Abstract
Current global efforts are focused on exploring alternative pneumococcal vaccine strategies, aimed at addressing the shortcomings of existing formulations, without compromising efficacy. One such strategy involves the use of one or more pneumococcal protein antigens common to all serotypes, to provide cheap, non-serotype-dependent protection. In this study, we evaluated the protective efficacy of immunization of mice with PdB (a pneumolysin toxoid), PspA, PspC (CbpA), PhtB, and PhtE in an invasive-disease model. The antigens were administered in alum adjuvant, either alone or in various combinations. Protection against intraperitoneal challenge with virulent type 2 and 6A strains was assessed in two murine strains. Our findings show that in some situations, different individual proteins gave the best (and worst) protection. However, in many cases, a synergistic/additive effect was seen by using multiple proteins even where the individual proteins showed little value by themselves. For instance, the median survival times for mice immunized with combinations of PdB and PspA, PdB and PspC, or PspA and PspC were significantly longer than those for mice immunized with any of the single antigens. To date, the combination of PdB, PspA, and PspC offers the best protection.
Streptococcus pneumoniae (the pneumococcus) is responsible for a wide spectrum of diseases, ranging from localized infections, such as otitis media and sinusitis, to life-threatening invasive diseases, such as pneumonia, meningitis, and bacteremia (22, 28, 40). In spite of the availability of antimicrobials, the capsular polysaccharide (PS) vaccine, and the 7-valent protein-PS conjugate vaccine, pneumococcal disease continues to cause high morbidity and mortality worldwide, especially in groups at high risk, such as children under 2 years of age, the elderly, and immunocompromised individuals (36). These continuing pneumococcal disease problems have arisen in part due to the increasing rates at which S. pneumoniae is acquiring resistance to multiple antimicrobials (27) but are largely due to the shortcomings associated with the current capsular-based vaccines, including high cost, serotype-specific protection, limited serotype coverage, and the likelihood of a concomitant increase in carriage, and subsequently disease, with nonvaccine serotypes. Consequently, concerted global efforts are currently focused on developing alternative pneumococcal-vaccine strategies that address these shortcomings, without compromising efficacy. One of these approaches involves the development of vaccines based on pneumococcal proteins that contribute to pathogenesis and are common to all serotypes. Such proteins, being T-cell-dependent antigens, should be highly immunogenic and are likely to elicit immunological memory in human infants. Furthermore, the proteins can be engineered for high-level expression at relatively low cost, and formulation is likely to be simpler, thereby making such vaccines more affordable for developing countries where the need is greatest.
The development of an effective protein-based vaccine requires a thorough understanding of the roles and relative contributions to pathogenesis of the various putative virulence proteins. Over the years, a number of candidate pneumococcal protein antigens have been evaluated for their virulence and vaccine potential. These include pneumolysin (Ply) toxoid (PdB) (3, 38); choline binding proteins PspA (15, 49) and PspC (13), also called CbpA or SpsA (21, 41); a 37-kDa metal-binding lipoprotein referred to as PsaA (6, 18); iron uptake ABC transporters PiuA and PiaA (14); heat shock protein ClpP (25); neuraminidases A and B; LytA and hyaluronidase (37); and pneumococcal histidine triad (Pht) proteins PhtB and PhtE (2, 19). Each of these proteins has been shown to elicit a statistically significant level of protection in animal models against systemic challenge with one or more S. pneumoniae serotypes (for a review, see reference 36). However, not all individual proteins have been fully characterized or directly compared, and results are highly dependent on the mouse strain and challenge strain used. Moreover, to date, no single pneumococcal protein has been able to elicit protection comparable to that achieved using protein-PS conjugate vaccines. Therefore, we hypothesized that a combination of virulence proteins should elicit enhanced protection, and this has been proven to be true for a combination of PdB and PspA (PdB+PspA) in murine models of pneumonia and systemic disease (12, 31), for a combination of PiuA and PiaA against systemic challenge (14), and for a combination of PspA and PsaA in a murine model of nasopharyngeal carriage (10). Interestingly, a combination of PdB, PspA, and PsaA did not result in enhanced protection over a combination of PdB and PspA (31). A combination of PdB and PspC was also tested in a murine model of systemic disease (33); however, the protection elicited by PspC alone in that study was so strong that it complicated the assessment of any additive protection that could be achieved with the combination of the two antigens. Furthermore, immunization of mice with a chimeric protein comprising the carboxyl-terminal regions of PhtB and PhtE provided superior protection against pneumonia relative to immunization with the individual counterparts (19).
