Abstract
Urinary tract infection (UTI) is an important health problem worldwide, with many millions of cases each year, and Escherichia coli is the most common organism causing UTI in humans. Also, E. coli is responsible for most infections in patients with chronic indwelling bladder catheter. The two asymptomatic bacteriuria (ABU) E. coli strains 83972 and VR50 are significantly better biofilm formers in their natural growth medium, human urine, than the two uropathogenic E. coli isolates CFT073 and 536. We used DNA microarrays to monitor the expression profile during biofilm growth in urine of the two ABU strains 83972 and VR50. Significant differences in expression levels were seen between the biofilm expression profiles of the two strains with the corresponding planktonic expression profiles in morpholinepropanesulfonic acid minimal laboratory medium and human urine; 417 and 355 genes were up- and down-regulated, respectively, during biofilm growth in urine of 83972 and VR50. Many genes involved in transcription and stress were up-regulated in biofilm-grown cells. The role in biofilm formation of four of the up-regulated genes, i.e., yceP, yqgA, ygiD, and aaeX, was investigated by creating single-knockout mutant versions of 83972 and VR50; all mutants showed reduced biofilm formation in urine by 18 to 43% compared with the wild type (P < 0.05). Also, the expression profile of strain 83972 in the human urinary tract partially overlaps with the biofilm expression profile.
Bacteria generally live attached to surfaces rather than as planktonic isolated cells. Often adhered bacteria form sessile communities, also referred to as biofilms, and these are commonly associated with many health problems (10). Biofilms can form on virtually any type of surface. In the medical field bacterial biofilms have attracted particular attention, because many persistent and chronic bacterial infections, including periodontitis, otitis media, biliary tract infection, and endocarditis, are now believed to be linked to the formation of biofilms. Also, virtually all medical implants are prone to colonization by bacteria, and the resultant biofilms often serve as a source for recurrent infections. Bacterial biofilm infections are particularly problematic, because sessile bacteria can withstand host immune defense mechanisms and are extremely resistant to antibiotics, biocides, and hydrodynamic shear forces that can efficiently clear corresponding planktonic bacteria (9, 10).
Urinary tract infection (UTI) is a serious health problem that affects millions of people each year (49). The recurrence rate is high, and often the infections are particularly troublesome and become chronic, with multiple episodes. UTI usually starts as a bladder infection but often ascends to affect the kidneys and ultimately can result in renal failure or dissemination to the blood. UTI is the most common infection in patients with indwelling bladder catheters, and bacteriuria is essentially unavoidable in this patient group, with an infection rate of ∼100% within 1 month (20). UTI is classified into disease categories by the site of infection: cystitis (the bladder), pyelonephritis (the kidney), and bacteriuria (the urine). The colonization of urine in the absence of clinical symptoms is called asymptomatic bacteriuria (ABU). ABU occurs in up to 6% of healthy individuals and 20% of elderly individuals. As the name implies, ABU strains generally do not cause symptoms; most patients with ABU do not need treatment, and in many cases the colonizing organism actually helps to prevent infection by other more virulent bacteria (12, 24).
Escherichia coli is responsible for more than 80% of all UTIs and causes both ABU and symptomatic UTI (22, 51). E. coli 83972, a clinical isolate capable of long-term bladder colonization, has become a model ABU organism. It was originally isolated from a girl with ABU who had carried it for at least 3 years without symptoms (1, 31). The strain is well adapted for growth in the human urinary tract (UT), where it establishes long-term bacteriuria (1, 24). Strain 83972 has been used for prophylactic purposes in numerous studies; as such it has been used as an alternative treatment in patients with recurrent UTI who are refractory to conventional therapy (12, 24). The mechanisms for bladder colonization used by ABU E. coli are not known. Low abundance of adhesins could explain to a large degree why ABU strains do not cause symptoms in the host; however, it does not explain how these strains are capable of efficient bladder colonization. Meanwhile, since E. coli strains are responsible for most infections in patients with chronic indwelling bladder catheter (56), we suspected that biofilm formation could contribute to this.
DNA microarray-assisted functional genomics provides a genome-wide portrait of the transcriptome of an organism and discloses how a bacterium adapts to a particular environmental niche at the transcriptional level. Here we analyze the global expression profile of two E. coli ABU strains, 83972 and VR50, during biofilm formation on abiotic surfaces in human urine.
MATERIALS AND METHODS
Bacterial strains.
E. coli 83972 (OR:K5:H−) is a prototype ABU strain and lacks defined O and K surface antigens (1). VR50 (OR:K1:H−) is a recently isolated ABU E. coli strain which grows very well in urine and is capable of expressing type 1 fimbriae (41). Creation of the fluorescently tagged versions of 83972 and VR50, 83972yfp and VR50yfp, has been described previously (21).
