Abstract
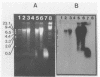
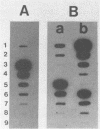
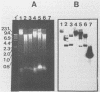
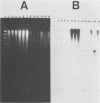
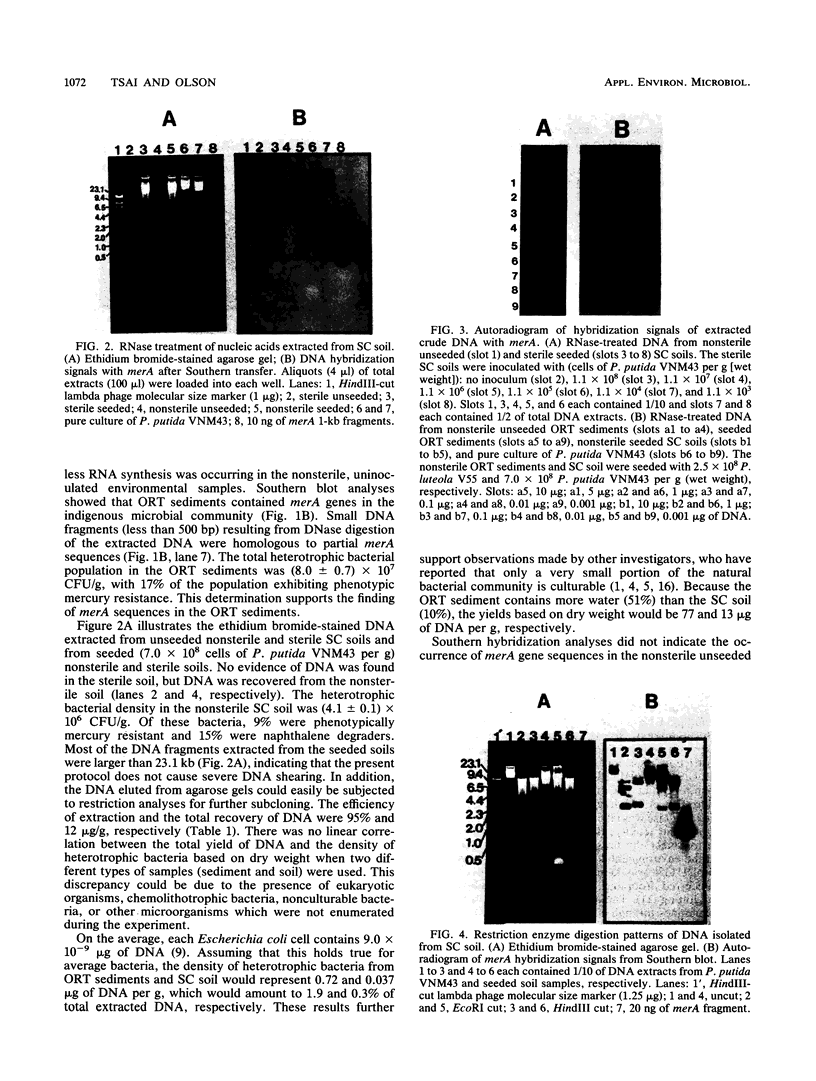
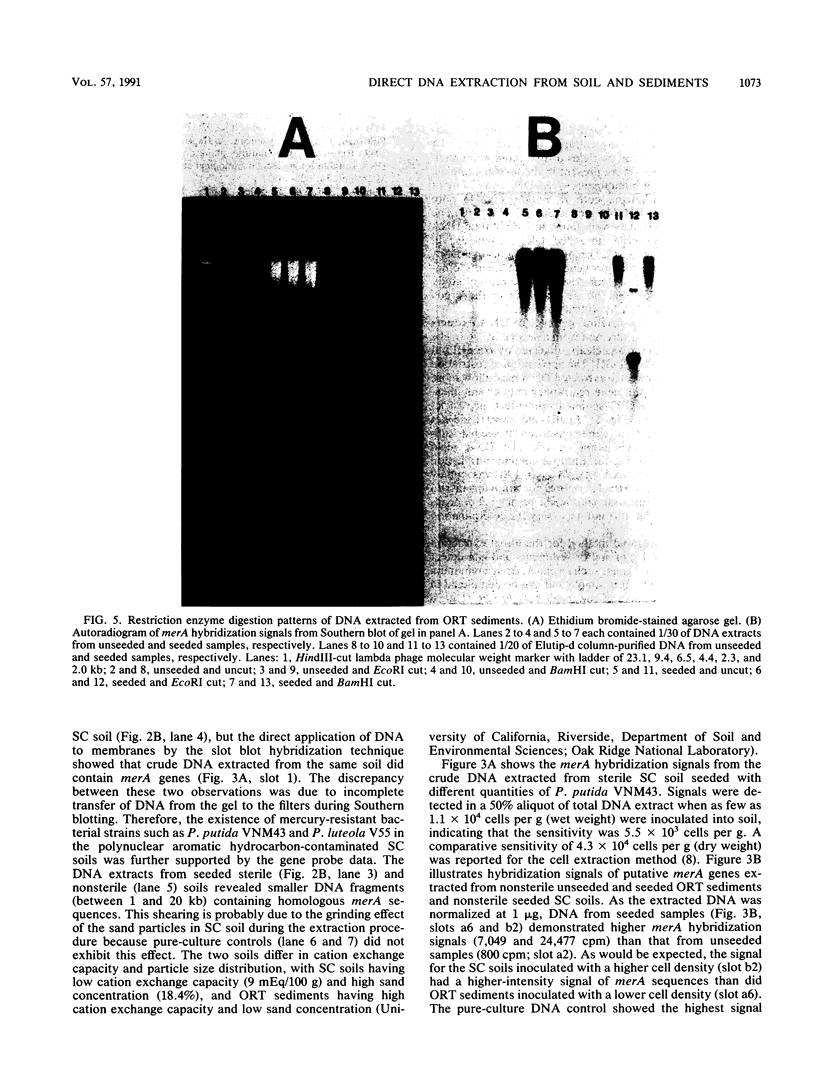
A rapid method for the direct extraction of DNA from soil and sediments was developed. The indigenous microorganisms in the soil and sediments were lysed by using lysozyme and a freeze-thaw procedure. The lysate was extracted with sodium dodecyl sulfate and phenol-chloroform. In addition to a high recovery efficiency (greater than 90%), the yields of DNA were high (38 and 12 micrograms/g [wet weight] from sediments and soil, respectively). This method generated minimal shearing of the extracted DNA. The crude DNA could be further purified with an Elutip-d column if necessary. An additional advantage of this method is that only 1 g of sample is required, which allows for the analysis of small samples and the processing of many samples in a relatively short (7 h) period.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson R. L., Buckley E. N., Palumbo A. V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984 Jan;47(1):49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco D. E., Mah R. A., Rabin A. C. Acridine orange-epifluorescence technique for counting bacteria in natural waters. Trans Am Microsc Soc. 1973 Jul;92(3):416–421. [PubMed] [Google Scholar]
- Fuhrman J. A., Comeau D. E., Hagström A., Chan A. M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988 Jun;54(6):1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson J. K., Holben W. E., Tiedje J. M. Detection in soil of a deletion in an engineered DNA sequence by using DNA probes. Appl Environ Microbiol. 1989 Nov;55(11):3022–3025. doi: 10.1128/aem.55.11.3022-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Shields M. S., Tedford E. T., Breen A., Hooper S. W., Sirotkin K. M., Davis J. W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985 May;49(5):1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Goksøyr J., Bej A. K., Atlas R. M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988 Dec;54(12):2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Salte K., Sørheim R., Goksøyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia S., Khan A., Rosenthal N. Construction and applications of DNA probes for detection of polychlorinated biphenyl-degrading genotypes in toxic organic-contaminated soil environments. Appl Environ Microbiol. 1990 Jan;56(1):254–259. doi: 10.1128/aem.56.1.254-259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]