To minimize the cost of a pneumococcal protein vaccine, it would be necessary to restrict the number of different antigens that might be included in the formulation to the most important virulence determinants. So far, the most promising and well-characterized vaccine candidates are Ply, PspA, and PspC. Ply, a cholesterol-dependent cytotoxin, is implicated in multiple steps of pneumococcal pathogenesis, including activation of complement, inhibition of ciliary beating in the human respiratory epithelium, and disruption of tight junctions between epithelial cells (8, 35, 39, 43). PspA is believed to play a pivotal role in preventing complement-mediated opsonization (1, 47, 49) and is also capable of binding to, and preventing killing by, lactoferrin (20, 42). PspC, on the other hand, has putative roles in adherence to the nasopharyngeal and lung epithelia and the brain microvascular endothelium (17, 40, 41). There is also evidence that PspC may mediate invasion of host cells at these locations (16, 34, 51). Interestingly, the genes encoding Ply, PspA, and PspC were upregulated and differentially expressed in vivo in mouse intranasal and intraperitoneal (i.p.) challenge models of infection (26, 32, 44). However, the precise role of PhtB and PhtE (and other Pht proteins) in pneumococcal pathogenesis is yet to be determined. Indirect evidence for the contribution of Pht proteins could be derived from signature-tagged-mutagenesis studies (23), which suggested a role for PhtA, PhtB, and PhtD in the progression to lung disease. Passive immunization studies (19) also suggested that antibody-mediated opsonophagocytosis might be the major mechanism of protection imparted by PhtB and PhtE.
It is imperative that pneumococcal protein vaccine candidates for clinical trials be tested and compared in various combinations, in different animal strains, and using various challenge strains to obtain the best protein vaccine formulation. In the present study, we carried out direct comparative analysis of the protective efficacies of PhtB and PhtE versus those of the well-characterized candidate protein antigens, namely, PdB, PspA, and PspC, against systemic disease. In addition, we examined whether a combination of two or three of these proteins resulted in superior protection over any of the antigens alone or previously tested combinations. This was carried out using two mouse strains (BALB/c and CD1 [Swiss]) and two virulent pneumococcal challenge strains belonging to serotypes 2 and 6A.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were D39, a virulent type 2 strain (4), and WCH16, a virulent type 6A clinical (blood) isolate from the Women's and Children's Hospital, North Adelaide, South Australia, Australia. Opaque-phase variants of the two strains, selected on Todd-Hewitt broth-1% yeast extract-catalase plates (48), were used in these experiments. For the challenge, the bacteria were grown at 37°C overnight on blood agar in 95% air and 5% CO2 and then inoculated into serum broth (7). They were then grown statically for 3 h at 37°C to give approximately 108 CFU/ml. Serotype-specific capsule production was confirmed by the Quellung reaction, as described previously (7). Bacteria for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and enzyme-linked immunosorbent assay (ELISA) analysis were grown in Todd-Hewitt broth-1% yeast extract (7) to an A600 of 0.25. The cells were pelleted by centrifugation at 20,000 × g for 5 min and lysed in a French pressure cell (SLM Aminco Inc.) at 12,000 lb/in2. Lysates for SDS-PAGE and Western blotting were then resuspended in sample loading buffer before electrophoresis.
Cloning and expression of His6-tagged PhtB and PhtE fusion proteins.
The cloning, expression, and purification of pneumolysin toxoid (PdB), PspA fragments, and PspC fragments from serotype 2 variants of pneumococci have been described elsewhere (11, 30, 33, 38). The cloning and expression of phtB and phtE from D39 were carried out as follows. Oligonucleotides AD37B (5′-GCTTATGAACTAGGCATGCATCAAGCTC-3′) and AD38B (5′-GATCAAGCTTGCTGCTACCTTTACTACTCTC-3′), incorporating an SphI and a HindIII restriction site (underlined), respectively, were used as primers for high-fidelity PCR amplification of an approximately 2.5-kb fragment from the 5′ end of phtB from D39 chromosomal DNA. Oligonucleotides AD35B (5′-CTATGTGCGCATGCACTAAACCAGCAT-3′) and AD36B (5′-ACTTTTTCTACTGCAGTTCCTTACGCT-3′), incorporating an SphI and a PstI restriction site (underlined), respectively, were used as primers for high-fidelity PCR amplification of an approximately 3.1-kb fragment from the 5′ end of phtE from D39 chromosomal DNA. The restriction sites were incorporated to allow in-frame cloning of the PCR products into the corresponding restriction sites in the polylinkers of pQE32 and pQE31, respectively (QIAGEN Inc.). The resultant recombinant plasmids were predicted to express N-terminal His6-tagged PhtB (814 amino acids) and PhtE (1,018 amino acids) fusion proteins lacking their respective signal peptides. Correct in-frame fusion of the fragments into the QIAexpress vectors was confirmed by automated dye terminator sequencing. The recombinant plasmids were then used to transform Escherichia coli BL21(DE3). High-level expression of the recombinant fusion proteins was achieved by the addition of isopropyl-β-d-thiogalactoside (IPTG) at a final concentration of 2 mM to a Terrific Broth (46) culture of each of the expression constructs in the presence of 200 μg of ampicillin/ml for 3 h at 37°C with vigorous shaking. The cells were then harvested by centrifugation at 6,000 × g for 10 min and resuspended in loading buffer (10 mM sodium phosphate [pH 7.0]). Afterwards, the cells were lysed in a French pressure cell at 12,000 lb/in2, and the resultant lysate was centrifuged at 100,000 × g for 1 h.