Growth conditions and stabilization of RNA for microarray experiments of E. coli grown in planktonic cultures.
Human urine was collected from four healthy men and women volunteers who had no history of UTI or antibiotic use in the prior 2 months. The urine was pooled, filter sterilized, stored at 4°C, and used within the following 2 days. Overnight cultures of VR50 were grown in triplicates in pooled human urine or morpholinepropanesulfonic acid (MOPS) minimal media (supplemented with 0.2% glucose and 1 mg/liter pantothenic acid) until reaching exponential phase and then used for inoculation of 25 ml urine or MOPS to an optical density at 600 nm (OD600) of 0.05. The cultures were grown at 37°C and 130 rpm, and 5-ml samples for isolation of RNA were extracted from three individual cultures at mid-exponential phase (corresponding to an OD600 of approximately 0.4 and 0.5 in urine and MOPS, respectively). Extracted samples were immediately mixed with two volumes of RNAprotect Bacteria Reagent (QIAGEN AG), incubated for 5 min at room temperature to stabilize RNA, and centrifuged. The pellets were then stored at −80°C. The samples for E. coli 83972 grown in MOPS-glucose and human urine were grown similarly, with the exception of no addition of pantothenic acid to the MOPS cultures, and have been analyzed and described previously (43).
Growth conditions and stabilization of RNA for microarray experiments of E. coli grown in biofilms.
Freshly grown urine cultures of E. coli 83972 and VR50 were used for inoculation of 25 ml pooled human urine in 94-mm petri dishes (Greiner Bio-One), each strain in triplicate. The cultures were grown statically at 37°C for 42 h, and the medium was carefully poured off and replaced by fresh preheated medium twice during incubation. After 42 h the bacterial lawn on each petri dish was quickly washed twice with 20 ml urine preheated to 37°C, and immediately thereafter 2 ml of a 1:2 mixture of phosphate-buffered saline (PBS) and RNAprotect Bacteria Reagent (QIAGEN AG) was poured on the lawn of attached cells and incubated for 5 min at room temperature to stabilize RNA. The stabilized mixture was then centrifuged, and pellets were stored at −80°C. The same batch of urine was used for growth of 83972 and VR50 in petri dishes and for growth of VR50 in planktonic cultures (see the above paragraph).
RNA isolation and cDNA labeling.
Total RNA was isolated using the RNeasy Mini kit (QIAGEN AG), and on-column DNase digestion was performed using an RNase-Free DNase set (QIAGEN AG). The quality of the total RNA was examined by agarose gel electrophoresis and by measuring the absorbance at 260 and 280 nm to ensure intact high-quality RNA. Purified RNA was precipitated with ethanol and stored at −80°C until further use. Conversion of RNA (10 μg per sample) to cDNA and microarray analysis were performed according to GeneChip Expression Analysis Technical Manual 701023, Rev. 4 (Affymetrix, Inc., Santa Clara, CA).
DNA microarray hybridization.
GeneChip E. coli Genome 2.0 arrays (Affymetrix) were used for hybridization of the labeled cDNA. In total, 18 samples were hybridized to 18 chips, i.e., 9 biologically independent samples from each of the two strains were hybridized to 1 chip each. For each strain, i.e., 83972 and VR50, three chips were hybridized with samples grown in three individual flasks in MOPS, three chips were hybridized with samples from cells grown in pooled human urine in three individual flasks, and three chips were hybridized with samples from biofilm cells grown in pooled human urine in three individual petri dishes. No pooling was performed, and all samples were treated separately. Hybridization, washing, and staining were performed according to GeneChip Expression Analysis Technical Manual 701023, Rev. 4 (Affymetrix), and the microarrays were scanned using a GeneChip Scanner 3000.
Data analysis.
