Abstract
Pathogenic Leptospira species adapt to a wide range of environmental conditions during disease transmission and infection. While the proteome of in vitro cultivated Leptospira has been characterized in several studies to date, relatively little is known of the proteome as expressed by Leptospira during disease processes. Isolates of Leptospira obtained from patients suffering the severe pulmonary form of leptospirosis cause acute lethal infection in guinea pigs and chronic asymptomatic infection in rats. Recent studies have demonstrated that protein and lipopolysaccharide constituents of Leptospira recovered from acutely infected guinea pig tissue differ from that of Leptospira in chronically infected rat tissue and in vitro cultivated Leptospira (J. E. Nally, E. Chow, M. C. Fishbein, D. R. Blanco, and M. A. Lovett, Infect. Immun. 73:3251-3260, 2005). In the current study, the proteome of Leptospira expressed during disease processes was characterized relative to that of in vitro cultivated Leptospira (IVCL) after enrichment for hydrophobic membrane proteins with Triton X-114. Protein samples were separated by two-dimensional gel electrophoresis, and antigens expressed during infection were identified by immunoblotting with monospecific antiserum and convalescent rat serum in addition to mass spectrometry. Results suggest a significant increase in the expression of the outer membrane protein Loa22 during acute infection of guinea pigs relative to other outer membrane proteins, whose expression is generally diminished relative to expression in IVCL. Significant amounts of LipL32 are also expressed by Leptospira during acute infection of guinea pigs.
Pathogenic species of Leptospira cause the global zoonotic disease leptospirosis. Chronically infected domestic and wild animal species harbor in their renal tubules Leptospira which is shed into the environment upon urination. Excreted Leptospira continues to survive in suitable moist environments until contact and penetration of new hosts via skin abrasions or mucosal surfaces such as conjunctival tissue of the eye (1, 4).
Dissemination of Leptospira organisms throughout the infected host can result in a wide range of clinical manifestations of disease, ranging from a self-limiting fever to acute lethal forms to asymptomatic chronic carriage. The severe pulmonary form of leptospirosis (SFPL) results in high mortality rates (18, 20, 21, 23-25). Isolates of Leptospira recovered from patients suffering from SPFL have been used to develop acute and chronic experimental animal models of disease (13, 14). Experimental infection of guinea pigs emulates the acute lethal form of the disease with pulmonary hemorrhage, as observed in human patients suffering SPFL. While few Leptospira organisms were found in infected guinea pig lung tissue, large numbers were present in liver, kidney, spleen, and intestines (13). In contrast, experimental infection of rats results in an asymptomatic chronic infection with Leptospira organisms shed in urine, thus reproducing the natural mode of transmission of the disease (14).
We have previously reported the development of techniques to extract intact, motile Leptospira organisms from infected animal tissue (14). Characterization of Leptospira organisms recovered from infected host tissue (host tissue Leptospira, or HTL) indicated that the lipopolysaccharide O-antigen content of leptospires in guinea pig liver was markedly reduced compared to that of organisms found in rat renal tubules or cultivated in vitro (14). Thus, there is an association between diminished O-antigen content and acute lethal infection, while O-antigen content during renal tubular colonization approximates that of in vitro cultivated leptospires.
In this report, we present an approach to characterize proteins of Leptospira expressed during relevant disease processes compared to those expressed during in vitro growth.
MATERIALS AND METHODS
Bacteria.
An isolate of Leptospira interrogans serovar Copenhageni, designated RJ16441, was obtained from blood cultures of a patient suffering from the severe pulmonary form of leptospirosis who was admitted to Antonio Pedro University Hospital, Rio de Janeiro, Brazil (13, 23). Cultures were maintained in Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid (Becton Dickinson, MD) or EMJH semisolid medium (EMJH liquid medium containing 0.2% noble agar). Isolates were passaged through guinea pigs to maintain virulence as previously described (13).
Animals.
Hartley male guinea pigs (Charles River Laboratories, Kingston, NY), 12 to 15 days of age and weighing 200 g, were injected intraperitoneally with 105 cells of low-passage RJ16441 in a final volume of 500 μl as previously described (13). Sprague-Dawley rats (Charles River Laboratories, Kingston, NY), ∼6 weeks of age and weighing 130 to 150 g, were injected intraperitoneally with 107 cells of low-passage RJ16441 in a final volume of 500 μl as previously described (14). Negative-control animals were injected with EMJH medium alone. Animals were monitored daily for signs of illness including weight loss and loss of mobility. Guinea pigs were euthanized on day 5 or 6. All animal studies were approved by the Animal Research Committee of the University of California Los Angeles.