Purification of His6-tagged PhtB and PhtE fusion proteins.
The His6-tagged PhtB and PhtE fusion proteins were initially purified by anion exchange chromatography on a DEAE-Sepharose column (Amersham Biosciences) previously equilibrated with a 10 mM sodium phosphate (pH 7.0) loading buffer. Proteins were eluted with a 10 to 250 mM sodium phosphate (pH 7.0) gradient and fractions analyzed by denaturing SDS-PAGE. Fractions containing His6-PhtB/PhtE were identified, pooled, and adjusted to pH 8.0. The pooled material was then loaded onto a 2-ml nickel-nitrilotriacetic acid column (QIAGEN Inc.) and purified as described previously (33).
Identification of the PspA family.
Strain Rx1, an unencapsulated derivative of D39, has been shown previously to belong to PspA family type 1 (24). The PspA family type for WCH16 was determined by PCR using primers LSM12/SKH63 (for PspA family 1) and primers LSM12/SKH52 (for PspA family 2). Genomic DNA from strains Rx1 and EF3296, respectively, were used as controls as described previously (9).
Mice.
Male 5- to 6-week-old BALB/c and CD1 mice were used in all experiments. The ethics committee of the University of Adelaide approved all animal experiments.
Immunization of mice.
In one experiment, two replicate immunization schedules were performed; each immunization schedule consisted of eight groups of 5- to 6-week-old male BALB/c mice (12 per group). The mice were immunized i.p. with either PdB alone, PspA alone, PspC alone, PdB+PspA, PdB+PspC, PspA+PspC, PdB+PspA+PspC, or a placebo. In another experiment, two replicate immunization schedules were performed, with each immunization schedule consisting of 12 groups of 5- to 6-week-old male CD1 mice (10 to 12 per group). The mice were immunized i.p. with either PdB alone, PspA alone, PspC alone, PhtB alone, PhtE alone, PdB+PspA, PdB+PspC, PdB+PhtB, PspA+PhtB, PhtB+PhtE, PdB+PspA+PhtB, or a placebo. In all experiments, each mouse received three doses of 10 μg of each antigen alone (or in combination) in 100 μg of alum adjuvant (Imject Alum no. 77161; Pierce, Rockford, IL) at 14-day intervals. The mice given the placebo received an identical course of saline plus alum. Sera were collected from individual mice by retro-orbital bleeding 1 week after the third immunization.
ELISA and Western blotting.
Aliquots of sera from individual mice were pooled on a group-by-group basis and assayed for protein-specific antibodies by ELISA, using 96-well polystyrene microtiter trays (Nunc) coated with purified antigens as described previously (31). Sera were analyzed for total immunoglobulin G (IgG), IgG subclass (IgG1, IgG2a, and IgG2b), and IgA antibodies generated against the vaccine antigens. Bound antibodies were detected by using alkaline phosphatase-conjugated anti-mouse IgA or anti-mouse IgG (heavy plus light chains) and subclass antibodies (Invitrogen), with disodium p-nitrophenol phosphate as the substrate. The sera were also analyzed for reactivity against purified proteins or whole-cell lysates of D39 and WCH16 by Western immunoblotting and ELISA.
Challenge.
Mice were challenged i.p. 2 weeks after the third immunization with either the highly virulent capsular type 2 strain D39 or a serotype 6A (WCH16) strain. In the first set of challenge experiments, groups of immunized BALB/c mice were challenged with either 2.5 × 105 CFU of D39 or 7.5 × 106 CFU of WCH16, representing approximately 103 50% lethal doses (LD50s) or 102 LD50s, respectively, for BALB/c mice. In the second set of challenge experiments, groups of immunized CD1 mice were challenged with either 5 × 102 CFU of D39 or 3 × 106 CFU of WCH16, corresponding to approximately 50 LD50s or 103 LD50s, respectively, for CD1 mice. The mice were closely monitored for 21 days, and the survival time of each mouse was recorded. Differences between the median survival times for groups were analyzed by the Mann-Whitney U test (two tailed).
RESULTS
Purification of His6-tagged PhtB and PhtE fusion proteins.
