydrochloride and reprobed with antibody to dolichol-phosphate mannose synthase (Dpm1p; Invitrogen, Carlsbad, CA) (1:4,000). Glycosylated and deglycosylated purified RTA standard was obtained from Sigma Aldrich (St. Louis, MO).
Analysis of growth rate.
Yeast cells were grown in SD-Leu medium containing 2% glucose to an A600 of 0.3 and were then transferred to SD-Leu containing 2% galactose. Aliquots were taken every 2 h, and the A600 was recorded. Doubling times were calculated based on exponential growth between 4 and 10 h postinduction.
Cell viability analysis.
Yeast cells expressing pre-RTA or pre-RTA mutants were grown on SD-Leu containing 2% glucose to an A600 of 0.3 and then transferred to SD-Leu medium containing 2% galactose to induce pre-RTA expression. A serial dilution of cells was plated on SD-Leu plates containing 2% glucose at 0, 4, 6, 10, and 12 h postinduction. Plates were incubated at 30°C for approximately 48 h.
rRNA depurination assay.
Dual primer extension analysis was conducted to quantify rRNA depurination as previously described (37). Briefly, 2 μg of total yeast RNA from cells expressing RTA was hybridized with 106 cpm of end-labeled depurination primer (5′-AGCGGATGGTGCTTCGCGGCAATG-3′). The second primer hybridized upstream of the depurination site close to the 5′ end of the 25S rRNA. To quantify the extent of depurination, the target RNA was initially hybridized in the presence of excess amounts (700 pmol) of the two [γ-32P]ATP-end-labeled negative-strand primers. The depurination primer described above annealed 73 nucleotides (nt) 3′ of the depurination site (A3137) on the 25S rRNA. The 25S control primer (5′-TTCACTCGCCGTTACTAAGG-3′) annealed 100 nt 3′ of the 25S rRNA 5′ end. To allow for accurate quantification, the labeled 25S control primer was diluted 1:4 with unlabeled 25S control primer. Superscript II reverse transcriptase was used in the primer extension assay as described above. Extension products for the control and depurination fragments (100 nt and 73 nt, respectively) were separated on a 7 M urea-5% polyacrylamide denaturing gel and visualized and quantified on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). The amount of total yeast RNA and rRNA used was previously determined to be in the linear range of detection.
Extraction of proteins from yeast and in vitro depurination assay.
Yeast cells (50 ml) containing pre-RTA or nontoxic mutants were induced on galactose for 6 h. Cells were resuspended in 1× low-salt buffer (20 mM HEPES-KOH, pH 7.6, 100 mM potassium acetate, 5 mM magnesium acetate, 1 mM EDTA, 2 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride) and lysed using glass beads. Samples were centrifuged briefly to remove cell debris and glass beads. The supernatant was transferred to a new tube and centrifuged at 100,000 × g for 30 min to remove cell membranes and ribosomes. The resulting supernatant (100 μl) was collected. Yeast ribosomes were isolated as previously described (44). Yeast ribosomes (15 μl) were incubated with RTA protein extracted from yeast (10 μl) in 10× RIP buffer (600 mM KCl, 100 mM Tris-HCl, pH 7.4, and 100 mM MgCl2) at 30°C for 30 min (44). One hundred microliters of 2× extraction buffer (240 mM NaCl, 50 mM Tris-HCl, pH 8.8, 20 mM EDTA, and 2% SDS) was added, and rRNA was extracted with phenol:chloroform and precipitated with ethanol. The rRNA was analyzed using the dual primer extension assay (37) as described above.
In vivo [35S]methionine incorporation.
Translation inhibition was measured by in vivo [35S]methionine incorporation. Yeast cells were grown to an A600 of 0.3 in SD-Leu-Met containing 2% glucose. Cells were then resuspended in SD-Leu-Met containing 2% galactose for 6 h to induce the expression of either wild-type pre-RTA or the mutant forms. At time zero, [35S]methionine was added to induced cells. After 30 min, 400 μl of yeast cells was removed for growth measurements, and additional aliquots of 400 μl were assayed for methionine incorporation in duplicate as previously described (37). The cpm was normalized to the A600 reading, and rates of translation were determined as cpm/A600/minute. Final results were displayed as percentages of total translation in yeast harboring the empty vector.
ROS production, cell death, and nuclear fragmentation.
Yeast cells were sampled at 0, 2, 4, 6, 10, and 24 h postinduction, stained with 0.05% Evans blue for 30 min, and then destained with water for 10 min. Cells were counted using a Zeiss Axiovert 200 inverted microscope. The percentage of cell death was calculated by counting ∼800 total cells as described by Xu et al. (47). All experiments were assayed in triplicate.
To detect nuclear fragmentation, cells were resuspended in phosphate-buffered saline buffer (20 mM sodium phosphate, 140 mM NaCl, pH 7.4) and stained with DAPI (4′,4-diaminido-2-phenylindole) (1 μg/ml) for 5 min at room temperature. After being stained, cells were washed with water five times and observed under a Zeiss Axiovert 200 inverted microscope with the epifluorescence setting. The digital images were acquired with a Zeiss Axiocam digital camera and software for image archival and management (Axiovision 3.0; Carl Zeiss Vision GmbH). ROS staining was carried out with diaminobenzidine (DAB) (1 mg/ml) for 10 min and was followed by washing with water three times (42). The stained cells were observed under a Zeiss Axiovert 200 inverted microscope as described above.
Intracellular production of H2O2 was detected using the oxidant-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (DCDHF-DA) (Invitrogen, Carlsbad, CA) (4). Two microliters of fresh 5 mM DCDHF-DA was added to 1 ml of yeast cell culture (107 cells) and incubated at 28°C for 30 min. The cells were then washed twice in sterile distilled water and resuspended in 1 ml of 50 mM Tris-HCl, pH 7.5. After 20 μl of chloroform and 10 μl of 0.1% SDS were added, the cells were incubated for 15 min and pelleted. The fluorescence of the supernatant was measured using an HTS700 Perkin Elmer bioassay reader (Wellesley, MA) with excitation at 490 nm and emission at 518 nm.
RESULTS
Random mutagenesis.