Purification of Leptospira from infected guinea pig liver.
Intact motile Leptospira organisms were extracted from infected guinea pig liver over Percoll density gradients as previously described (14).
Triton X-114 extraction of Leptospira organisms.
In vitro cultivated Leptospira (IVCL) or Percoll-purified Leptospira organisms from infected guinea pig liver (HTL)were extracted with Triton X-114 (TX-114) (Sigma) as previously described (14). The Percoll-purified HTL fraction was diluted 1:8 in phosphate-buffered saline before extraction with TX-114 to dilute the gradient in the extraction process.
Gel electrophoresis and immunoblotting.
Two-dimensional (2-D) gel electrophoresis was performed from pH 3 to 10 and from pH 4 to 7, as previously described (16). Total protein of 2-D gels was visualized after staining with Deep Purple (Amersham Biosciences) as per the manufacturer's instructions. For immunoblotting, samples were transferred to Immobilon-P transfer membrane (Millipore, Bedford, MA) and blocked with 5% (wt/vol) nonfat dry milk in phosphate-buffered saline-0.1% Tween 20. Membranes were individually incubated with indicated antisera for 1 h at indicated concentrations (chronic rat serum, 1:1,000; anti-outer membrane vesicle [OMV], 1:2,000; anti-OmpL1, 1:4,000; anti-LipL32, 1:5,000; anti-LipL21, 1:4,000; anti-LipL41, 1:4,000; and anti-LipL31, 1:2,500), followed by incubation with appropriate secondary antibodies: horseradish-peroxidase donkey anti-rabbit immunoglobulin G (IgG) conjugate (1:2,500; Amersham Biosciences, Piscataway, NJ), horseradish-peroxidase goat anti-rat IgG conjugate (1:2,000; Amersham Biosciences, Piscataway, NJ), or horseradish-peroxidase sheep anti-mouse IgG conjugate (1:2,500; Amersham Biosciences, Piscataway, NJ). Bound conjugates were detected with SuperSignal West Dura extended duration substrate (Pierce Biotechnology Inc., Rockford, IL). Since different proteins have different levels of antigenicity with each serum, optimal protein amounts for immunoblotting were determined to facilitate the identification of as many antigens as possible; 2 μg (see Fig. 2 and 3) or 4 μg (see Fig. 4) of processed IVCL cells was used for each immunoblot, and 8 μg (see Fig. 2 and 3) or 16 μg (see Fig. 4) of processed HTL cells was used for each immunoblot. Predicted molecular masses and isoelectric points of mature proteins described in results were based on submission of the amino acid sequences (without signal peptide) to ExPASy (http://au.expasy.org/tools/pi_tool.html).
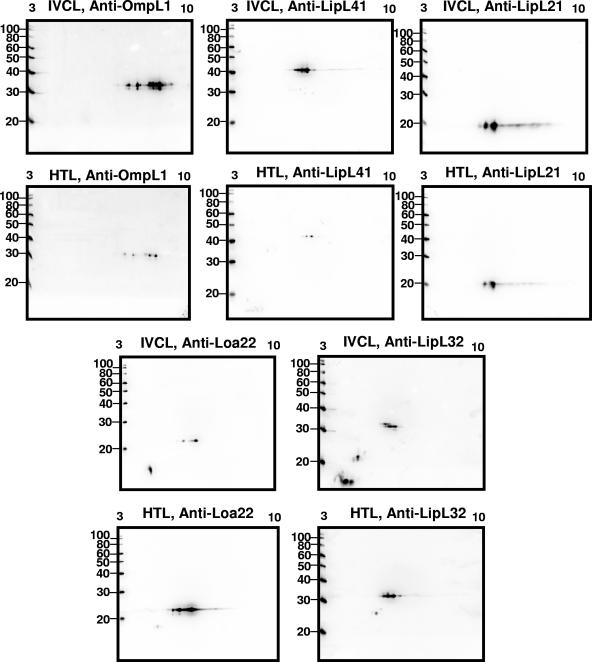
FIG. 2.
Two-dimensional immunoblots, pH 3 to 10, of TX-114-extracted IVCL (2 μg) or HTL (8 μg) sample. Samples were probed with monospecific antiserum specific for OmpL1, LipL41, LipL21, Loa22, and LipL32. Molecular mass markers (kDa) are provided on the left.