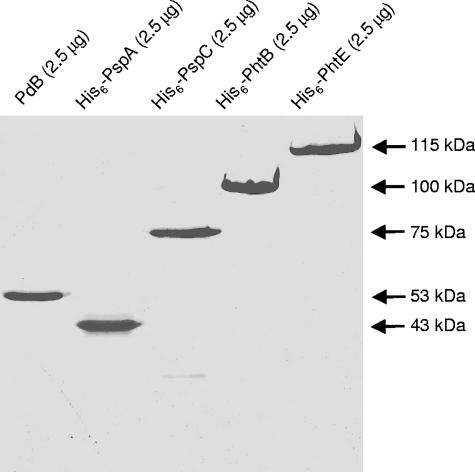
A two-step procedure was employed to purify recombinant His6-tagged PhtB and PhtE fusion proteins (see Materials and Methods). The purified His6-tagged PhtB and PhtE proteins were analyzed by SDS-PAGE and Coomassie brilliant blue staining (Fig. 1). The proteins migrated with apparent molecular sizes of approximately 100 kDa and 115 kDa, respectively, consistent with their predicted sizes based on the DNA sequence, excluding the residues removed by cleavage of the N-terminal signal peptidase motif. The three other vaccine antigens used in this study, namely, PdB and the N-terminal fragments of His6-PspA and His6-PspC, were analyzed in parallel and also migrated at their expected sizes of approximately 53 kDa, 43 kDa, and 75 kDa, respectively. All vaccine antigens were judged to be >95% pure.
FIG. 1.
SDS-PAGE analysis (12% gel) of recombinant protein antigens used in this study. The samples were prepared as described in Materials and Methods. The apparent molecular mass of each protein is indicated on the right. The proteins were cloned and expressed from capsular type 2 variants of pneumococci.
Analysis of sera.
ELISA analysis of pooled sera from groups of mice immunized with the purified antigens either singly or in combination shows that strong, antigen-specific antibody responses were generated (Table 1) . Furthermore, antigen-specific antibody titers were not diminished when the antigens were administered in combination, indicating that there was no detectable antagonistic effect of combining these antigens. Antibodies elicited by PspA and PspC elicited a degree of cross-reaction, consistent with known sequence similarity in the proline-rich region and in the alpha-helical regions of some molecules (13), as did antibodies elicited by PhtB and PhtE, which exhibit 75% N-terminal amino acid sequence identity (19). As expected, the IgG1 response was predominant, followed by the IgA, IgG2b, and IgG2a antibody responses, in that order (data not shown).
TABLE 1.
Antibody titers (total IgG) obtained from mice immunized with PdB, PspA, PspC, PhtB, and PhtE
| Immunization group | Antibody titera response to indicated protein antigen
|
||||
|---|---|---|---|---|---|
| PdB | PspA | PspC | PhtB | PhtE | |
| Alum | −(<100)b | ||||
| PdB | 5,000 | ||||
| PspA | 2,750 ± 250 | 1,800 ± 200c | |||
| PspC | 3,000 ± 500c | 5,000 ± 1,000 | |||
| PhtB | 10,000 ± 2,000 | 3,000 ± 500c | |||
| PhtE | 2,000 ± 300c | 5,500 ± 1,500 | |||
| PdB+PspA | 5,250 ± 750 | 2,750 ± 250 | ND | ND | ND |
| PdB+PspC | 4,500 ± 500 | ND | 4,500 ± 500 | ND | ND |
| PdB+PhtB | 5,750 ± 1,250 | ND | ND | 8,500 ± 1,500 | ND |
| PspA+PspC | ND | 3,500 ± 500 | 4,200 ± 500 | ND | ND |
| PspA+PhtB | ND | 2,750 ± 250 | ND | 10,000 ± 2,000 | ND |
| PhtB+PhtE | ND | ND | ND | 8,000 ± 1,500 | 7,000 ± 3,000 |
| PdB+PspA+PspC | 5,000 ± 500 | 3,000 ± 500 | 4,000 ± 500 | ND | ND |
| PdB+PspA+PhtB | 4,000 | 2,750 ± 250 | ND | 8,500 ± 1,500 | ND |
The antibody titer is the reciprocal of the dilution of serum giving 50% of the highest absorbance value above the background level at 405 nm, as determined by ELISA. ND, not determined.
−(<100) denotes no reactivity.
Denotes cross-reactive antibodies (see text).
