Abstract
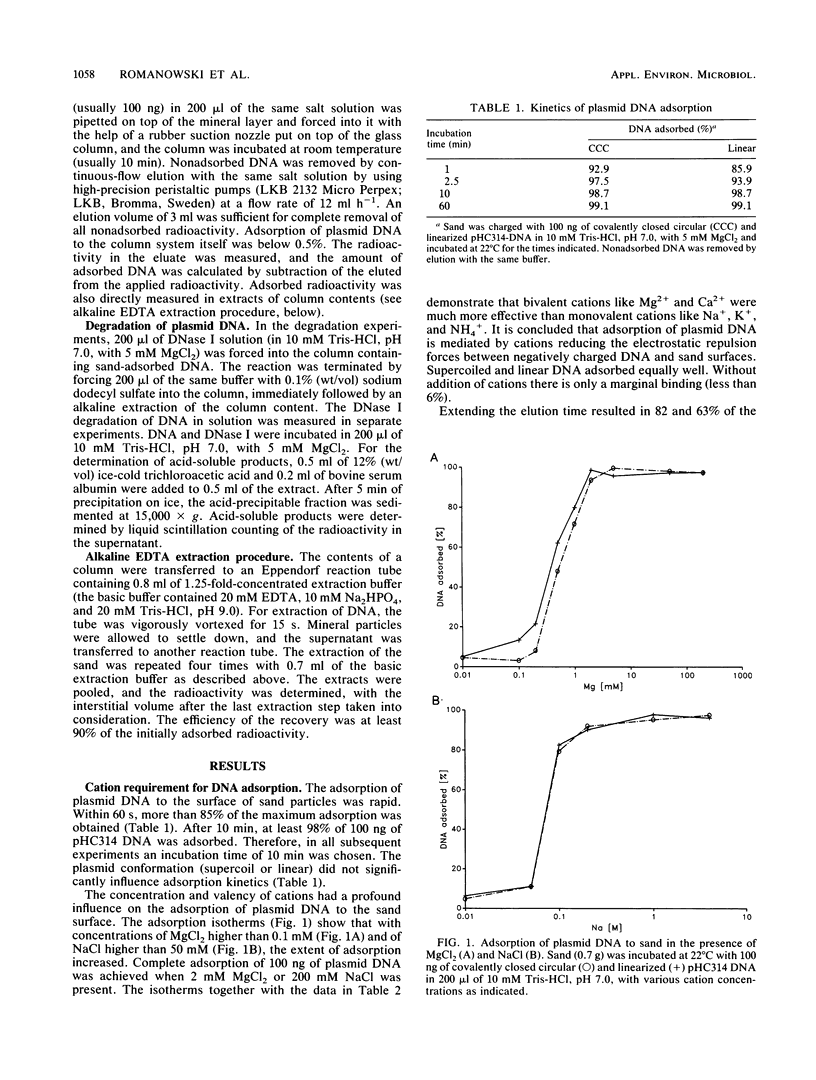
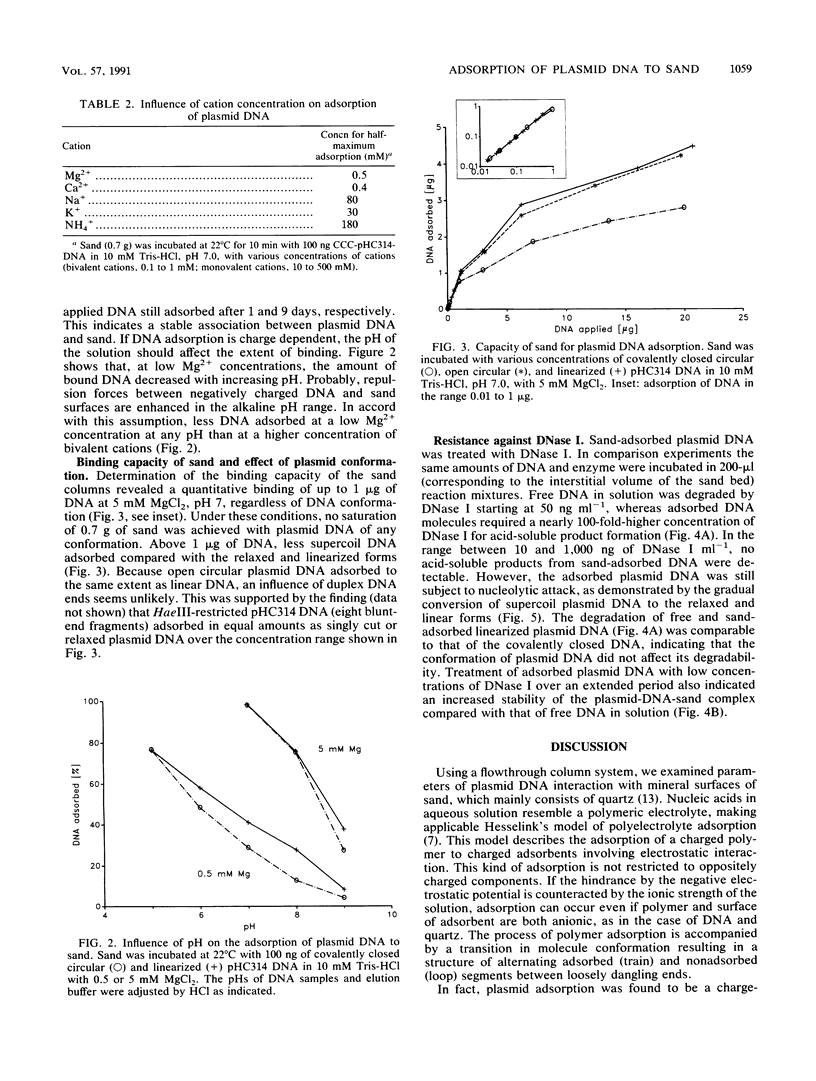
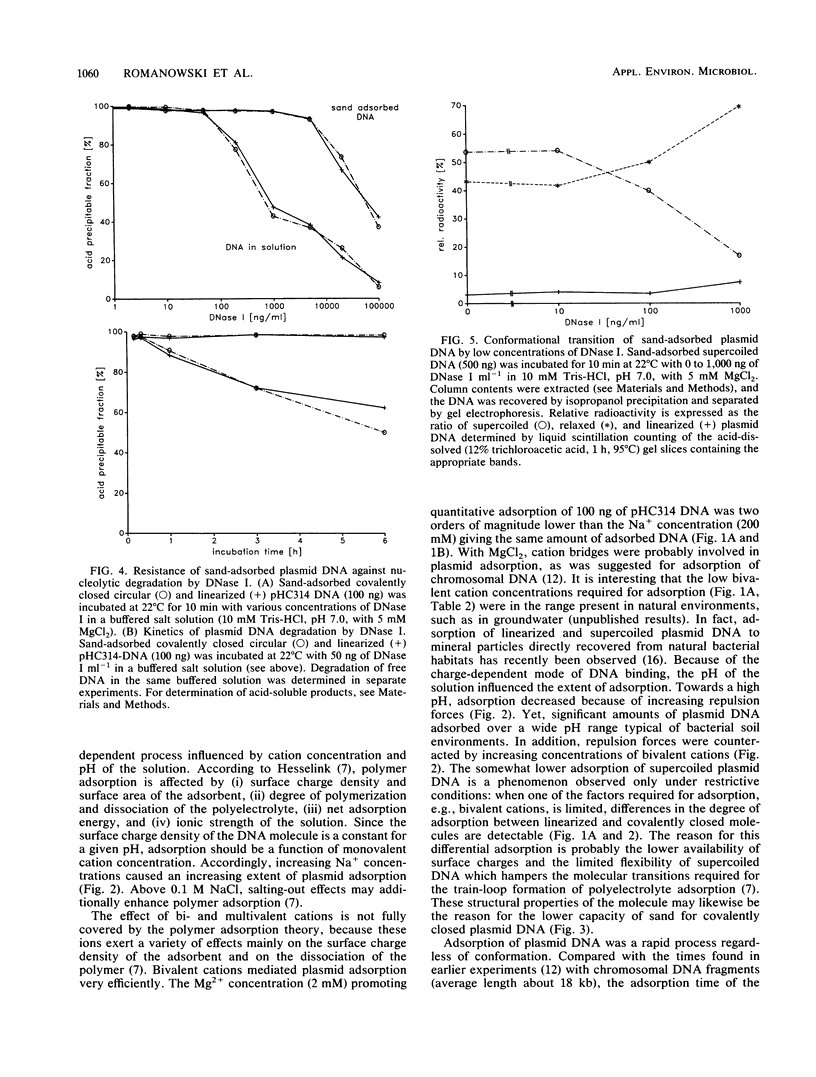
The adsorption of [3H]thymidine-labeled plasmid DNA (pHC314; 2.4 kb) of different conformations to chemically pure sand was studied in a flowthrough microenvironment. The extent of adsorption was affected by the concentration and valency of cations, indicating a charge-dependent process. Bivalent cations (Mg2+, Ca2+) were 100-fold more effective than monovalent cations (Na+, K+, NH4+). Quantitative adsorption of up to 1 microgram of negatively supercoiled or linearized plasmid DNA to 0.7 g of sand was observed in the presence of 5 mM MgCl2 at pH 7. Under these conditions, more than 85% of DNA adsorbed within 60 s. Maximum adsorption was 4 micrograms of DNA to 0.7 g of sand. Supercoil molecules adsorbed slightly less than linearized or open circular plasmids. An increase of the pH from 5 to 9 decreased adsorption at 0.5 mM MgCl2 about eightfold. It is concluded that adsorption of plasmid DNA to sand depends on the