The full-length cDNA corresponding to pre-RTA, which consists of a 35-residue N-terminal extension and the 267-residue mature RTA, was cloned into the yeast expression vector downstream of the GAL1 promoter, mutagenized using hydroxylamine, and transformed into yeast. Cells were plated on medium containing glucose and replica plated on galactose-containing plates. Out of a total of 15,000 transformants screened, 128 (0.82%) were able to grow on galactose-containing medium. Immunoblot analysis showed that RTA expression was detected in 87 (68%) out of 128 colonies. Of the 87 colonies that showed detectable RTA expression, 37 expressed a protein of the same molecular weight as the wild-type RTA and 50 expressed smaller forms of RTA. All 87 plasmids isolated were retransformed into yeast to confirm that the loss of cytotoxicity was due to the plasmid. Nucleotide sequence analysis identified a total of 35 different mutations that led to the loss of cytotoxicity (Table 1). The majority of the mutations were isolated multiple times from colonies present on different plates, indicating that the mutagenesis screen using hydroxylamine was saturated. The mutants were divided into three groups. Group I (NT1001 to NT1016) contained 16 different mutations with a premature termination codon, resulting in a truncated form of the protein. Group II (NT1021 to NT1029) contained nine different frameshift mutations. In this group, the N termini of the proteins were the same as that of pre-RTA, but the C termini were different depending on the position of the frameshift mutation. The number of amino acids added to the C termini before the stop codon are indicated in Table 1. Group III (NT1031 to NT1044) consisted of 14 different point mutations that resulted in single amino acid changes in the protein. Only two mutants in this group (NT1039 and NT1040) contained double point mutations. To determine which mutation was necessary for the loss of cytotoxicity, single mutations were generated by site-directed mutagenesis. As shown in Table 1, the expression of pre-RTA containing the single point mutations was toxic to yeast, indicating that both mutations are required simultaneously for the loss of cytotoxicity.
TABLE 1.
Characterization of nontoxic RTA mutants obtained by random mutagenesis
| Plasmid | Protein change | No. of occurrences | Cytotoxicity | Depurination (% of wild-type) | Translation (% of vector control)b | Doubling time (h) |
|---|---|---|---|---|---|---|
| Pre-RTA | Yes | 100 | 35 | 18 | ||
| Vector control | No | 2.0 | 100 | 6.3 | ||
| Group I | ||||||
| NT1001 | Q19 stop | 1 | No | 5.0 | ND | ND |
| NT1002 | Q55 stop | 2 | No | 1.0 | ND | ND |
| NT1003 | Q112 stop | 2 | No | 1.0 | ND | ND |
| NT1004 | Q128 stop | 3 | No | 1.0 | ND | ND |
| NT1005 | G140 stop | 1 | No | 1.0 | ND | ND |
| NT1006 | S149 stop | 1 | No | 2.0 | ND | ND |
| NT1007 | Q160 stop | 3 | No | 12.3 | ND | ND |
| NT1008 | Q173 stop | 3 | No | 4.3 | ND | ND |
| NT1009 | S176 stop | 2 | No | 4.5 | ND | ND |
| NT1010 | Q182 stop | 2 | No | 3.4 | ND | ND |
| NT1011 | W211 stop | 4 | No | 3.7 | ND | ND |
| NT1012 | Q219 stop | 2 | No | 7.9 | ND | ND |
| NT1013 | Q223 stop | 3 | No | 4.3 | ND | ND |
| NT1014 | Q231 stop | 3 | No | 9.6 | 60 | 9.0 |
| NT1015 | Q233 stop | 6 | No | 6.7 | 59 | 8.7 |
| NT1016 | L248 stop | 2 | No | 15.8 | 58 | 7.0 |
| Group II | ||||||
| NT1021 | T77P + 4a | 1 | No | 0.4 | ND | ND |
| NT1022 | Y84T + 48 | 1 | No | 5.6 | ND | ND |
| NT1023 | F92S + 40 | 1 | No | 2.4 | ND | ND |
| NT1024 | R114D + 18 | 1 | No | 2.8 | ND | ND |
| NT1025 | P202L + 1 | 2 | No | 2.3 | ND | ND |
| NT1026 | R213D + 31 | 1 | No | 2.8 | ND | ND |
| NT1027 | R213D + 31c | 1 | No | 3.2 | ND | ND |
| NT1028 | S215F + 6 | 1 | No | 5.5 | ND | ND |
| NT1029 | P250L + 1 | 1 | No | 5.1 | ND | ND |
| Group III | ||||||
| NT1031 | G83D | 6 | No | 41 | 62 | 12 |
| NT1032 | G140R | 2 | No | 5 | 93 | 9.1 |
| NT1033 | A147P | 3 | No | 33 | 69 | 10 |
| NT1034 | E177K | 3 | No | 5.6 | 73 | 9.8 |
| NT1035 | Δ I 184 | 1 | No | 8.2 | 69 | 9.0 |
| NT1036 | E208K | 2 | No | 29 | 58 | 10 |
| NT1037 | G212E | 9 | No | 19 | 88 | 6.9 |
| NT1038 | S215F | 2 | No | 110 | 32 | 15 |
| NT1039 | P95L-E145K | 1 | No | 115 | 41 | 10 |
| NT1041 | P95L (by PCR) | Yes | 149 | 34 | 26 | |
| NT1042 | E145K (by PCR) | Yes | 108 | 27 | 18 | |
| NT1040 | P250L-A253V | 1 | No | 5.2 | 100 | 7.7 |
| NT1043 | P250L (by PCR) | Yes | 158 | 31 | 20 | |
| NT1044 | A253V (by PCR) | Yes | 175 | 30 | 24 |
The numbers after the mutations indicate the numbers of amino acids added to the C termini before a stop codon is generated.
ND, not determined.
The amino acids added to the C terminus are different from those added for the mutation listed immediately above.
Table 2 shows the frequency of the base pair changes, including the silent mutations. As expected for hydroxylamine mutagenesis, C-to-T and G-to-A transitions accounted for 82% of the total base pair changes. The frequency of other base pair changes was relatively low. The frequency of the deletions or additions was approximately 12%. Due to the high frequency of C-to-T changes, 11 out of 14 glutamines encoded by CAA/G in pre-RTA were changed to stop codons (TAA/G) resulting in premature termination.
TABLE 2.
Frequency of the mutations in pre-RTA
| Base pair change | No. of occurrences | Percentage (%) |
|---|---|---|
| C-to-T | 43 | 47 |
| C-to-A | 2 | 2 |
| C-to-G | 1 | 1 |
| G-to-A | 32 | 35 |
| G-to-C | 2 | 2 |
| T-to-A | 2 | 2 |
| Deletion of T | 3 | 3 |
| Deletion of A | 1 | 1 |
| Deletion of C | 2 | 2 |
| Deletion of G | 2 | 2 |
| Deletion of TAT | 1 | 1 |
| Addition of T | 1 | 1 |
Wild-type pre-RTA and the nontoxic mutants are expressed in yeast.