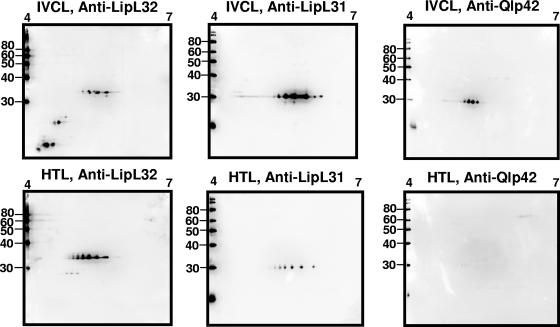
FIG. 3.
Two-dimensional immunoblots, pH 4 to 7, of TX-114-extracted IVCL (2 μg) or HTL (8 μg) sample. Samples were probed with monospecific antiserum specific for LipL32, LipL31, and Qlp42. Molecular mass markers (kDa) are provided on the left.
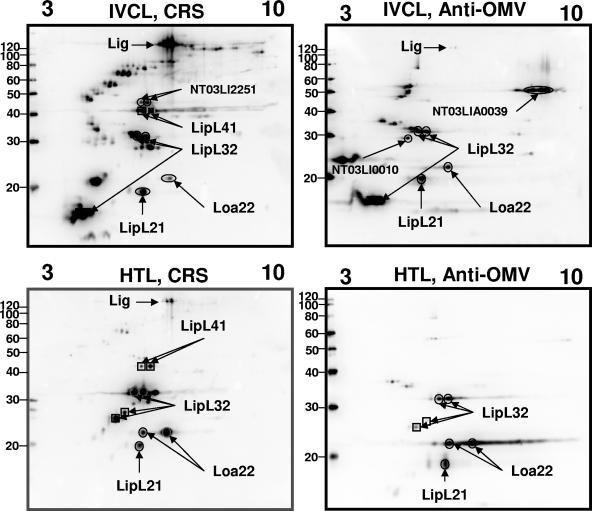
FIG. 4.
Two-dimensional immunoblots, pH 3 to 10, of TX-114-extracted IVCL (4 μg) or HTL (16 μg) sample. Samples were probed with CRS or anti-OMV. The antigens identified by mass spectrometry or immunoblotting with monospecific antiserum are indicated. Molecular mass markers (kDa) are provided on the left.
Analysis of tryptic peptide sequence tags by micro-liquid chromatography MS/MS.
Protein spots in 2-D gels identified by total protein staining were excised and digested with trypsin for analysis by mass spectrometry as previously described (16). Samples were analyzed by micro-liquid chromatography tandem mass spectrometry (MS/MS) with data-dependent acquisition (Q STAR XL; Applied Biosystems, Foster City, CA) after dissolution in 10 μl of 0.1% formic acid-5% acetonitrile (vol/vol). A reverse-phase PLRP-S column (200 μm by 10 cm, 5 μm, 300 Å; Michrom Biosciences, San Jose, CA) was equilibrated for 20 min at 2 μl/min with 100% eluent A (0.1% formic acid, 5% acetontrile in water) prior to sample injection (5 μl). A compound linear gradient was initiated 3 min after sample injection ramping to 80% eluent A and 20% eluent B (0.1% formic acid in acetonitrile) at 8 min; 65% A and 35% B at 13 min; 25% A and 75% B at 23 min, and 90% A and 10% B at 23.1 min. Column eluent was directed to a stainless steel nano-electrospray emitter (ES301; Proxeon, Odense, Denmark) at 4.4 kV for ionization without nebulizer gas. The mass spectrometer was operated in the IDA (information dependent acquisition) mode with a survey scan (400 to 1,500 m/z), data-dependent MS/MS on the two most abundant ions with exclusion after two MS/MS experiments. Individual sequencing experiments were matched to either a custom L. interrogans serovar Copenhageni strain Fiocruz L1-130 sequence database downloaded from NCBI (www.ncbi.nlm.nih.gov) (19) or to the global mass spectrometry database provided at the Matrix Science server using Mascot software (http://www.matrixscience.com; Matrix Sciences, London, United Kingdom). The search was run under the mode “no enzyme” to identify semi- and nontryptic peptides. MS/MS spectra matched to peptide sequences with Mascot scores exceeding 30 were examined manually, specifically with respect to calculated parent and product ion mass accuracy as well as to whether the return was fully or partially tryptic. Where two or more peptides were matched reliably, a strong hit was reported. Where a single good quality peptide hit was returned, a potential hit was reported. The mass spectral data were interpreted without knowledge of the isoelectric points or molecular masses predicted from the 2-D analysis.
RESULTS
Identification of TX-114-extracted proteins of IVCL by mass spectrometry.