Array normalization and expression value calculation were performed using the DNA-Chip Analyzer (dChip) 1.3+ software program (http://www.dchip.org/) (29). The Invariant Set Normalization method (30) was used to normalize arrays at the probe cell level to make them comparable, and the model-based (PM-only) method was used for probe selection and computing expression values. These expression levels were attached with standard errors as measurement accuracy, which were subsequently used to compute 90% confidence intervals of fold changes in two-group comparisons. All 18 chips (12 chips generated in this study and 6 chips [83972 grown in MOPS and urine] described earlier [43]) were normalized together, and a number of different comparisons were performed in order to filter out genes differently expressed during biofilm growth in urine compared with planktonic cultures grown in both MOPS and urine. (i) The three arrays of each strain (83972 and VR50) hybridized with samples grown in MOPS were used as the baseline for calculation of fold changes in each strain grown in biofilms in urine. (ii) Calculation of fold changes in each strain grown in biofilms in urine was performed in the same way as in the first comparison but with the three arrays hybridized with samples grown planktonically in pooled human urine as the baseline. (iii) In one comparison, the fold changes in biofilm were calculated on all 6 biofilm arrays from both strains using all 12 planktonic arrays as the baseline. The comparison criteria were carefully chosen to make sure that the estimation of the percentages of genes identified by chance, the empirical false discovery rate, was kept low (≤0.6% for all comparisons in our study). Permuting our samples randomly 200 times resulted in false discovery rates between 0.0% and 0.6%, or 0 to 11 false-positive genes, for the different comparisons.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed to confirm DNA microarray gene expression data. RNA, from the DNase-treated RNA samples used for microarray hybridization and analysis, was converted to cDNA using SuperScript II (Invitrogen). cDNA was used directly as template for PCR, and a negative control on the RNA sample (not converted to cDNA) was run in parallel to confirm that all DNA had been removed in the earlier step. The total number of cycles used in PCR ranged from 15 to 40. RT-PCR products were examined by agarose gel electrophoresis.
Motility assay.
The motility ability was tested for the two ABU strains. Two microliters of biofilm urine cultures were stabbed into urine plates (4:1 ratio of human urine and 0.9% NaCl) containing 0.3% agar and incubated at 37°C for 16 h. The assay was repeated thrice in different batches of urine.
Biofilm formation in flow-cell chambers in human urine.
Flow-chamber experiments were performed at 37°C in human urine, essentially as described previously (8). Briefly, biofilms were grown on a microscope glass coverslip (Knittel 24 × 50 mm st1; Knittel Gläser) in three-channel chambers (channel dimension, 1 by 4 by 40 mm). Two hundred fifty microliters of an overnight culture standardized to an OD600 of 0.05 was inoculated in each channel, and the cells were allowed to attach to the substratum for 1 h before the flow was turned on at a rate of approximately 3 ml/h. Biofilm formation was followed by microscopic observations with an LSM510 scanning confocal laser microscope (Carl Zeiss). Images were obtained using a 40×/1.3 Plan-Neofluar oil objective.
Construction of knockout mutants of 83972 and VR50.
Mutants of 83972 and VR50 were constructed using the λRed recombinase gene replacement system (13). Briefly, the npt gene from plasmid pKD4 was amplified using four different primer sets containing 40-nucleotide homology extensions of the four target genes (unpublished data), i.e., yceP, yqgA, ygiD, and aaeX. The PCR products were purified from agarose gels and transformed into 83972(pKD46) and VR50(pKD46), and kanamycin-resistant colonies were selected. The λRed helper plasmid pKD46 was cured by growth at 37°C, and the correct double-crossover and recombination event was confirmed by PCR.
Biofilm formation in microplates in human urine.
Cells were grown in pooled human urine for 5 to 6 h, and 10 μl was used for inoculation of 1.0 ml urine in 24-well flat-bottom microplates (Iwaki). The microplates were incubated statically at 37°C for 16 h. Unbound cells were removed by inversion of the microplate and tapping on absorbent paper; adhered cells were then stained with 0.1% crystal violet for 15 min. Excess stain was removed by washing three times with PBS. Crystal violet was then solubilized by the addition of ethanol-acetone (80:20, vol/vol), and the A590 was measured (Ultrospec III; Pharmacia). Each strain (wild-type and deletion mutant strains) was assayed in four wells on each plate, and the whole experiment was repeated three times in different batches of urine. In each plate, four wells were used as blanks containing sterile urine and four wells were inoculated with the wild-type strain (83972 or VR50) for reference. For the longer incubation time of 42 h used in the experiment comparing wild-type ABU and uropathogenic E. coli (UPEC) strains, the medium was changed twice during incubation, mimicking the growth conditions used for biofilm microarray analysis.
Microarray data accession number.
The supporting microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-926.
RESULTS
Biofilm formation of ABU strains 83972 and VR50.
We have recently found that many ABU strains form significantly more biofilm when grown in pooled human urine on abiotic surfaces than UPEC strains (21). Also, ABU strains E. coli 83972 and VR50 form about 10- to 20-fold better biofilm in polystyrene microtiter plates after 42 h than UPEC strains CFT073 and 536 (paired two-tailed t test, P < 0.001) (Fig. 1). To study the gene expression in ABU strains during growth in biofilm, we used DNA microarray technology. The two ABU strains were grown in polystyrene petri dishes in pooled human urine for 42 h to allow biofilm formation. The bacterial biofilm was washed twice to remove planktonic cells before immediate stabilization of RNA using RNAprotect Bacteria Reagent (QIAGEN AG).
FIG. 1.