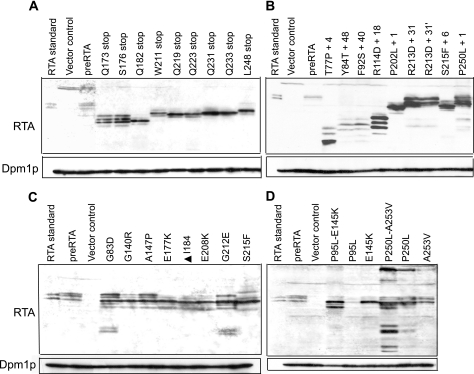
Immunoblot analysis using polyclonal antibodies against RTA was used to examine protein expression in each mutant at 6 h postinduction. As shown in Fig. 1, purified RTA from Ricinus communis contains two bands. Based on comparison with the deglycosylated RTA standard, the upper band corresponds to the glycosylated from of RTA (data not shown). The ER membrane fraction isolated from yeast harboring the pre-RTA plasmid contained two bands that comigrated with the purified RTA (Fig. 1), indicating that pre-RTA synthesized in yeast is processed the in same way as RTA synthesized in plants. A very low level of protein was detected in the cytosolic fraction, indicating that the majority of RTA expressed in yeast is associated with the ER membranes (data not shown).
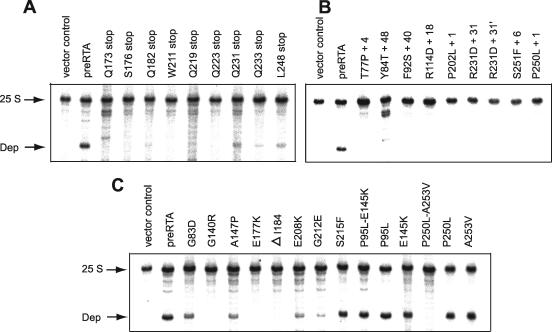
FIG. 1.
Immunoblot analysis of RTA expression. Membrane fractions (15 μg) isolated from cells (an amount corresponding to an optical density at 600 nm of 1) expressing pre-RTA or mutants containing a premature termination codon (A), a frameshift mutation (B), a single point mutation (C), or a double point mutation (D) were separated on a 12% SDS-polyacrylamide gel and probed with polyclonal anti-RTA (1:3,000). The RTA standard (1.5 ng) was purified RTA (Sigma, St. Louis, MO). The blots were probed with the ER membrane marker Dpm1p as a loading control.
Immunoblot analysis indicated that all 39 mutants that contained premature termination codons (Fig. 1A), frameshift mutations (Fig. 1B), or point mutations (Fig. 1C and D) expressed detectable levels of RTA. The blot was reprobed with antibody against the ER membrane protein Dpm1p as a loading control. The majority of the mutant proteins migrated on the SDS-polyacrylamide gels according to their predicted sizes. In several mutants, different forms of the protein were observed. The P250L-A253V double mutantcontained both larger and smaller forms of the protein, suggesting possible aggregation and breakdown. In general, yeast cells carrying the nontoxic forms of RTA expressed higher levels of protein than cells carrying wild-type or toxic (P95L) forms of pre-RTA. These results demonstrated that the loss of cytotoxicity was not due to the loss of protein expression.
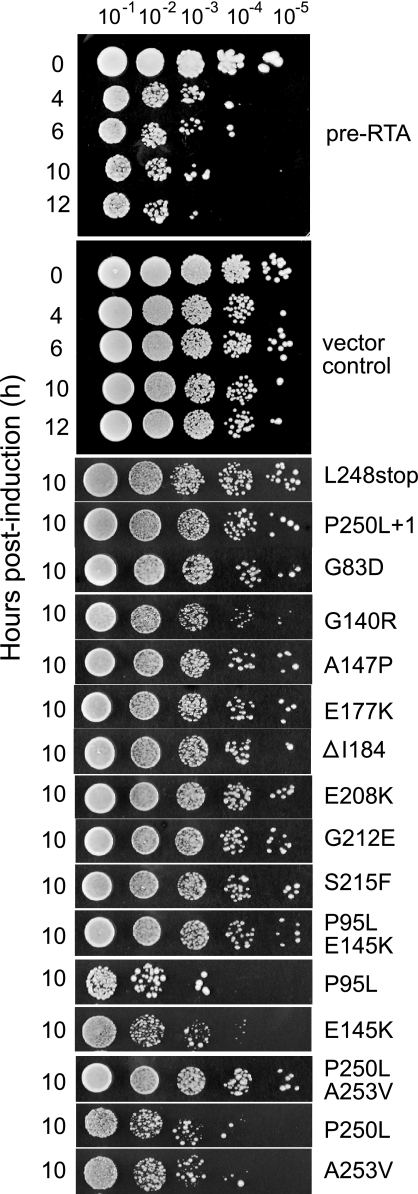
Pre-RTA mutants are not toxic to yeast cells.
Irreversible growth inhibition was examined by conducting viability assays. Cells expressing pre-RTA or the nontoxic mutants were plated on glucose after induction in galactose for the indicated times (Fig. 2). Upon induction in yeast, the wild-type RTA reduced the viability of cells by almost 3 logs at 10 h (Fig. 2, top panel). In contrast, the nontoxic RTA mutants exhibited minimal loss of viability at 10 h postinduction. All nontoxic mutants analyzed exhibited viability similar to that seen with cells harboring the empty vector. Only the L248stop mutant in group I and the P250L + 1 mutant in group II are shown because they had the shortest deletions at their C termini. The two double mutants, the P95L-E145K and P250L-A253V mutants, were nontoxic and did not reduce viability. However, the single mutants corresponding to each double mutant (the P95L, E145K, P250L, and A253V mutants) reduced the viability of yeast cells (Fig. 2).
FIG. 2.
Viability of cells expressing pre-RTA and the mutant forms of RTA. Yeast cells were first grown in SD-Leu medium supplemented with 2% glucose to an optical density at 600 nm of 0.3 and then transferred to SD-Leu supplemented with 2% galactose. At the indicated hours postinduction on SD-Leu medium containing galactose (left), serial dilutions were spotted on SD-Leu plates supplemented with 2% glucose. The top two panels show the cell viability up to 12 h in cells expressing the wild-type pre-RTA or harboring the empty vector.
Nontoxic RTA mutants depurinate the rRNA.
To determine if the reduced toxicity of the pre-RTA mutants was due to reduced depurination of ribosomes, total RNA was isolated from yeast cells expressing the wild-type or mutant forms of RTA, and depurination of the rRNA was examined by a dual primer extension assay (37). As shown in Fig. 3A, ribosomes were depurinated in cells expressing pre-RTA. Cells expressing the Q231stop, Q233stop, and L248stop mutants showed a weak depurination band, indicating that these mutants retained a low level of ribosome depurination (Fig. 3A). The rest of the C-terminal deletion mutants did not depurinate ribosomes. The frameshift mutants did not show any depurination (Fig. 3B). In contrast, 5 of the 10 point mutants depurinated yeast ribosomes in vivo (Fig. 3C). The depurination assay was repeated several times with all mutants, and the levels of depurination calculated from independent experiments are averaged in Table 1. As shown in Table 1, the S215F mutant and the P95L-E145K double mutant depurinated ribosomes at 110% and 115%, respectively. These results indicated that both mutants depurinated ribosomes at a level similar to that seen for the wild-type pre-RTA in vivo, but unlike the wild-type pre-RTA, they were nontoxic (Table 1) and did not reduce the viability of yeast cells (Fig. 2).