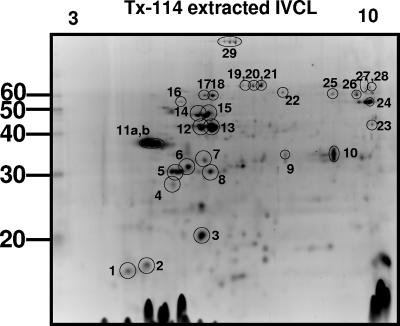
Fractions of IVCL enriched for hydrophobic membrane proteins were separated by 2-D gel electrophoresis and visualized by staining with Deep Purple, (Fig. 1). Table 1 provides a list of those proteins in Fig. 1 that were excised and identified by mass spectrometry. In addition to several known outer membrane proteins, including LigA, OmpL1, LipL41, LipL21, LipL36, LipL32 (also known as Hap-1), LipL31, and Qlp42 (also known as LipL45), several additional proteins were identified that confirm annotation of the completed genomes of Leptospira and confirm the expression of these proteins during in vitro culture. These include a putative outer membrane protein (spot number 2), four putative lipoproteins (spot numbers 4, 6, 14, 15, and 23) in addition to the annotated lipoprotein LipL71 (spot number 16), two conserved hypothetical proteins (spot numbers 17, 18, and 24), two outer membrane efflux proteins (spot numbers 25 to 28), a penicillin G acylase precursor (spot number 22), and a carboxy-terminal processing protease precursor (spot numbers 19 to 21). Not all proteins were amenable to identification by mass spectrometry, as previously discussed (16).
FIG. 1.
Total protein staining of TX-114-extracted IVCL sample separated by 2-D gel electrophoresis, pH 3 to 10. Protein spots excised for identification by mass spectrometry are indicated and correspond to identifications presented in Table 1. Molecular mass markers (kDa) are provided on the left.
TABLE 1.
TX-114-extracted proteins of L. interrogans serovar Copenhageni identified by mass spectrometry from Fig. 1a
| Spot no. | TIGR locus | TIGR annotation | Mass | pI | Score | No. of peptides |
|---|---|---|---|---|---|---|
| 1 | NT03LI1637 | LipL32 | 29612.88 | 6.8072 | 202 | 17 |
| 2 | NT03LI2344 | Putative outer membrane protein | 17836.21 | 5.9774 | 164 | 10 |
| 3 | NT03LI0012 | LipL21 | 19660.97 | 7.7989 | 194 | 13 |
| 4 | NT03LI0591 | Putative lipoprotein | 23476.71 | 5.2288 | 139 | 7 |
| 5 | NT03LI1973 | LipL45 | 42101.00 | 7.5931 | 431 | 28 |
| 512 | 15 | |||||
| 6 | NT03LI0010 | Putative lipoprotein | 27700.76 | 6.7327 | 426 | 24 |
| 236 | 8 | |||||
| 7 | NT03LI1637 | LipL32 | 29612.88 | 6.8072 | 113 | 8 |
| 31 | 1 | |||||
| 8 | NT03LI1763 | LipL31 | 27620.73 | 7.3454 | 564 | 43 |
| 234 | 9 | |||||
| 9 | NT03LI1168 | OmpL1 | 33457.00 | 8.8726 | 148 | 3 |
| 10 | NT03LI1168 | OmpL1 | 33457.00 | 8.8726 | 280 | 13 |
| 305 | 8 | |||||
| 11A | NT03LI1850 | Flagellin/FlaB1 | 31470.84 | 7.9096 | 141 | 5 |
| 11B | NT03LI3663 | LipL36 | 36949.31 | 4.4751 | 136 | 10 |
| 118 | 4 | |||||
| 159 | 5 | |||||
| 116 | 3 | |||||
| 95 | 3 | |||||
| 12 | NT03LI3555 | LipL41 | 37384.65 | 5.7378 | 763 | 41 |
| 558 | 17 | |||||
| 13 | NT03LI3555 | LipL41 | 37384.65 | 5.7378 | 921 | 61 |
| 750 | 25 | |||||
| 14 | NT03LI2251 | Putative lipoprotein | 44713.71 | 6.9004 | 965 | 47 |
| 511 | 14 | |||||
| 15 | NT03LI2251 | Putative lipoprotein | 44713.71 | 6.9004 | 1054 | 65 |
| 675 | 21 | |||||
| 16 | NT03LI1213 | LipL71 | 62027.26 | 5.3356 | 110 | 2 |
| 17 | NT03LI0139 | Conserved hypothetical protein | 61989.42 | 5.5294 | 221 | 6 |
| 18 | NT03LI0139 | Conserved hypothetical protein | 61989.42 | 5.5294 | 552 | 18 |
| 19 | NT03LI2676 | Carboxy-terminal processing protease precursor | 66174.27 | 7.0959 | 134 | 3 |
| 20 | NT03LI2676 | Carboxy-terminal processing protease precursor | 66174.27 | 7.0959 | 311 | 9 |
| 21 | NT03LI2676 | Carboxy-terminal processing protease precursor | 66174.27 | 7.0959 | 657 | 15 |