Biofilm formation in polystyrene microplates in human urine of two UPEC isolates, 536 and CFT073, and two ABU strains, VR50 and 83972. The cells were incubated for 42 h at 37°C, and the urine was changed twice during incubation; the plate was scanned after staining with crystal violet and washing three times with PBS. The two ABU strains formed significantly more biofilm than the two UPEC strains (paired two-tailed t test, P < 0.001).
Global gene expression of two ABU E. coli strains in biofilm.
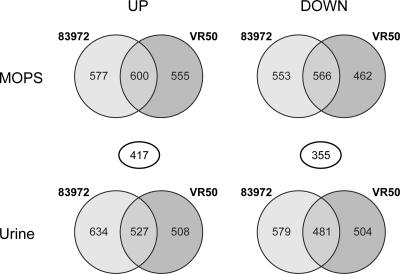
The genomic expression profiles of E. coli 83972 and VR50 obtained during biofilm formation in urine were compared with those during exponential growth in MOPS-glucose medium and pooled human urine (all in triplicates). The GeneChip E. coli Genome 2.0 Array was employed, which contains probe sets for all genes present in E. coli strains MG1655, CFT073, EDL933, and O157:H7-Sakai. In total, of 8,716 E. coli transcripts, 2,296 and 2,183 were significantly changed during biofilm growth for 83972 and VR50, respectively, compared with MOPS. The corresponding numbers of genes compared with planktonic cells in urine were 2,221 and 2,020, respectively. Of these significantly changed genes, about half (51 to 53%) were up-regulated. Approximately half of the differentially expressed genes during biofilm growth compared with the gene profile during planktonic growth were shared between the two ABU strains (Fig. 2). We have chosen mainly to focus on these genes.
FIG. 2.
Venn diagrams showing numbers of significantly up- and down-regulated genes in E. coli 83972 and VR50 during biofilm growth in urine compared with that during planktonic growth in MOPS-glucose and urine. The numbers displayed in circles between the Venn diagrams show the number of genes shared between the two overlapping areas; 417 and 355 genes were up- and down-regulated, respectively, during biofilm growth in both strains compared to that during planktonic growth in both MOPS and urine.
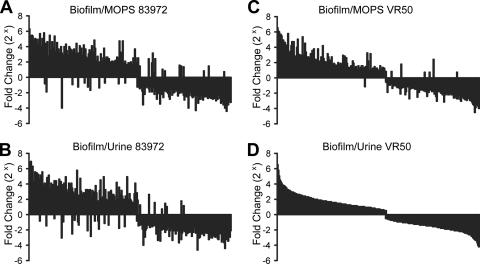
In total, 815 genes were found to be significantly changed during biofilm formation, in both E. coli 83972 and VR50, compared with the gene profile of planktonic growth in MOPS and urine (Fig. 3). Expression of the majority of the genes (94.7%) was affected in the same way, i.e., 417 and 355 genes were up- and down-regulated, respectively, during biofilm growth in both 83972 and VR50. Taken together, a large number of genes was found significantly changed during biofilm growth compared with the gene profile of planktonic growth, revealing a large similarity of the expression profiles in the two strains and indicating very different gene expression profiles during the two growth conditions.
FIG. 3.
Comparison of significantly changed genes during biofilm growth of E. coli 83972 and VR50 compared with the gene profile during planktonic growth. The 815 genes, which were significantly changed during biofilm growth in urine in both strains compared to that during planktonic growth in both MOPS and urine, have been ordered from the most up-regulated to the most down-regulated gene in VR50 compared with that in planktonic urine (D). The diagrams display the fold change of the genes in 83972 and VR50 during biofilm growth in urine compared with that during planktonic growth in MOPS (A and C, respectively) and urine (B and D, respectively).
Functional analysis of MG1655 transcripts of ABU E. coli in biofilm.
Classification of the significantly changed MG1655 genes into functional groups (52, 53) reveals which groups, categories of genes, are most affected during biofilm formation in urine compared to the gene profile during planktonic growth in MOPS and urine. Of a total of 417 and 355 up- and down-regulated genes in biofilm, 327 and 310, respectively, were MG1655 transcripts and are classified in Table 1. The functional analysis revealed that the group containing genes involved in transcription was by far the most up-regulated during growth in biofilm. The most down-regulated groups were those including genes involved in “translation” and “cell cycle control.” Other E. coli biofilm array studies have shown no change in expression of ribosomal genes during biofilm growth compared with exponentially grown cells (5, 39, 44) and up-regulation compared with stationary-phase cells (44). Of 54 ribosomal genes, 63% were significantly down-regulated, up to 8.9-fold, in biofilm in both ABU strains compared to that of exponentially grown cells. This indicates that our biofilm cells have a slower growth rate than exponential-phase cells, reflecting a growth rate somewhere between that of stationary and exponential phases. Interestingly, the least affected group was “cell motility,” where only 1 of 91 genes was found to be changed during biofilm growth compared to that during planktonic growth.