FIG. 3.
Ribosome depurination in yeast expressing pre-RTA and the mutant forms in vivo. Total RNA isolated after 6 h of growth on galactose was analyzed by dual primer extension analysis using two different end-labeled primers: the depurination primer (Dep), which was used to measure the extent of depurination, and the 25S rRNA primer (25 S), which was used to measure the total amount of 25S rRNA (37). Primer extension analysis of the mutants with a change corresponding to a premature termination codon (A), a frameshift mutation (B), or a point mutation (C) is shown. Primer extension analysis of cells harboring the empty vector is shown as a control.
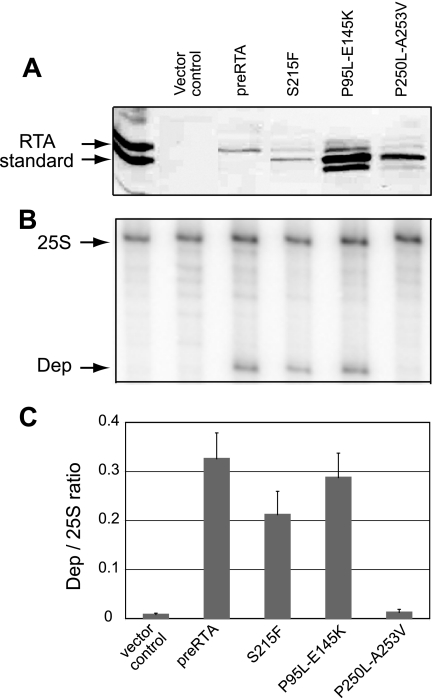
To determine if the mutant proteins were enzymatically active in vitro, we extracted the S215F and P95L-E145K mutants from the cytosolic fraction of yeast cells. These mutants were selected since they depurinated ribosomes at a level similar to that seen for the wild-type RTA in vivo. Since the P250L-A253V mutant was nontoxic and did not depurinate ribosomes in vivo, we used it as a control for the in vitro depurination experiments. Purified yeast ribosomes were treated with similar amounts of the wild-type RTA and the mutant proteins isolated from yeast (Fig. 4A), and ribosome depurination was examined by dual primer extension analysis. As shown in Fig. 4B, the wild-type RTA extracted from yeast depurinated yeast ribosomes in vitro. Both the S215F and the P95L-E145K mutants depurinated yeast ribosomes in vitro, while the P250L-A253V mutant was not able to depurinate ribosomes. The levels of ribosome depurination quantified from three independent depurination experiments are shown in Fig. 4C. The in vitro depurination results were similar to those obtained in vivo and demonstrated that the S215F and the P95L-E145K mutants were catalytically active, while the P250L-A253V mutant was not active.
FIG. 4.
Ribosome depurination by wild-type RTA and mutants in vitro. (A) Total protein extracted from the cytosolic fraction of 10 ml of yeast cells expressing pre-RTA or the mutants was analyzed on a 12% SDS-polyacrylamide gel and probed with polyclonal anti-RTA (1:3,000). The first lane is purified RTA standard (10 ng). (B) Ribosomes isolated from yeast cells were treated with either wild-type RTA or the S215F, P95L-E145K, and P250L-A253V mutants extracted from the cytosolic fractions of yeast cells in vitro, and the extents of depurination were determined by dual primer extension analysis (37). The first lane corresponds to the untreated ribosomes, and the second lane corresponds to primer extension analysis with protein extracted from cells harboring the empty vector. (C) The extents of ribosome depurination were quantified using a PhosphorImager from three independent depurination experiments with the wild-type and mutant proteins extracted from yeast in vitro.
Ribosome depurination results in translation inhibition.
To determine if ribosome depurination correlated with translation inhibition, we examined total translation in cells expressing pre-RTA and compared it to that in control cells harboring the empty vector. Translation rates were determined by measuring the slope of the [35S]methionine incorporation curve and expressed as percentages of the translation rate in cells harboring the empty vector. As shown in Table 1, in cells expressing the wild-type pre-RTA, the rate of translation was reduced to 35% of the rate of translation in cells harboring the empty vector (100%). Total translation was not inhibited in yeast expressing the RTA mutants that did not depurinate ribosomes. In contrast, total translation was inhibited in cells expressing the S215F mutant or the P95L-E145K double mutant, which depurinated ribosomes (Table 1). These results demonstrated that translation inhibition correlated well with ribosome depurination, consistent with the inability of the depurinated ribosomes to translate protein. In contrast, translation inhibition did not correlate with cytotoxicity, indicating that translation inhibition does not entirely account for the cytotoxicity of RTA. These results were different from those observed with yeast expressing a single-chain RIP, pokeweed antiviral protein (PAP). Ribosome depurination did not lead to translation inhibition in yeast expressing several nontoxic PAP mutants, including the N70A mutant (36), suggesting possible differences in the ways that translation is inhibited by PAP and ricin.
Cell growth rate does not always correlate with ribosome depurination.
The rate of growth was measured by examining the doubling time of the mutants. As shown in Table 1, the doubling time of cells expressing pre-RTA was 18 h, while cells harboring the vector had a doubling time of 6.3 h. The doubling times of cells expressing the E177K active-site mutant or the nontoxic G140R and ΔI184 mutants were longer than that of cells harboring the empty vector, even though these mutants did not depurinate ribosomes (Fig. 3C) or inhibit translation. Although ribosomes were depurinated and translation was inhibited in cells expressing the P95L-E145K double mutant, the doubling time of cells expressing this mutant (10 h) was similar to the doubling time of cells expressing the E177K active-site mutant (9.8 h). In contrast, the doubling time of cells expressing the S215F mutant (15 h), which depurinated ribosomes and inhibited translation, was similar to that of cells expressing the wild-type pre-RTA, although this mutant was nontoxic. These results demonstrated that the rate of growth of RTA mutants did not always correlate with the extent of ribosome depurination, indicating that the reduction in growth is not entirely due to ribosome depurination.
Characteristic markers of apoptosis are observed for cells expressing the pre-RTA.
The previous results indicated that the reduction in growth observed for cells expressing pre-RTA was not entirely due to ribosome depurination or translation inhibition. To assess whether cell death induced by the expression of pre-RTA was accompanied by morphological features of apoptosis, we examined apoptotic markers in yeast expressing several RTA mutants. Cells expressing the wild-type pre-RTA, the S215F mutant, and the P95L-E145K mutant were analyzed, since these mutants depurinated ribosomes at similar levels. Cells expressing the E177K active-site mutant and the double P250L-A253V mutant were used as negative controls, since these mutants were not toxic and did not depurinate ribosomes. The single mutants corresponding to each double mutant were used as positive controls, since they were toxic and depurinated ribosomes.