| 22 | NT03LI0330 | Penicillin G acylase precursor | 95417.05 | 8.6793 | 78 | 2 |
| 23 | NT03LI4002 | Putative lipoprotein | 46340.41 | 9.7852 | 226 | 9 |
| 24 | NT03LIA0039 | Conserved hypothetical protein | 49905.83 | 8.9696 | 600 | 39 |
| 257 | 9 | |||||
| 25 | NT03LI3081 | Outer membrane efflux protein | 59860.32 | 9.5055 | 212 | 6 |
| 26 | NT03LI3081 | Outer membrane efflux protein | 59860.32 | 9.5055 | 354 | 9 |
| 27 | NT03LI3222 | Outer membrane efflux protein | 63512.25 | 9.5550 | 70 | 3 |
| 28 | NT03LI3222 | Outer membrane efflux protein | 63512.25 | 9.5550 | 143 | 4 |
| 29 | NT03LI0545 | LigA | 128128.55 | 6.8598 | 1332 | 53 |
| 217 | 7 | |||||
| 231 | 6 | |||||
| 807 | 21 |
Proteins of TX-114-extracted IVCL identified by mass spectrometry. Spot numbers correspond to those shown in Fig. 1. Locus, annotation, mass, and isoelectric point information is as provided by The Institute for Genomic Research based on the annotation of L. interrogans Copenhageni Fiocruz L1-130 (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=ntli03). A Mascot search score of mass spectrometry results is also provided, as is the number of peptides used to provide the score.
Identification of antigens expressed in IVCL and HTL using monospecific antiserum.
Since several proteins of the outer membrane of in vitro cultivated Leptospira have been identified and characterized at the molecular level, including OmpL1, LipL41, LipL21, Loa22, LipL32, LipL31, and Qlp42 (3, 5, 6, 8, 10-12, 22), immunoblots of TX-114-extracted IVCL and HTL samples were probed with monospecific antiserum to determine whether these proteins were expressed during acute infection of guinea pigs (Fig. 2).
Immunoblot analysis with anti-OmpL1 indicated that OmpL1, an outer membrane porin of Leptospira (31.06 kDa; pI 7.9) is expressed in both IVCL and HTL. Expression of both LipL41 (36.82 kDa; pI 6.0) and LipL21 (17.83 kDa; pI 6.5), surface-exposed lipoproteins, is detected in IVCL and HTL. Loa22, a surface-exposed lipoprotein (18.73 kDa, pI 6.83), is also expressed in IVCL and HTL, but larger levels of Loa22 are detected in HTL compared to IVCL, relative to an apparent diminution in levels of OmpL1, LipL41, and LipL21.
LipL32 has a predicted mass of 27.654 kDa and a pI of 5.74 although actual mass ranges from 28.47 to 28.58 kDa due to the presence of lipoforms (16). LipL32 is detected in both IVCL and HTL (Fig. 2). In addition, anti-LipL32 is reactive with lower-molecular-mass antigens, indicating the presence of lower mass products of LipL32 in each sample. Interestingly, the lower-molecular-mass products reactive in IVCL are different from lower-molecular-mass products detected in HTL. Overall, and relative to an apparent diminution of expression by OmpL1, LipL41, and LipL21, LipL32 is easily detected in HTL samples at levels comparable to those detected in IVCL samples.
In order to provide improved resolution of LipL32 and the antigens which have a molecular mass and pI similar to that of LipL32, immunoblot assays were also performed over a pH range of 4 to 7 (Fig. 3). LipL31, a cytoplasmic membrane lipoprotein (25.45 kDa; pI 5.92) was detected in IVCL and HTL. While Qlp42 has a predicted mass of 39.8 kDa and pI of 6.1, it is actually detected as two proteins with masses of 28.41 and 26.46 kDa (11, 12). Immunoblotting confirmed the expression of Qlp42 in IVCL but not in HTL at this level of detection.
Identification of antigens expressed in IVCL and HTL using chronic rat serum and anti-OMV serum.