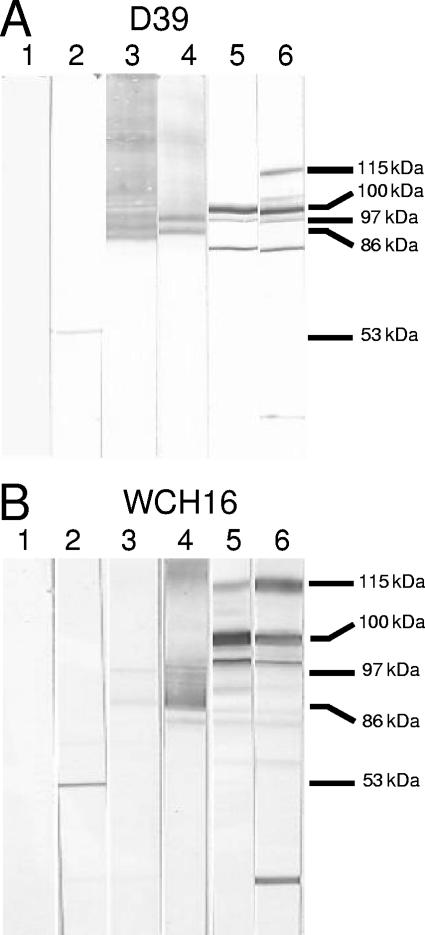
Western immunoblot analysis of whole-cell lysates of D39 (Fig. 2A) and WCH16 (Fig. 2B) also demonstrated protein-specific antibody responses to each of the serotype 2-derived antigens (11, 30, 33, 38). Anti-PdB sera specifically labeled pneumolysin, but a labeled smear was observed with anti-PspA sera, reflecting the peculiar electrophoretic mobility of PspA. However, anti-PspA reacted very weakly against WCH16 lysate, possibly a result of the heterogeneity between the N termini of PspA from serotype 2 and 6A strains, although both serotypes belong to PspA family 1 as determined by PCR. Similarly, a labeled smear was observed when the lysates were reacted with anti-PspC sera. However, the sera reacted strongly with another protein, likely to be PspA, as the reactive band was absent when a lysate of a PspA-negative D39 mutant was used as the antigen (not shown), as demonstrated previously (33). This is also consistent with the known sequence similarity in the proline-rich regions of PspA and PspC (13). PhtB antiserum reacted with the mature protein (approximately 100 kDa) but also reacted strongly with three other bands of approximately 115 kDa, 105 kDa, and 96 kDa, which could be PhtE, PhtD, and PhtA, respectively. Furthermore, anti-PhtE reacted with the full-length protein (approximately 115 kDa) and other bands of approximately 105 kDa (PhtD), 100 kDa (PhtB), 96 kDa (PhtA), and 52 kDa (possibly active mature PhtE) in both lysates. A smaller band of approximately 30 kDa was also detected in the anti-PhtE blots, possibly a truncated form of PhtA or PhtD. The specificity of the reactivity of the antisera to PhtB or PhtE was confirmed, as the reactive band was absent when a lysate of a PhtB- or PhtE-negative D39 mutant was used as the antigen (unpublished observations). These results are comparable to those obtained previously (2, 19). Moreover, the reactivity of sera raised against a combination of each of the proteins was a sum of the reactivities of the individual sera (not shown). ELISA analysis of whole-cell lysates of D39 and WCH16 also indicated specific reactivities, but lower titers, to each of the proteins (data not shown).
FIG. 2.
Western blots of whole-cell lysates of S. pneumoniae serotype 2 strain D39 (A) and serotype 6A strain WCH16 (B) showing the reactivities of the antibodies raised against the various protein antigens. The lysates were reacted with specific antisera generated from mice immunized with the various antigens cloned and expressed from capsular type 2 variants of pneumococci. Results for nitrocellulose membranes reacted with sera from mice immunized with alum (lane 1), anti-PdB (lane 2), anti-PspA (lane 3), anti-PspC (lane 4), anti-PhtB (lane 5), and anti-PhtE (lane 6) are shown. The corresponding molecular mass of each of the proteins is indicated: pneumolysin (53 kDa), PspA (86 kDa), PspC (97 kDa), PhtB (100 kDa), and PhtE (115 kDa). See text for full description.
Protection studies.
In the first series of experiments, groups of BALB/c mice immunized with PdB, PspA, or PspC, and combinations thereof, were challenged with either D39 or WCH16. Each of the proteins has been shown previously to elicit protection by using moderate challenge doses of S. pneumoniae (for a review, see reference 36). In the D39 challenge experiment (Fig. 3), a very high challenge dose was employed since we were principally interested in detecting additive protection. Consequently, none of the single antigens elicited significant protection over that of the placebo group. However, mice immunized with PdB+PspC survived significantly longer than mice immunized with either PdB alone (P = 0.05) or PspC alone (P = 0.02). Furthermore, mice immunized with PspA+PspC survived significantly longer than mice immunized with either PspA alone (P = 0.05) or PspC alone (P = 0.02). Most interestingly, mice immunized with PdB+PspA+PspC survived significantly longer than those that received PdB+PspA (P = 0.05).
FIG. 3.
Survival times for mice after intraperitoneal challenge. Groups of 12 BALB/c mice were immunized with the indicated antigens and challenged 2 weeks after the third immunization with approximately 2.5 × 105 CFU of D39. The broken lines denote the median survival time for each group.
In the WCH16 challenge experiment (Fig. 4), mice immunized with PspA alone survived significantly longer than those that received the alum placebo (P = 0.002). Mice immunized with PdB+PspA, PdB+PspC, or PdB+PspA+PspC survived significantly longer than those that received PdB alone (P = 0.002 in all cases). Moreover, mice that received PdB+PspA or PdB+PspA+PspC survived significantly longer than those that received PspA alone (P = 0.05 and P = 0.02, respectively). Furthermore, mice that were immunized with PspA+PspC or PdB+PspA+PspC survived significantly longer than those that received PspC alone (P = 0.002 in both cases). Finally, mice that received PdB+PspA+PspC survived significantly longer than those that received PdB+PspC (P = 0.002).