neutralization of negative charges on the DNA molecules and the mineral surfaces by cations. The results are discussed on the grounds of the polyelectrolyte adsorption model. Sand-adsorbed DNA was 100 times more resistant against DNase I than was DNA free in solution. The data support the idea that plasmid DNA can enter the extracellular bacterial gene pool which is located at mineral surfaces in natural bacterial habitats.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aardema B. W., Lorenz M. G., Krumbein W. E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl Environ Microbiol. 1983 Aug;46(2):417–420. doi: 10.1128/aem.46.2.417-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros I., Pósfai G., Venetianer P. High-copy-number derivatives of the plasmid cloning vector pBR322. Gene. 1984 Oct;30(1-3):257–260. doi: 10.1016/0378-1119(84)90130-6. [DOI] [PubMed] [Google Scholar]
- Doran J. L., Bingle W. H., Roy K. L., Hiratsuka K., Page W. J. Plasmid transformation of Azotobacter vinelandii OP. J Gen Microbiol. 1987 Aug;133(8):2059–2072. doi: 10.1099/00221287-133-8-2059. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Lindow S. E., Panopoulos N. J., McFarland B. L. Genetic engineering of bacteria from managed and natural habitats. Science. 1989 Jun 16;244(4910):1300–1307. doi: 10.1126/science.2660261. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Aardema B. W., Wackernagel W. Highly efficient genetic transformation of Bacillus subtilis attached to sand grains. J Gen Microbiol. 1988 Jan;134(1):107–112. doi: 10.1099/00221287-134-1-107. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987 Dec;53(12):2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol. 1990;154(4):380–385. doi: 10.1007/BF00276535. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Schold M., Johnson M. J., Dembek P., Itakura K. Oligonucleotide directed mutagenesis of the human beta-globin gene: a general method for producing specific point mutations in cloned DNA. Nucleic Acids Res. 1981 Aug 11;9(15):3647–3656. doi: 10.1093/nar/9.15.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]