TX-114-extracted antigens from HTL and IVCL samples were detected with serum from chronically infected rats (chronic rat serum [CRS]) (14) or with serum raised against OMVs prepared from IVCL (16). Alignment of immunoblots of TX-114-extracted HTL samples with immunoblots of TX-114-extracted IVCL samples indicated that both CRS and anti-OMV serum recognize antigens expressed by both IVCL and HTL, as shown in Fig. 4. However, several antigens are differentially expressed in IVCL and HTL sample preparations.
Alignment of immunoblots in Fig. 4 with those in Fig. 2 confirms the identity of LipL41, LipL21, LipL32, and Loa22 proteins as antigens reactive with CRS and anti-OMV serum. However, it was also noted that CRS and anti-OMV, at the level of detection presented in Fig. 1, do not react with a protein of similar mass and pI to OmpL1. As shown in Fig. 2, larger levels of Loa22 are detected with CRS in HTL compared to IVCL (Fig. 4). Similarly, LipL32 is readily detected in HTL and IVCL relative to apparently diminished levels of LipL21 and LipL41. Further, the lower-molecular-mass products of LipL32 shown in Fig. 2 correspond in mass and pI to lower-molecular-mass antigens reactive with CRS and anti-OMV.
To identify antigens of IVCL and HTL samples that were reactive with CRS and anti-OMV, but not with serum against known outer membrane proteins, immunoblots with CRS and anti-OMV (Fig. 4) were aligned with total protein stains of IVCL (Fig. 1). This allowed for the excision of protein spots with the same molecular mass and pI values as antigens recognized by CRS and anti-OMV and their identification by mass spectrometry. This approach was also used to corroborate the identity of the antigens identified by immunoblotting with monospecific antiserum.
Immunoblots of TX-114-extracted IVCL and HTL with antiserum specific for LigA, a putative lipoprotein (126.4 kDa; pI 6.26), were inconclusive for detection of LigA expression in 2-D immunoblot analysis (data not shown). However, mass spectrometry provided conclusive evidence for its presence in the TX-114 detergent phase of IVCL (spot number 29). LigA of IVCL was highly reactive with CRS (Fig. 4), confirming expression during in vitro growth and chronic infection of rats. LigA of HTL was also reactive with CRS, though to a lesser extent than that detected in IVCL and relative to the amounts loaded for each sample.
Additional antigens reactive with OMV that were identified include the putative lipoprotein NT03LI0010 (Fig. 1, spot number 6) and the conserved hypothetical protein NT03LIA0039 (Fig. 1, spot numbers 17 and 18) (proteins are identified by Institute for Genomic Research [TIGR] locus numbers in Fig. 4). Both antigens were detected in IVCL but not HTL. CRS was reactive with the putative lipoprotein NT03LI2251 (spot numbers 14 and 15) which was detected in IVCL but not HTL, suggesting that this is a lipoprotein expressed at some stage during chronic infection of rats but not at the time point examined during acute lethal infection of guinea pigs.
Loa22 was not identified by mass spectrometry, nor was any protein spot with a similar molecular mass and pI to Loa22 identified on 2-D gels of TX-114-extracted IVCL. However, it was identified with the added sensitivity provided by immunoblotting with Loa22-specific antiserum. Loa22 has previously been identified by mass spectrometry in 2-D gels of outer membrane vesicles, which corresponds in molecular mass and pI to the Loa22 antigen detected in Fig. 2 (16).
DISCUSSION
It has long been appreciated that Leptospira species adapt to and survive in vastly different environments, but little is known about the molecular nature of these adaptations. The leptospiral outer membrane lipoprotein LipL36 has provided one example of environmentally regulated protein expression. Immunohistochemical staining demonstrated expression of LipL36 during in vitro growth at 30°C but not in infected tissue or at culture temperatures of 37°C, indicating an adaptive response by the organism to infection which included the diminution of expression of LipL36 (7, 15).