TABLE 1.
Numbers of significantly up- and down-regulated genes in each functional group of E. coli 83972 and VR50 during biofilm growth in urine compared to that of planktonic growth in MOPS-glucose and urine
| Description | Up-regulated genes in biofilm
|
Down-regulated genes in biofilm
|
No. of genes | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Amino acid transport and metabolism | 5 | 2 | 36 | 12 | 296 |
| Carbohydrate transport and metabolism | 13 | 4 | 23 | 8 | 298 |
| Cell cycle control, mitosis, and meiosis | 3 | 10 | 6 | 19 | 31 |
| Cell motility | 1 | 1 | 0 | 0 | 91 |
| Cell wall/membrane biogenesis | 4 | 2 | 25 | 13 | 200 |
| Coenzyme transport and metabolism | 9 | 8 | 14 | 13 | 108 |
| Defense mechanisms | 1 | 3 | 0 | 0 | 35 |
| Energy production and conversion | 12 | 5 | 34 | 14 | 240 |
| Function unknown | 31 | 13 | 15 | 6 | 247 |
| General function prediction only | 26 | 10 | 16 | 6 | 269 |
| Inorganic ion transport and metabolism | 14 | 9 | 7 | 4 | 160 |
| Intracellular trafficking and secretion | 2 | 6 | 6 | 17 | 36 |
| Lipid transport and metabolism | 3 | 4 | 8 | 11 | 71 |
| Nucleotide transport and metabolism | 6 | 8 | 8 | 10 | 78 |
| Posttranslational modification, protein turnover, chaperones | 13 | 11 | 11 | 9 | 119 |
| Replication, recombination, and repair | 17 | 10 | 14 | 8 | 165 |
| Secondary metabolites biosynthesis, transport, and catabolism | 2 | 4 | 7 | 13 | 54 |
| Signal transduction mechanisms | 12 | 10 | 2 | 2 | 116 |
| Transcription | 44 | 19 | 8 | 3 | 235 |
| Translation | 12 | 8 | 45 | 29 | 156 |
| Not in COGsa | 97 | 9 | 25 | 2 | 1,065 |
| MG1655 (Total) | 327 | 8 | 310 | 8 | 4,070 |
| CFT073 | 67 | 3 | 37 | 1 | 2,486 |
| EDL933 | 15 | 1 | 7 | 0 | 1,787 |
| SAKAI | 8 | 2 | 1 | 0 | 373 |
| Total | 417 | 5 | 355 | 4 | 8,716 |
COGs, clusters of orthologous groups of proteins.
Biofilm-related genes.
Of particular interest are the genes that have previously been suggested to be involved in E. coli biofilm formation, albeit with different strains and growth conditions (5, 39, 44). Among these are genes encoding cell surface structures, e.g., Ag43, type 1 fimbriae, extracellular polysaccharides, curli, and flagella, as well as genes found differently expressed in other biofilm microarray studies, e.g., genes involved in quorum sensing (luxS and tqsA) and regulation (rfaH and rpoS). The flu gene, encoding Ag43, has been shown to promote biofilm formation and has been reported to be up-regulated in E. coli K-12 biofilms (44). In the present study, the flu gene showed low expression and no change between the different growth conditions in either 83972 or VR50. Neither the antigen 43 precursor genes c1273 and c3655 nor any of the other known and putative autotransporter genes (b2647, b1169, b1170, ycgV, and aidA) showed any up-regulation during biofilm growth of the two strains. The sat gene, encoding the secreted autotransporter toxin (Sat) of uropathogenic E. coli, was down-regulated 4.3-fold in VR50 and showed no change in 83972.
Down-regulation of fimbriae.
The fim genes, encoding type 1 fimbriae, have been reported to be up-regulated in several E. coli biofilm studies (5, 39, 44). Strain 83972 is not able to express type 1 fimbriae, due to a large deletion affecting the fimEAIC region (26), while VR50 is capable of expressing type 1 fimbriae (41). All the remaining fim genes present in 83972, fimBDFGH, were down-regulated (2.2- to 9.3-fold) during biofilm growth compared to that with MOPS. In VR50 the regulatory fimB gene, encoding a site-specific recombinase capable of conferring off-to-on inversion of the phase switch, was down-regulated 3.3-fold compared to growth in MOPS, while the other fim genes showed low expression levels and no change between the different growth conditions.