FIG. 4.
Survival times for mice after intraperitoneal challenge. Groups of 12 BALB/c mice were immunized with the indicated antigens and challenged 2 weeks after the third immunization with approximately 7.5 × 106 CFU of WCH16. The broken lines denote the median survival time for each group.
In the second series of experiments, groups of CD1 mice immunized with PdB, PspA, PspC, PhtB, or PhtE, and combinations thereof, were challenged with either D39 or WCH16. In the D39 challenge experiment (Fig. 5), mice immunized with PdB alone survived significantly longer than those that received the alum placebo (P = 0.05), but no other single antigen afforded significant protection. Moreover, with the exception of PhtE versus PhtB+PhtE (P = 0.05) and PspC versus PdB+PspC (P = 0.002), the various combinations of the antigens did not result in statistically significant additive protection over that of any of the antigens alone. This was most likely due to the high virulence of D39 combined with the high susceptibility of CD1 mice to pneumococcal challenge.
FIG. 5.
Survival times for mice after intraperitoneal challenge. Groups of 10 to 12 CD1 mice were immunized with the indicated antigens and challenged 2 weeks after the third immunization with approximately 5 × 102 CFU of D39. The broken lines denote the median survival time for each group.
In the corresponding WCH16 challenge experiment (Fig. 6), mice immunized with PdB alone, PspA alone, PhtB alone, or PhtE alone survived significantly longer than those that received the alum placebo (P = 0.02, P = 0.002, P = 0.002, or P = 0.002, respectively). In addition, mice that were immunized with PdB+PspA or PdB+PspC survived significantly longer than those that received PdB alone (P = 0.02 and P = 0.02, respectively). Mice that were immunized with PdB+PspA survived significantly longer than those that received PspA alone (P = 0.002); those that received PdB+PspC survived significantly longer than those that received PspC alone (P = 0.002), while mice that were immunized with PdB+PhtB, PspA+PhtB, or PhtB+PhtE survived significantly longer than those that received PhtB alone (P = 0.002, P = 0.02, and P = 0.02, respectively). Lastly, mice that were immunized with PdB+PspA+PhtB survived significantly longer than those that received PspA+PhtB (P = 0.02).
FIG. 6.
Survival times for mice after intraperitoneal challenge. Groups of 10 to 12 CD1 mice were immunized with the indicated antigens and challenged 2 weeks after the third immunization with approximately 3 × 106 CFU of WCH16. The broken lines denote the median survival time for each group.
DISCUSSION
Extensive efforts are currently geared toward the development of effective alternative vaccination strategies against pneumococcal disease to address the shortcomings associated with the current capsule-based vaccines. One of these approaches is focused on evaluating the potential of pneumococcal virulence proteins as vaccine candidates in murine models of systemic disease and nasopharyngeal carriage (10, 12, 14, 31, 36). In this work, we carried out direct comparative analysis of the protective efficacies of the new PhtB and PhtE vaccine candidates versus those of the well-characterized candidate protein antigens, PdB, PspA, and PspC, against our established virulent challenge strains D39 and WCH16 in an i.p. model of invasive disease. The i.p. challenge route was employed in our effort to evaluate the protective abilities of these proteins against pneumococcal bacteremia and does not address or predict effects on colonization or nonbacteremic pneumonia, which could be assessed by the intranasal route.
The results of the i.p. challenge experiments indicate that the relative protection afforded by single proteins is challenge strain and mouse strain dependent. For instance, after challenge with serotype 2 and serotype 6A strains in CD1 mice, the median survival times for mice immunized with the pneumolysin toxoid (PdB) were longer than those for mice immunized with the other proteins, corroborating previous findings (3, 33; unpublished observations). Interestingly, PspA gave the best protection against heterologous serotype 6A challenge in BALB/c mice, although anti-PspA serum reacted very weakly against WCH16 lysate due to the heterogeneity between the N termini of PspA proteins from serotype 2 and 6A strains. This finding supports previous studies, which showed that PspA elicits cross-protective immunity against diverse capsular and PspA types (11, 29, 45). Furthermore, we had previously observed that D39, being a highly virulent strain, is difficult to protect against using PspA alone in systemic challenge models (31; unpublished data), as is the case in this study. Surprisingly, in this study, neither PhtB nor PhtE was able to elicit the level of protection reported by Hamel et al. (19).