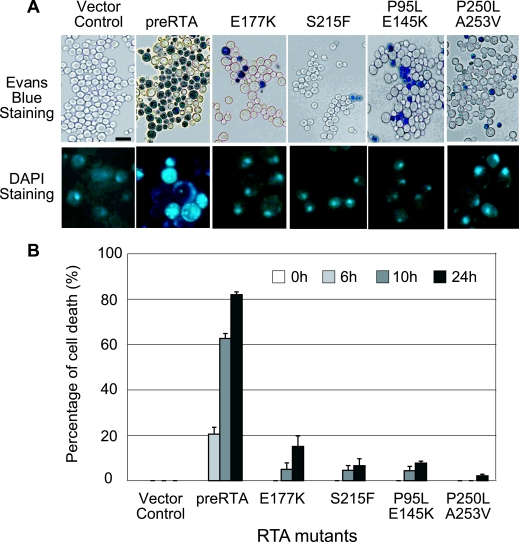
Cells growing in liquid culture were stained with Evans blue at different times after induction. The extent of staining at 24 h postinduction is shown in Fig. 5A and is quantified in Fig. 5B. In cells expressing the wild-type pre-RTA or the toxic mutants, cell death was observed at 6 h after induction and gradually increased up to 24 h (Fig. 5A and B; also, see Fig. S1 in the supplemental material). In contrast, minimal loss of cell viability was observed for cells expressing the nontoxic mutants or for cells harboring the empty vector up to 24 h after induction (Fig. 5A and B; also, see Fig. S1 in the supplemental material). These results correlated well with the viability assay results (Fig. 2).
FIG. 5.
Analysis of cell death and nuclear fragmentation in yeast expressing pre-RTA and the mutants. (A) Cells were stained with Evans blue or DAPI at 24 h after induction and visualized using a Zeiss Axiovert 200 inverted microscope (magnification, ×40). The DAPI-stained nuclei are shown enlarged 40 times relative to the yeast cells. (B) The percentages of cell death at different hours after induction were quantified and are represented as the means ± standard deviations (n = 3).
Chromatin condensation and DNA fragmentation are typical markers for apoptosis in yeast (26). DAPI staining of the cells expressing the nontoxic mutants or harboring the vector showed normal and single round-shaped nuclei, whereas cells expressing the wild-type pre-RTA or the toxic mutants revealed an abnormal shape and a fragmented nuclear phenotype at 24 h postinduction (Fig. 5A; also, see Fig. S2 in the supplemental material).
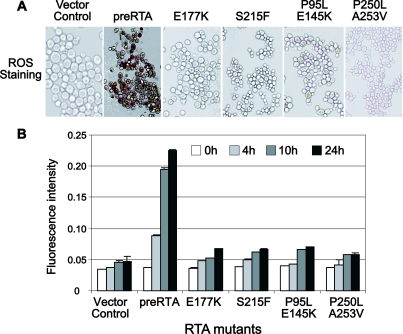
The accumulation of ROS is a significant trigger of apoptosis in yeast (25). To determine whether yeast cell death induced by ricin is accompanied by the production of ROS, cells were stained with DAB and visualized under a Zeiss Axiovert 200 inverted microscope at 24 h postinduction (Fig. 6). In cells expressing the nontoxic mutants or harboring the vector, there was no staining for ROS up to 24 h postinduction (Fig. 6A; also, see Fig. S3 in the supplemental material). In contrast, DAB staining became visible at 6 h after induction in cells expressing pre-RTA or the toxic E145K, P250L, and A253V mutants and increased up to 24 h (see Fig. S3 in the supplemental material). These results suggested that expression of the wild-type pre-RTA resulted in increased ROS accumulation and promoted apoptosis-like cell death in yeast.
FIG. 6.
Production of ROS in cells expressing pre-RTA or the mutants. (A) Yeast cells were stained using DAB at 24 h after induction. (B) The H2O2 production was quantified using DCDHF-DA. The results are represented as the means ± standard deviations (n = 3).
To quantify intracellular ROS production, DCDHF-DA oxidation was used as a marker to measure intracellular levels of H2O2. As shown in Fig. 6B, the increased level of H2O2 observed in cells expressing the pre-RTA up to 24 h postinduction correlated well with cell death and ROS staining (Fig. 6A; also, see Fig. S3 in the supplemental material). In contrast, the H2O2 levels did not increase in cells expressing the nontoxic RTA mutants up to 24 h after induction. These results indicated that RTA expression induces oxidative damage in yeast cells, leading to increased ROS levels. Taken together, our results indicate that apoptosis-like cell death is induced in yeast expressing pre-RTA and correlates well with the increased generation of ROS. In contrast, apoptosis-like cell death and production of ROS are not observed for yeast expressing the nontoxic forms of RTA.
DISCUSSION
We have established Saccharomyces cerevisiae as a simple and genetically tractable system to investigate the biological activity of ricin and demonstrated that expression of pre-RTA from the galactose-inducible GAL1 promoter is lethal to yeast. To investigate the relationship between ribosome depurination and the cytotoxicity of ricin, we conducted large-scale mutagenesis of pre-RTA in yeast and isolated nontoxic RTA mutants on the basis of their inability to kill yeast cells. Since RTA isolated from the castor bean is glycosylated, we used pre-RTA instead of the mature RTA to allow glycosylation of RTA in vivo in yeast. In previous studies, various random mutagenesis methods have been used to isolate nontoxic RTA mutants from yeast (1, 10). Systematic deletion analysis has been used to identify amino acids critical for the activity of RTA in vitro (18, 31). Our study is the first report which determines the precise effects of each mutation on cytotoxicity, ribosome depurination, and translation inhibition and provides evidence that cytotoxicity of RTA is not entirely due to ribosome depurination.
In a recent study using PCR-based mutagenesis of the mature RTA gene, 80% of the changes observed were T-to-C and A-to-G transitions (1). In contrast, 80% of the changes observed in our study using chemical mutagenesis were either C-to-T or G-to-A transitions (Table 2). Multiple mutations were obtained with the PCR-based mutagenesis, and single mutations had to be generated to determine if they were responsible for the phenotype (1). In contrast, only 2 of the 35 mutants generated in our study using hydroxylamine contained double mutations (Table 1). Analysis of the single point mutant constructs corresponding to each double point mutant indicated that both mutations were necessary for the loss of cytotoxicity.
Of the nine frameshift mutations isolated here, seven were caused by a single base pair deletion. Two had deletions of two base pairs and were isolated only once. The 25 mutations with stop codons or single amino acid changes were caused by single base pair changes. Most of these mutations were isolated more than twice, and some were isolated nine times from different plates, indicating that the mutation screen using hydroxylamine was saturated. The codons encoding 11 out of the 14 glutamines in pre-RTA were changed to stop codons, providing further evidence that our mutation screen was saturated. Mutations were not isolated at positions corresponding to Gln5, Gln98, and Gln266, since changing the codon encoding Gln5 to a stop codon would result in a four-amino-acid peptide that would not be detected by immunoblot analysis. Changing the codon encoding Gln266 to a stop codon would not affect the cytotoxicity of RTA (18); thus, a mutant with this characteristic would not have been isolated by our screen. Therefore, the only mutation in a Gln residue codon not isolated here was the change of a codon encoding Gln98 into a stop codon, which would encode a nontoxic form of RTA.