P and F1C fimbriae are two other fimbriae associated with UPEC strains and UTI (27, 34). All pap genes, encoding P fimbriae, showed low expression levels and no change in both strains. The foc genes, encoding F1C fimbriae, showed low expression and no change in VR50, while focACG genes were down-regulated (1.8- to 6.9-fold) during biofilm growth in 83972 compared with that during planktonic growth in MOPS and urine. The aufA gene of the recently characterized Auf fimbriae associated with UPEC (7) was up-regulated 2.7- and 11.3-fold in 83972 during biofilm growth compared with that of planktonic growth in MOPS and urine, respectively. The yadC gene, encoding a putative fimbria-like protein, was up-regulated in 83972 6.6- and 10-fold in biofilm compared to that with MOPS and urine, respectively. Other putative fimbrial genes (sfmACDHF, yraH, yadKLMN, ygiL, eaeH, and c4424) showed no change between the different growth conditions.
Taken together, no fimbria-encoding genes were seen up-regulated in VR50 biofilm, while 83972 biofilm showed up-regulation of two fimbriae-associated genes, aufA and yadC, encoding a major fimbrial subunit and a putative fimbria-like protein, respectively.
Down-regulation of extracellular matrix components.
Production and secretion of polysaccharide polymers, such as colanic acid and cellulose, into the extracellular biofilm matrix is often recognized as important in the development of bacterial biofilms (6). It has been shown that colanic acid production is required for development of E. coli biofilm architecture (11). In our strains the genes involved in biosynthesis of colanic acid were all down-regulated (galEKTU) or showed no change (cpsBG, fcl, wca, wza, rcsABCF, gmd, and ugd) in biofilm. The genes involved in lipid A and 2-keto-3-deoxyoctulosonic acid biosynthesis were down-regulated (lpxABD and kdsABD) or showed no change. The murACDEFG, mraY, and ddlAB genes involved in peptidoglycan biosynthesis were all down-regulated 2.1- to 6.2-fold. The genes involved in biosynthesis of enterobacterial common antigen, rffACDEHGMT, were down-regulated 2.3- to 4.6-fold in biofilm growth of both strains. The kpsEDCSMT genes, involved in transport of polysaccharide to the cell surface, were all down-regulated 2.7- to 7.0-fold during biofilm growth.
Cellulose synthesis has recently been shown to be involved in biofilm formation in the commensal E. coli strain 1094 (36). The bcsABZC and bcsEFG genes, involved in cellulose synthesis, were all down-regulated in 83972 biofilm, while the bcsEFG genes were down-regulated in VR50 biofilm.
Curli expression has also been shown to affect biofilm formation in E. coli (57). None of the curli genes, csgABCDEFG or crl, were differentially expressed in the two ABU biofilms, and all csg genes showed very low expression levels.
Motility has been shown to influence biofilm architecture in E. coli (58), and other biofilm array studies of E. coli have reported no change in expression of genes encoding motility (5, 39, 44). In this study, some genes encoding flagellar proteins were down-regulated in VR50, while all flagellar biosynthesis genes showed no change and low expression levels in 83972. In agreement with this, the two ABU strains proved nonmotile on low-agar urine plates.
Taken together, no genes associated with production of the extracellular matrix components or flagella were up-regulated in the two biofilm-forming ABU strains, suggesting that the biofilms formed by these strains might be smooth and flat, lacking a highly structured morphology.
Down-regulation of the AI-2 quorum-sensing system.
LuxS is involved in biosynthesis of autoinducer 2 (AI-2), a hormone-like signal that mediates cell-cell communication during quorum sensing, the response to increased cell density (50). Furthermore, LuxS is involved in regulation of pathogenicity genes in enterohemorrhagic and enteropathogenic E. coli strains (47). The luxS gene was down-regulated 7.9- and 3.7-fold in 83972 and VR50, respectively. Consistent with this, the yhbH gene, a sigma 54-modulating protein that is induced by AI-2 (14), was also down-regulated in both strains, 4.7- and 2.0-fold. Furthermore, deletion of luxS has been shown to affect methionine metabolism and regulation in K-12 by decreased expression of metE (55); metE was repressed (11-fold) in 83972 and VR50 together with metBCHKL, which were down-regulated 2.3- to 4.1-fold, while the major repressor of the met genes, metJ, was up-regulated 2.0-fold. YdgG (TqsA) has recently been shown to control biofilm formation in E. coli through transport of AI-2: deletion of tqsA resulted in a major increase in biofilm thickness and cell motility of E. coli K-12 (23). The tqsA gene was up-regulated 4.0-fold in 83972 (the 2.2-fold up-regulation in VR50 was not significant). YceP and YliH have recently been shown to regulate biofilm formation through quorum sensing in E. coli K-12; deletion of either yceP or yliH increased biofilm formation and extracellular AI-2 concentrations 50-fold (18). The yceP gene was up-regulated 6.3- and 18.6-fold, and the yliH gene 5.8- and 94-fold, in 83972 and VR50 biofilms, respectively. AaeR is thought to be a virulence regulator that is activated by quorum sensing and to be involved in activation of expression of locus of enterocyte effacement pathogenicity genes in enterohemorrhagic and enteropathogenic E. coli (46). It has been shown that AaeR is a positive transcription factor for the divergently transcribed aaeXAB operon, and more recently it has been shown that the aaeAB genes encode an aromatic carboxylic acid efflux pump whose physiological role may be as a metabolic relief valve to alleviate toxic effects of imbalanced metabolism (54). The aaeR gene showed no change in our biofilm cells, while the aaeXAB operon belonged to one of the most up-regulated in 83972 and VR50 biofilms (5.5- to 21.2-fold compared with planktonic growth).