We also demonstrated that nearly all combinations of the protein antigens provided higher degrees of protection than any of the antigens alone, corroborating previous findings (12, 31). A combination of PdB and PspA consistently elicited enhanced protection against challenge, followed by a combination of PdB and PspC and a combination of PspA and PspC. This is not unexpected, as these proteins have been shown previously to contribute to different stages of the pathogenic process and, as such, have complementary or synergistic roles (5, 11-13, 31, 33, 37, 42, 47, 50). For both PspA and PspC, the conserved central proline-rich region contains cross-protective epitopes, as demonstrated previously (13). It is highly likely that the protection afforded by immunization of mice with the various protein antigens and combinations thereof was, at least in part, antibody mediated, as demonstrated previously using antisera raised against PdB and/or PspA (29, 31). However, combinations involving Pht proteins were generally inferior, especially in the presence of PspA. This was surprising, as there was no obvious diminution in antigen-specific antibody titers when the antigens were administered in combination with each other. The protection imparted by immunization with PspA has been shown to be a consequence of the blocking of the ability of PspA to inhibit complement fixation and killing by lactoferrin (42, 47). Although the biological function of PhtB (or PhtE) is unknown, passive-immunization studies by Hamel et al. (19) established that surface-labeling antibodies are biologically linked to survival and suggested that antibody-mediated opsonophagocytosis may be the major mechanism of protection imparted by PhtB and PhtE. If antibodies against PhtB are indeed primarily opsonic, then a combination with known complement-inhibiting immunogens, such as PspA, should have resulted in additive or synergistic protection, which is clearly not the case in this work. We speculate that there could be interference with PspA-mediated protection due to PhtB antibodies either by steric hindrance of PspA-antibody binding or by some unknown mechanism(s), and more work is warranted to clarify this phenomenon.
The enhanced protection obtained with the triple combination of PdB, PspA, and PspC, albeit marginal over that obtained with double combinations, is very encouraging and is the best result obtained with triple protein combinations so far. However, this is limited to BALB/c mice and two challenge strains to date and as such needs to be demonstrated with other mouse and challenge strains.
Taken together, our findings clearly show that in some situations, different individual proteins gave the best (and worst) protection. This provides an additional rationale for combining the proteins, as particular proteins might not work against some strains in some people. However, by using more than one protein, this risk is minimized. Moreover, there is clearly a synergistic/additive effect achieved in many cases by using multiple proteins, even in cases where the individual proteins showed little value by themselves. Overall, our results imply that rational decision regarding the formulation of multicomponent pneumococcal protein vaccines will require rigorous comparisons of individual antigens and all possible combinations thereof, using multiple mouse and challenge strains.
Acknowledgments
We thank Susan Hollingshead for providing primer sequences and control DNA for PspA family typing.
This work was supported by National Health and Medical Research Council of Australia Program grant 284214.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Abeyta, M., G. G. Hardy, and J. Yother. 2003. Genetic alteration of capsule type but not PspA type affects accessibility of surface-bound complement and surface antigens of Streptococcus pneumoniae. Infect. Immun. 71:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamou, J. E., J. H. Heinrichs, A. L. Erwin, W. Walsh, T. Gayle, M. Dormitzer, R. Dagan, Y. A. Brewah, P. Barren, R. Lathigra, S. Langermann, S. Koenig, and S. Johnson. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, J. E., R. A. Lock, C. C. A. M. Peeters, J. T. Poolman, P. W. Andrew, T. J. Mitchell, D. Hansman, and J. C. Paton. 1994. Immunization of mice with pneumolysin toxoid confers a significant degree of protection against at least nine serotypes of Streptococcus pneumoniae. Infect. Immun. 62:5683-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balachandran, P., A. Brooks-Walter, A. Virolainen-Julkunen, S. K. Hollingshead, and D. E. Briles. 2002. The role of pneumococcal surface protein C (PspC) in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kDa putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64:5255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, A. M., and J. C. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulnois, G. J., J. C. Paton, T. J. Mitchell, and P. W. Andrew. 1991. Structure and function of pneumolysin, the multifunctional, thiol-activated toxin of Streptococcus pneumoniae. Mol. Microbiol. 5:2611-2616. [DOI] [PubMed] [Google Scholar]
- 9.Brandileone, M. C., A. L. Andrade, E. M. Teles, R. C. Zanella, T. I. Yara, J. L. Di Fabio, and S. K. Hollingshead. 2004. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil: towards novel pneumococcal protein vaccines. Vaccine 22:3890-3896. [DOI] [PubMed] [Google Scholar]