We have recently reported that a human SPFL isolate causes the severe pulmonary form of leptospirosis in guinea pigs and chronic asymptomatic carriage in rats (13, 14). Since large numbers of Leptospira organisms can be found in the livers of infected guinea pigs, a procedure was developed for extracting intact motile Leptospira organisms from infected host tissue. The completed genomes of L. interrogans serovar Lai and serovar Copenhageni are predicted to have 3,728 and 4,727 protein-coding genes, respectively (17). In order to reduce the complexity of proteins for sample analysis, both IVCL and HTL samples were extracted with 2% TX-114 which also enriches for hydrophobic protein antigens, as previously described (9, 15, 26). This has the added advantage of identifying putative vaccinogen and diagnostic antigens associated with the outer membrane of Leptospira during infection. Two-dimensional immunoblotting of TX-114-extracted IVCL indicated that CRS reacts with several antigens of the outer membrane of IVCL, which is confirmed by their reactivity with monospecific antiserum for LipL32, LipL21, LipL41, and Loa22. Mass spectrometry confirmed the identity of two additional CRS-reactive antigens as the putative lipoprotein NT03LI2251. Several other antigens have yet to be identified. By definition, all of these antigens are expressed during the chronic infection process of rats and in sufficient amounts to generate an antibody response. Similarly, CRS detected several antigens in HTL samples derived from the liver tissue of acutely infected guinea pigs. These antigens include several also expressed in IVCL including the aforementioned LipL32, LigA, LipL21, LipL41, and Loa22.
While absolute quantification of the amounts of each antigen present in each sample is not provided, these findings do provide an accurate view of the expression of antigens relative to each other in HTL and IVCL preparations. For example, Loa 22 is expressed in large amounts in HTL and is the only antigen whose expression appears to be significantly up-regulated during disease relative to detection of other antigens in the same sample. In contrast to Loa22, the relative amounts of LigA, LipL41, and LipL21 are reduced in HTL compared to IVCL. There are also a number of unidentified antigens detected with CRS or OMV serum, and their expression appears reduced in HTL relative to their representation in IVCL. Significant amounts of LipL32 are detected both during in vitro growth and disease relative to the diminution of several other known outer membrane antigens.
Expression of LipL32, LipL21, and Loa22 was detected with antiserum specific for OMVs of IVCL and serum from chronically infected rats. Some antigens were detected only with anti-OMV, including the putative lipoprotein NT03LI0010 and the conserved hypothetical protein NT03LIA0039, indicating that they are expressed in IVCL but not in sufficient amounts during chronic infection of rats to generate a detectable antibody response. LigA is reactive with CRS but only slightly reactive with anti-OMV. Similar amounts of LigA are present in each IVCL sample, suggesting that the greater reactivity with CRS is due to the expression of LigA during chronic infection of rats. As with CRS, anti-OMV generally reacts with a smaller set of antigens in the HTL sample but does react with LipL32, LipL21, and Loa22. Reactivity with the putative lipoprotein NT03LI0010 and the conserved hypothetical protein NT03LIA0039 is not detected in the HTL sample, confirming their diminished expression in HTL, as already demonstrated by a lack of reactivity with CRS.
While LipL32 is expressed in both IVCL and HTL, it was noted that several lower-molecular-mass antigens are specifically reactive with LipL32 monospecific antiserum, indicating that these fragments are likely derived from the mature LipL32. It is of interest that different lower-molecular-mass fragments of LipL32 are detected in IVCL and HTL samples. The meaning of this finding, observed with both CRS and anti-OMV antiserum, is unclear at this time. However, breakdown products of LipL32 have previously been noted during experimental preparations of outer membranes of IVCL (2, 26).
In this report, we have described the hydrophobic proteome of guinea pig liver-derived HTL recovered during the course of acute lethal infection. The relative amounts of Loa22 and LipL32 were enhanced in HTL compared to IVCL samples. There is also a striking reduction in the relative content of other hydrophobic protein antigens in HTL relative to their representation in IVCL. We have recently demonstrated that the lipopolysaccharide O-antigen content of HTL found in guinea pig liver is markedly reduced compared to that of IVCL. Taken together, these findings indicate that the surface antigen structure of HTL differs markedly from that of IVCL. The role of these compositional changes in pathogenesis remains to be determined.
Acknowledgments
These studies were supported by National Institutes of Health grant AI056258 to M.A.L. and a Ruth L. Kirschenstein National Research Service Award AI055235 to J.E.N. from the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
We thank David Haake for kindly providing antiserum specific for LipL32, LipL41, Qlp42, OmpL1, LipL21, and LipL31. We thank Nobuo Koizumi and Haruo Watanabe for kindly providing antiserum for Loa22.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Bharti, A. R., J. E. Nally, J. N. Ricaldi, M. A. Matthias, M. M. Diaz, M. A. Lovett, P. N. Levett, R. H. Gilman, M. R. Willig, E. Gotuzzo, and J. M. Vinetz. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757-771. [DOI] [PubMed] [Google Scholar]
- 2.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, and B. Adler. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen, P. A., D. A. Haake, D. M. Bulach, R. L. Zuerner, and B. Adler. 2003. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 71:2414-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis, 2 ed. MediSci, Melbourne, Australia.