Taken together, down-regulation of genes involved in synthesis of AI-2 and up-regulation of tqsA, yceP, and yliH, which all negatively affect biofilm formation through AI-2, indicate that the AI-2 system is repressed in our biofilm-grown cells.
Regulation of genes involved in transcription.
The most up-regulated group was the group including genes involved in transcription, and many genes encoding transcriptional regulators were significantly up-regulated in biofilm-grown cells (Table 2) and among the most highly expressed (Table 3). The two regulatory genes rfaH and rpoS are believed to play a role in biofilm formation. The transcriptional activator RfaH participates in controlling several genes involved in the biosynthesis, assembly, and export of the lipopolysaccharide core. Inactivation of rfaH was reported to result in increased initial adhesion and biofilm formation by UPEC strain 536 (4). It is therefore interesting that this gene was significantly up-regulated in 83972 and VR50 biofilms. Although many genes involved in transcription were up-regulated, eight were down-regulated (Table 1), and five of these encoded RNA polymerase and sigma factors. Thus, the rpoS and rpoN genes, encoding the sigma S and 54 factors involved in general stress response and quorum sensing, respectively, were down-regulated (44).
TABLE 2.
Top 30 most up-regulated genes during biofilm growth in urine of E. coli 83972 and VR50 compared to that during planktonic growth in MOPS and urine
| Gene | Code | Function and/or product | FC in BFa | Signal intensity in BFb |
|---|---|---|---|---|
| cspG | b0990 | Homolog of Salmonella cold shock protein | 80.7 | 4,659 |
| cspH | b0989 | Cold shock-like protein | 56.5 | 2,532 |
| ibpB | b3686 | Heat shock protein | 33.6 | 5,787 |
| marR | b1530 | Multiple antibiotic resistance protein; repressor of mar operon | 31.5 | 4,859 |
| yqiJ | b3050 | Putative oxidoreductase | 30.6 | 1,522 |
| oxyS | b4458 | Global regulatory RNA OxyS | 30.5 | 3,916 |
| c2422 | c2422 | Putative inner membrane ABC transporter | 28.1 | 1,096 |
| ddg | b2378 | Palmitoleoyl-acyl carrier protein-dependent acyltransferase, cold-induced gene | 26.2 | 1,859 |
| ygiD | b3039 | Putative enzyme with dioxygenase domain | 23.4 | 3,863 |
| c3685 | c3685 | Hypothetical protein | 22.5 | 791 |
| aaeX | b3242 | Hypothetical protein | 21.2 | 3,905 |
| grxA | b0849 | Glutaredoxin 1 redox coenzyme for glutathione-dependent ribonucleotide reductase | 21.1 | 4,675 |
| c2436 | c2436 | Putative pesticin receptor precursor, homologous to fyuA | 19.8 | 1,196 |
| c2424 | c2424 | Putative peptide synthetase | 19.6 | 1,408 |
| yfiP | b2583 | Hypothetical protein | 19.0 | 1,863 |
| yqjF | c3859 | Hypothetical protein | 18.9 | 3,664 |
| mdtL | b3710 | Drug/chloramphenicol transport protein (MFS family) | 18.3 | 1,862 |
| yhcN | b3238 | Hypothetical protein | 17.3 | 5,790 |
| yqgA | b2966 | Putative transport membrane protein | 17.2 | 1,010 |
| rydB | b4430 | Small toxic membrane polypeptide | 17.1 | 1,167 |
| sgaB | b4194 | Putative PTS family enzyme II component, possibly ascorbate-specific | 16.5 | 2,167 |
| pflE | b0824 | Putative pyruvate formate lyase activating enzyme | 16.4 | 891 |
| marB | b1532 | Multiple antibiotic resistance protein | 16.3 | 2,173 |
| sgaE | b4198 | l-ribulose 5-phosphate 4-epimerase | 16.2 | 1,405 |
| ybaO | Z0555 | Putative transcriptional regulator | 15.9 | 2,505 |
| ytfH | b4212 | Putative transcriptional regulator | 15.9 | 2,383 |
| cspA | b3556 | Cold shock protein 7.4, transcriptional activator of hns | 15.9 | 6,387 |
| yjfY | b4199 | Hypothetical protein | 15.2 | 960 |
| c3865 | c3865 | Hypothetical protein | 14.7 | 676 |
| fxsA | b4140 | Suppress F exclusion of bacteriophage T7 | 13.4 | 2,475 |
Fold change (FC) in biofilm (BF) was calculated from all 6 biofilm arrays using all 12 planktonic arrays as the baseline.