Mutations were not isolated in the N-terminal extension of pre-RTA, suggesting that these mutations did not affect the cytotoxicity of RTA. A mutation in the N-terminal extension may disrupt the ability of pre-RTA to translocate through the ER membrane without affecting its cytotoxicity, since expression of the mature RTA is toxic to yeast (1). Similarly, mutations were not recovered at Asn10 and Asn236, which are glycosylated in the mature RTA. These results provide further evidence that glycosylation does not affect the cytotoxicity of RTA (33).
The three-dimensional X-ray structure of ricin indicates that the amino-terminal 117 residues form six β-strands and two α-helices (28). The central domain is made up of five helices, of which the longest, helix E, runs through the center of the molecule and contains the key active-site residues, Glu177 and Arg180 (28). The third domain consists of a two-stranded antiparallel β-sheet and an α-helix, which is anchored to the first helix in the N-terminal domain. It forms part of the active-site cleft and interacts with RTB in the holotoxin (28).
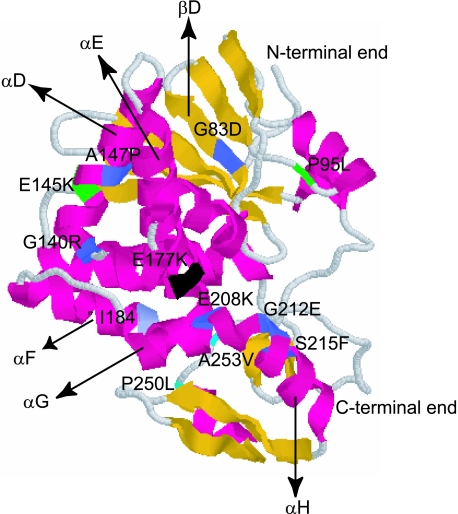
The results from three separate random mutagenesis studies and several systematic deletion experiments indicate that there are five regions important for the function of RTA: β-strand D; α-helices D, E, and G to H; and a hydrogen-bonded turn and β-strand region (Ile249 to Val256) close to the C-terminal end of the protein (Fig. 7) (1, 10, 18, 31). The α-helix E contains the active-site residues, Glu177 and Arg180. The E177K mutation was isolated several times in different studies (1, 10). Mutations in Arg180 (R180G) (1) in helix E and Ile184 (ΔI184) (31) at the beginning of helix F disrupted the enzymatic activity of RTA in vitro, emphasizing the critical nature of this region. In our study, deletion of Ile184 led to a loss of cytotoxicity and ribosome depurination in vivo (Table 1). Ile184 may be critical for ribosome depurination, since it contacts Phe181 and methylene carbons of Glu177, stabilizing the active center (31).
FIG. 7.
Three-dimensional structure of mature RTA showing the positions of the point mutations and the α-helices and β-sheets that contain these mutations. Coordinates of the crystal structure from the Protein Data Bank (1J1M; http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?form=6&db=t&Dopt=s&uid=25955) were used in conjunction with the Protein Explorer software (http://www.umass.edu/microbio/chime/pe_beta/pe/protexpl/frntdoor.htm) to create this figure. The point mutations are shown in blue. The active site mutation is shown in black. The double mutations are shown in green and cyan.
A significant number of mutations that eliminated the cytotoxicity of RTA resided within the α-helices G to H. Several mutations in this region, including those at Glu208 and Gly212, reduced the depurination activity and the cytotoxicity of RTA (Table 1). Previous studies indicated that Glu208, which is at the bottom of the active-site cleft, can substitute for Glu177 in the E177A mutant (11). Deletion of Gly212 led to the loss of enzymatic activity of RTA in vitro (31). In contrast, the point mutation at Ser215 (S215F) in helix H did not affect the ribosome depurination in vivo but significantly decreased the cytotoxicity of RTA (Table 1 and Fig. 3). Previous studies showed that Ser215 could be deleted from RTA without a complete loss of enzymatic activity in vitro (30). These results indicated that the role of Ser215 in cytotoxicity could be separated from its role in ribosome depurination.
The α-helix D crosses helix E in the middle (Fig. 7). Each of the amino acids in helix D could be singly deleted, provided that the deletion does not disrupt the amphipathicity of the helix (29). The A147P mutation reduced the depurination activity of RTA and led to the loss of its cytotoxicity (Table 1). The deletion of Ala147 in helix D abolished the activity of RTA in vitro (31). The A147P mutation likely disrupted the structure of helix D in the middle, destabilizing the active site. The point mutation at Gly140 (G140R), which is located at the beginning of helix D, abolished both cytotoxicity and depurination (Table 1 and Fig. 3C). In contrast, deletion of this glycine did not affect the activity of RTA in vitro (31). These results indicate that the structure of RTA might be affected more when Gly140 is changed to an arginine than when it is deleted.
Mutation G83D (NT1031), which is in β-strand D, eliminated the cytotoxicity of RTA in yeast and reduced its depurination activity (Table 1). Since Gly83 is relatively distant from the active site, it is unlikely that Gly83 participates in the catalysis. Previous studies indicated that RTA lost its depurination activity when Gly83 was deleted (18, 31). A point mutation in the corresponding Gly in PAP (G75D) led to a loss of depurination in vivo (15) and affected the binding of PAP to ribosomes (14). These results suggest that β-strand D might be important for the interaction of RTA with the ribosome, such that a mutation in Gly83 may affect binding of RTA to ribosomes.
The final important region is close to the C-terminal end of RTA. Nonsense mutations demonstrated that deleting 19 (L248 stop) amino acids from the C-terminal end of pre-RTA eliminated its cytotoxicity in yeast. The last frameshift mutation, P250L+S, which deleted 17 amino acids from the C terminus and changed Pro250 to Leu, also eliminated the ribosome depurination activity (Table 1). Previous studies indicated that deletions between Arg258 and Pro262 or Pro263 and Phe267 did not affect the cytotoxicity of RTA (18). However, point mutations upstream of Arg258, at Ile252, Leu254, and Val256, eliminated the cytotoxicity of RTA (1, 10). In our study, the single mutations at Pro250 (P250L) and at Ala253 (A253V) had little effect on the cytotoxicity of RTA or its ability to depurinate ribosomes. However, when they were combined (P250L-A253V), both cytotoxicity and ribosome depurination were eliminated. These results indicated that the C-terminal region of RTA is critical for ribosome depurination and cytotoxicity. Different forms of protein were observed for the membrane fraction from cells expressing P250L-A253V (Fig. 1), suggesting that the point mutations at the C terminus may have prevented the retrotranslocation of RTA from the ER to the cytosol (40).