- 5.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175:4225-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haake, D. A., C. Martinich, T. A. Summers, E. S. Shang, J. D. Pruetz, A. M. McCoy, M. K. Mazel, and C. A. Bolin. 1998. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect. Immun. 66:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haake, D. A., and J. Matsunaga. 2002. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect. Immun. 70:4936-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haake, D. A., E. M. Walker, D. R. Blanco, C. A. Bolin, M. N. Miller, and M. A. Lovett. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koizumi, N., and H. Watanabe. 2003. Molecular cloning and characterization of a novel leptospiral lipoprotein with OmpA domain. FEMS Microbiol. Lett. 226:215-219. [DOI] [PubMed] [Google Scholar]
- 11.Matsunaga, J., T. A. Young, J. K. Barnett, D. Barnett, C. A. Bolin, and D. A. Haake. 2002. Novel 45-kilodalton leptospiral protein that is processed to a 31-kilodalton growth-phase-regulated peripheral membrane protein. Infect. Immun. 70:323-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nally, J. E., S. Artiushin, and J. F. Timoney. 2001. Molecular characterization of thermoinduced immunogenic proteins Qlp42 and Hsp15 of Leptospira interrogans. Infect. Immun. 69:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nally, J. E., C. Chantranuwat, X. Y. Wu, M. C. Fishbein, M. M. Pereira, J. J. Da Silva, D. R. Blanco, and M. A. Lovett. 2004. Alveolar septal deposition of immunoglobulin and complement parallels pulmonary hemorrhage in a guinea pig model of severe pulmonary leptospirosis. Am. J. Pathol. 164:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nally, J. E., E. Chow, M. C. Fishbein, D. R. Blanco, and M. A. Lovett. 2005. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect. Immun. 73:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nally, J. E., J. F. Timoney, and B. Stevenson. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nally, J. E., J. P. Whitelegge, R. Aguilera, M. M. Pereira, D. R. Blanco, and M. A. Lovett. 2005. Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics 5:144-152. [DOI] [PubMed] [Google Scholar]
- 17.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwattayakul, K., J. Homvijitkul, O. Khow, and V. Sitprija. 2002. Leptospirosis in northeastern Thailand: hypotension and complications. Southeast Asian J. Trop. Med. Public Health 33:155-160. [PubMed] [Google Scholar]
- 19.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 20.Sehgal, S. C., M. V. Murhekar, and A. P. Sugunan. 1995. Outbreak of leptospirosis with pulmonary involvement in north Andaman. Indian J. Med. Res. 102:9-12. [PubMed] [Google Scholar]
- 21.Seijo, A., H. Coto, J. San Juan, J. Videla, B. Deodato, B. Cernigoi, O. G. Messina, O. Collia, D. de Bassadoni, R. Schtirbu, A. Olenchuk, G. D. de Mazzonelli, and A. Parma. 2002. Lethal leptospiral pulmonary hemorrhage: an emerging disease in Buenos Aires, Argentina. Emerg. Infect. Dis. 8:1004-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang, E. S., T. A. Summers, and D. A. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva, J. J., M. O. Dalston, J. E. Carvalho, S. Setubal, J. M. Oliveira, and M. M. Pereira. 2002. Clinicopathological and immunohistochemical features of the severe pulmonary form of leptospirosis. Rev. Soc. Bras. Med. Trop. 35:395-399. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, F. G., K. A. Green, G. J. Haug, and D. L. Brookes. 1998. Leptospirosis associated with severe pulmonary haemorrhage in Far North Queensland. Med. J. Aust. 169:151-153. [DOI] [PubMed] [Google Scholar]
- 25.Trevejo, R. T., J. G. Rigau-Perez, D. A. Ashford, E. M. McClure, C. Jarquin-Gonzalez, J. J. Amador, J. O. de los Reyes, A. Gonzalez, S. R. Zaki, W. J. Shieh, R. G. McLean, R. S. Nasci, R. S. Weyant, C. A. Bolin, S. L. Bragg, B. A. Perkins, and R. A. Spiegel. 1998. Epidemic leptospirosis associated with pulmonary hemorrhage-Nicaragua, 1995. J. Infect. Dis. 178:1457-1463. [DOI] [PubMed] [Google Scholar]
- 26.Zuerner, R. L., W. Knudtson, C. A. Bolin, and G. Trueba. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb. Pathog. 10:311-322. [DOI] [PubMed] [Google Scholar]