- 10.Briles, D. E., E. Ades, J. C. Paton, J. S. Sampson, G. M. Carlone, R. C. Huebner, A. Virolainen, E. Swiatlo, and S. K. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 12.Briles, D. E., S. K. Hollingshead, J. C. Paton, E. W. Ades, L. Novak, F. W. Van Ginkel, and W. H. Benjamin, Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339-348. [DOI] [PubMed] [Google Scholar]
- 13.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crain, M. J., W. D. Waltman II, J. S. Turner, J. Yother, D. F. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cundell, D. R., N. P. Gerald, C. Gerald, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 17.Cundell, D. R., and E. I. Tuomanen. 1994. Receptor specificity of adherence of Streptococcus pneumoniae to human type-II pneumocytes and vascular endothelial cells in vitro. Microb. Pathog. 17:361-374. [DOI] [PubMed] [Google Scholar]
- 18.Dintilhac, A., G. Alloing, C. Granadel, and J.-P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 19.Hamel, J., N. Charland, I. Pineau, C. Ouellet, S. Rioux, D. Martin, and B. R. Brodeur. 2004. Prevention of pneumococcal disease in mice immunized with conserved surface-accessible proteins. Infect. Immun. 72:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerschmidt, S., G. Bethe, P. H. Remane, and G. S. Chhatwal. 1999. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 67:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt, S., S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 25:1113-1124. [DOI] [PubMed] [Google Scholar]
- 22.Hausdorff, W. P., D. R. Feikin, and K. P. Klugman. 2005. Epidemiological differences among pneumococcal serotypes. Lancet Infect. Dis. 5:83-93. [DOI] [PubMed] [Google Scholar]
- 23.Hava, D., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 24.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon, H. Y., A. D. Ogunniyi, M. H. Choi, S. N. Pyo, D. K. Rhee, and J. C. Paton. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 72:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeMessurier, K. S., A. D. Ogunniyi, and J. C. Paton. 2006. Differential expression of key pneumococcal virulence genes in vivo. Microbiology 152:305-311. [DOI] [PubMed] [Google Scholar]
- 27.Lynch, J. P., III, and G. G. Zhanel. 2005. Escalation of antimicrobial resistance among Streptococcus pneumoniae: implications for therapy. Semin. Respir. Crit. Care Med. 26:575-616. [DOI] [PubMed] [Google Scholar]
- 28.McCullers, J. A., and E. I. Tuomanen. 2001. Molecular pathogenesis of pneumococcal pneumonia. Front. Biosci. 6:D877-D889. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular serotype. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabors, G. S., P. A. Braun, D. J. Herrmann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 31.Ogunniyi, A. D., R. L. Folland, D. E. Briles, S. K. Hollingshead, and J. C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 33.Ogunniyi, A. D., M. C. Woodrow, J. T. Poolman, and J. C. Paton. 2001. Protection against Streptococcus pneumoniae elicited by immunization with pneumolysin and CbpA. Infect. Immun. 69:5997-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661-1669. [DOI] [PubMed] [Google Scholar]
- 35.Paton, J. C. 1996. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol. 4:103-106. [DOI] [PubMed] [Google Scholar]
- 36.Paton, J. C. 2004. New pneumococcal vaccines: basic science developments, p. 382-402. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 37.Paton, J. C., A. M. Berry, and R. A. Lock. 1997. Molecular analysis of putative pneumococcal virulence proteins. Microb. Drug. Resist. 3:1-10. [DOI] [PubMed] [Google Scholar]
- 38.Paton, J. C., R. A. Lock, C.-J. Lee, J. P. Li, A. M. Berry, T. J. Mitchell, P. W. Andrew, D. Hansman, and G. J. Boulnois. 1991. Purification and immunogenicity of genetically obtained pneumolysin toxoids and their conjugation to Streptococcus pneumoniae type 19F polysaccharide. Infect. Immun. 59:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paton, J. C., B. Rowan-Kelly, and A. Ferrante. 1984. Activation of human complement by the pneumococcal toxin pneumolysin Infect. Immun. 43:1085-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 42.Shaper, M., S. K. Hollingshead, W. H. Benjamin, Jr., and D. E. Briles. 2004. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect. Immun. 72:5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinfort, C., R. Wilson, T. Mitchell, C. Feldman, A. Rutman, H. Todd, D. Sykes, J. Walker, K. Saunders, P. W. Andrew, G. J. Boulnois, and P. J. Cole. 1989. Effects of Streptococcus pneumoniae on human respiratory epithelium in vitro. Infect. Immun. 57:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiatlo, E., J. King, G. S. Nabors, B. Mathews, and D. E. Briles. 2003. Pneumococcal surface protein A is expressed in vivo, and antibodies to PspA are effective for therapy in a murine model of pneumococcal sepsis. Infect. Immun. 71:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tart, R. C., L. S. McDaniel, B. A. Ralph, and D. E. Briles. 1996. Truncated Streptococcus pneumoniae PspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J. Infect. Dis. 173:380-386. [DOI] [PubMed] [Google Scholar]
- 46.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 9:12. [Google Scholar]
- 47.Tu, A.-H. T., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuste, J., M. Botto, J. C. Paton, D. W. Holden, and J. S. Brown. 2005. Additive inhibition of complement deposition by pneumolysin and PspA facilitates Streptococcus pneumoniae septicemia. J. Immunol. 175:1813-1819. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, J. R., K. E. Mostov, M. E. Lamm, M. Nanno, S. Shimida, M. Ohwaki, and E. Tuomanen. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827-837. [DOI] [PubMed] [Google Scholar]