Ricin and other protein toxins can cause apoptotic cell death in animal cells that is associated with DNA fragmentation and target cell lysis (20). However, the detailed mechanism of ricin-induced apoptosis and in particular the mechanism by which protein synthesis inhibition by ricin results in apoptosis are unclear. Several lines of evidence suggest that the specific attack on the 28S rRNA of the large ribosomal subunit by RTA may cause the ribotoxic stress response, which in turn leads to apoptosis through a stress-mediated signaling pathway (17). The cleavage of DNA in the nucleus and nuclear fragmentation are typical apoptotic hallmarks in yeast (9, 21). Cells expressing the nontoxic RTA mutants or harboring the empty vector showed a normal and single round-shaped nuclei, whereas cells expressing the pre-RTA or the toxic mutants harbored abnormally shaped and fragmented nuclei (Fig. 5A; also, see Fig. S2 in the supplemental material).
The induction of ROS plays a major role in mediating cell death in yeast (26). Previous studies have demonstrated the accumulation of ROS in yeast cells exposed to oxidative stress or expressing mammalian Bax (22), as well as in the cell cycle mutant cdc48S565G and the gsh1 deletion mutant in the absence of glutathione (24). The involvement of ROS upstream of caspase-3 activation has been demonstrated for signal transduction pathways induced by ricin in various cell types (39). These studies have led to the conclusion that the production of ROS is a necessary and sufficient condition for the induction of apoptosis in yeast. To determine if ROS is produced in yeast expressing pre-RTA, cells were stained with DAB at different times after induction. A weak ROS staining was detected after 6 h of growth on galactose; this staining became significantly more intense thereafter in pre-RTA-expressing cells, as well as in cells expressing the toxic mutants (see Fig. S3 in the supplemental material). In contrast, no ROS staining was detected in cells harboring the vector or expressing the nontoxic mutants (Fig. 6A). Similar results were obtained when intracellular ROS levels were quantitatively determined. A nearly fivefold increase in ROS production was observed for cells expressing pre-RTA compared to controls at 24 h after the galactose shift (Fig. 6B). The induction of ROS correlated well with cell death (Fig. 5A), suggesting that ROS may act as an effector of apoptosis and trigger the subsequent apoptotic events. Taken together, our results strongly suggest that apoptosis-like cell death is induced in yeast expressing the wild-type or toxic forms of pre-RTA and correlates well with the increased generation of ROS. In contrast, apoptotic features are not observed in cells expressing the nontoxic RTA mutants. These results present the first evidence that apoptosis-like cell death induced by RTA is not entirely due to ribosome depurination and translation inhibition. The nontoxic forms of RTA with mutations outside the active site may be candidates for the development of safe and effective recombinant vaccines against ricin. They may also be of value in the development of more effective immunotoxins with reduced side effects.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health grant (AI59720) and a National Science Foundation grant (MCB0348299) to N. E. Tumer and a National Science Foundation GK-12 STEM Fellowship to M. Baricevic.
We thank Michael Lawton, Eric Lam, Bijal Parikh, and Wendie Cohick for critical reading of the manuscript, Rong Di for the construction of the RTA plasmid, Tiffany Kung and Jia-Chi Chiou for help with the analysis of the RTA mutants, and Ira Wool for the generous gift of pRAIBI30.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Allen, S. C., A. Byron, J. M. Lord, J. Davey, L. M. Roberts, and G. Ladds. 2005. Utilization of the budding yeast Saccharomyces cerevisiae for the generation and isolation of non-lethal ricin A chain variants. Yeast 22:1287-1297. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 2002. Bioterrorism: from threat to reality. Annu. Rev. Microbiol. 56:167-185. [DOI] [PubMed] [Google Scholar]
- 3.Audi, J., M. Belson, M. Patel, J. Schier, and J. Osterloh. 2005. Ricin poisoning: a comprehensive review. JAMA 294:2342-2351. [DOI] [PubMed] [Google Scholar]
- 4.Balzan, R., K. Sapienza, D. R. Galea, N. Vassallo, H. Frey, and W. H. Bannister. 2004. Aspirin commits yeast cells to apoptosis depending on carbon source. Microbiology 150:109-115. [DOI] [PubMed] [Google Scholar]
- 5.Bolognesi, A., and L. Polito. 2004. Immunotoxins and other conjugates: pre-clinical studies. Mini-Rev. Med. Chem. 4:563-583. [DOI] [PubMed] [Google Scholar]
- 6.Castelletti, D., G. Fracasso, S. Righetti, G. Tridente, R. Schnell, A. Engert, and M. Colombatti. 2004. A dominant linear B-cell epitope of ricin A-chain is the target of a neutralizing antibody response in Hodgkin's lymphoma patients treated with an anti-CD25 immunotoxin. Clin. Exp. Immunol. 136:365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endo, Y., and K. Tsurugi. 1987. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 262:8128-8130. [PubMed] [Google Scholar]
- 8.Endo, Y., and K. Tsurugi. 1988. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 263:8735-8739. [PubMed] [Google Scholar]
- 9.Fahrenkrog, B., U. Sauder, and U. Aebi. 2004. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 117:115-126. [DOI] [PubMed] [Google Scholar]
- 10.Frankel, A., D. Schlossman, P. Welsh, A. Hertler, D. Withers, and S. Johnston. 1989. Selection and characterization of ricin toxin A-chain mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel, A., P. Welsh, J. Richardson, and J. D. Robertus. 1990. Role of arginine 180 and glutamic acid 177 of ricin toxin A chain in enzymatic inactivation of ribosomes. Mol. Cell. Biol. 10:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths, G. D., C. D. Lindsay, A. C. Allenby, S. C. Bailey, J. W. Scawin, P. Rice, and D. G. Upshall. 1995. Protection against inhalation toxicity of ricin and abrin by immunization. Hum. Exp. Toxicol. 14:155-164. [DOI] [PubMed] [Google Scholar]
- 13.Hartley, M. R., and J. M. Lord. 2004. Cytotoxic ribosome-inactivating lectins from plants. Biochim. Biophys. Acta 1701:1-14. [DOI] [PubMed] [Google Scholar]
- 14.Hudak, K. A., A. B. Hammell, J. Yasenchak, N. E. Tumer, and J. D. Dinman. 2001. A C-terminal deletion mutant of pokeweed antiviral protein inhibits programmed +1 ribosomal frameshifting and Ty1 retrotransposition without depurinating the sarcin/ricin loop of rRNA. Virology 279:292-301. [DOI] [PubMed] [Google Scholar]
- 15.Hudak, K. A., B. A. Parikh, R. Di, M. Baricevic, M. Santana, M. Seskar, and N. E. Tumer. 2004. Generation of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: evidence that ribosome depurination is not sufficient for cytotoxicity. Nucleic Acids Res. 32:4244-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur, Y., D. J. Hwang, O. Zoubenko, C. Coetzer, F. M. Uckun, and N. E. Tumer. 1995. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc. Natl. Acad. Sci. USA 92:8448-8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iordanov, M. S., D. Pribnow, J. L. Magun, T. H. Dinh, J. A. Pearson, S. L. Chen, and B. E. Magun. 1997. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 17:3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitaoka, Y. 1998. Involvement of the amino acids outside the active-site cleft in the catalysis of ricin A chain. Eur. J. Biochem. 257:255-262. [DOI] [PubMed] [Google Scholar]
- 19.Knight, B. 1979. Ricin—a potent homicidal poison. Br. Med. J. 1(6159):350-351. [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu, N., T. Oda, and T. Muramatsu. 1998. Involvement of both caspase-like proteases and serine proteases in apoptotic cell death induced by ricin, modeccin, diphtheria toxin, and pseudomonas toxin. J. Biochem. (Tokyo) 124:1038-1044. [DOI] [PubMed] [Google Scholar]
- 21.Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 22.Ligr, M., F. Madeo, E. Frohlich, W. Hilt, K. U. Frohlich, and D. H. Wolf. 1998. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 438:61-65. [DOI] [PubMed] [Google Scholar]
- 23.Lord, J. M., E. Deeks, C. J. Marsden, K. Moore, C. Pateman, D. C. Smith, R. A. Spooner, P. Watson, and L. M. Roberts. 2003. Retrograde transport of toxins across the endoplasmic reticulum membrane. Biochem. Soc. Trans. 31:1260-1262. [DOI] [PubMed] [Google Scholar]
- 24.Madeo, F., E. Frohlich, and K. U. Frohlich. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. J. Sigrist, D. H. Wolf, and K. U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madeo, F., E. Herker, S. Wissing, H. Jungwirth, T. Eisenberg, and K. U. Frohlich. 2004. Apoptosis in yeast. Curr. Opin. Microbiol. 7:655-660. [DOI] [PubMed] [Google Scholar]
- 27.Montanaro, L., S. Sperti, A. Mattioli, G. Testoni, and F. Stirpe. 1975. Inhibition by ricin of protein synthesis in vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem. J. 146:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monzingo, A. F., and J. D. Robertus. 1992. X-ray analysis of substrate analogs in the ricin A-chain active site. J. Mol. Biol. 227:1136-1145. [DOI] [PubMed] [Google Scholar]
- 29.Morris, K. N., and I. G. Wool. 1994. Analysis of the contribution of an amphiphilic alpha-helix to the structure and to the function of ricin A chain. Proc. Natl. Acad. Sci. USA 91:7530-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, K. N., and I. G. Wool. 1992. Determination by systematic deletion of the amino acids essential for catalysis by ricin A chain. Proc. Natl. Acad. Sci. USA 89:4869-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munishkin, A., and I. G. Wool. 1995. Systematic deletion analysis of ricin A-chain function. Single amino acid deletions. J. Biol. Chem. 270:30581-30587. [DOI] [PubMed] [Google Scholar]
- 32.Narayanan, S., K. Surendranath, N. Bora, A. Surolia, and A. A. Karande. 2005. Ribosome inactivating proteins and apoptosis. FEBS Lett. 579:1324-1331. [DOI] [PubMed] [Google Scholar]
- 33.O'Hare, M., L. M. Roberts, P. E. Thorpe, G. J. Watson, B. Prior, and J. M. Lord. 1987. Expression of ricin A chain in Escherichia coli. FEBS Lett. 216:73-78. [DOI] [PubMed] [Google Scholar]
- 34.Olsnes, S., and J. V. Kozlov. 2001. Ricin. Toxicon 39:1723-1728. [DOI] [PubMed] [Google Scholar]
- 35.Osborn, R. W., and M. R. Hartley. 1990. Dual effects of the ricin A chain on protein synthesis in rabbit reticulocyte lysate. Inhibition of initiation and translocation. Eur. J. Biochem. 193:401-407. [DOI] [PubMed] [Google Scholar]
- 36.Parikh, B. A., U. Baykal, R. Di, and N. E. Tumer. 2005. Evidence for retro-translocation of pokeweed antiviral protein from endoplasmic reticulum into cytosol and separation of its activity on ribosomes from its activity on capped RNA. Biochemistry 44:2478-2490. [DOI] [PubMed] [Google Scholar]
- 37.Parikh, B. A., C. Coetzer, and N. E. Tumer. 2002. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J. Biol. Chem. 277:41428-41437. [DOI] [PubMed] [Google Scholar]
- 38.Piatak, M., J. A. Lane, W. Laird, M. J. Bjorn, A. Wang, and M. Williams. 1988. Expression of soluble and fully functional ricin A chain in Escherichia coli is temperature-sensitive. J. Biol. Chem. 263:4837-4843. [PubMed] [Google Scholar]
- 39.Rao, P. V., R. Jayaraj, A. S. Bhaskar, O. Kumar, R. Bhattacharya, P. Saxena, P. K. Dash, and R. Vijayaraghavan. 2005. Mechanism of ricin-induced apoptosis in human cervical cancer cells. Biochem. Pharmacol. 69:855-865. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, J. C., L. M. Roberts, K. Romisch, J. Davey, D. H. Wolf, and J. M. Lord. 1999. Ricin A chain utilizes the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 459:80-84. [DOI] [PubMed] [Google Scholar]
- 41.Stirpe, F., and M. G. Battelli. 2006. Ribosome-inactivating proteins: progress and problems. Cell. Mol. Life Sci. 63:1850-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thordal-Christensen, H., Z. Zhang, Y. Wei, and D. B. Collinge. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11:1187-1194. [Google Scholar]
- 43.Tommasi, M., D. Castelletti, M. Pasti, G. Fracasso, I. Lorenzetti, S. Sartoris, C. Pera, G. B. Ferrara, G. Tridente, and M. Colombatti. 2001. Identification of ricin A-chain HLA class II-restricted epitopes by human T-cell clones. Clin. Exp. Immunol. 125:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumer, N. E., D. J. Hwang, and M. Bonness. 1997. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA 94:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumer, N. E., B. A. Parikh, P. Li, and J. D. Dinman. 1998. The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae. J. Virol. 72:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitetta, E. S., J. E. Smallshaw, E. Coleman, H. Jafri, C. Foster, R. Munford, and J. Schindler. 2006. A pilot clinical trial of a recombinant ricin vaccine in normal humans. Proc. Natl. Acad. Sci. USA 103:2268-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, Q., J. M. Jurgensmeier, and J. C. Reed. 1999. Methods of assaying Bcl-2 and Bax family proteins in yeast. Methods 17:292-304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.