Signal intensity was calculated from all six biofilm arrays treated as one single sample group.
TABLE 3.
Top 50 most expressed genes during biofilm growth in urine of E. coli 83972 and VR50 compared to that during planktonic growth in MOPS and urine
| Gene | Code | Function and/or product | Signal intensitya | FC in BFb |
|---|---|---|---|---|
| rrfH | b0205 | 5S rRNA | 11,223 | 1.2 |
| rrlG | Z3875 | 23S rRNA | 9,568 | −1.0 |
| rrsG | b2591 | 16S rRNA | 8,062 | 1.1 |
| ssrA | b2621 | 10Sa RNA (tmRNA); tags incomplete translation products for degradation | 7,924 | 1.1 |
| ssrS | b2911 | 6S RNA | 7,601 | 4.6 |
| rnpB | b3123 | RNase P, RNA component; M1 RNA | 7,559 | 1.2 |
| csrB | b4408 | CsrB regulatory RNA | 6,638 | 1.2 |
| ahpC | b0605 | Alkyl hydroperoxide reductase, detoxification of hydroperoxides | 6,622 | 1.3 |
| rhoL | b3782 | rho operon leader peptide | 6,476 | 1.8 |
| cspA | b3556 | Cold shock protein 7.4, transcriptional activator of hns | 6,387 | 15.9 |
| ibpA | b3687 | Heat shock protein | 6,120 | 8.7 |
| t44 | b4414 | Small toxic membrane polypeptide | 6,025 | 1.0 |
| folE | b2153 | GTP cyclohydrolase I | 5,942 | 1.2 |
| ybiJ | b0802 | Hypothetical protein | 5,817 | 8.2 |
| ycfR | b1112 | Hypothetical protein | 5,804 | 9.3 |
| yhcN | b3238 | Hypothetical protein | 5,790 | 17.3 |
| ibpB | b3686 | Heat shock protein | 5,787 | 33.6 |
| bfd | b3337 | Regulatory or redox component complexing with Bfr, in iron storage and mobility | 5,582 | 5.2 |
| rplU | b3186 | 50S ribosomal subunit protein L21 | 5,548 | 1.1 |
| rplN | b3310 | 50S ribosomal subunit protein L14 | 5,323 | −1.0 |
| dps | b0812 | Global regulator, starvation conditions | 5,274 | 2.6 |
| trxC | b2582 | Putative thioredoxin-like protein | 5,235 | 8.1 |
| marA | b1531 | Multiple antibiotic resistance; transcriptional activator of defense systems | 5,164 | 8.2 |
| fur | b0683 | Negative regulator | 5,145 | 2.7 |
| asnA | b3744 | Asparagine synthetase A | 5,129 | 5.5 |
| yhaK | b3106 | Predicted pirin-related protein | 5,103 | 12.1 |
| dnaK | b0014 | Chaperone Hsp70 in DNA biosynthesis/cell division | 5,101 | 1.7 |
| csrC | b4457 | CsrC regulatory RNA | 5,014 | 1.3 |
| fnr | b1334 | Transcriptional regulation of aerobic, anaerobic respiration, osmotic balance | 4,981 | 2.1 |
| rplM | b3231 | 50S ribosomal subunit protein L13 | 4,955 | −1.1 |
| marR | b1530 | Multiple antibiotic resistance protein; repressor of mar operon | 4,859 | 31.5 |
| rho | b3783 | Transcription termination factor Rho; polarity suppressor | 4,768 | 1.9 |
| ffs | b0455 | 4.5S RNA; component of ribonucleoprotein particle | 4,752 | 5.9 |
| rpoD | b3067 | RNA polymerase, sigma 70 factor | 4,751 | 2.1 |
| glgS | b3049 | Glycogen biosynthesis, rpoS dependent | 4,747 | 5.3 |
| groE | b4142 | GroES, binds to Hsp60 in presence of Mg-ATP, suppressing its ATPase activity | 4,731 | 1.2 |
| nlpI | b3163 | NlpI lipoprotein involved in cell division | 4,698 | 3.7 |
| infC | Z2747 | Initiation factor IF-3 | 4,677 | −1.1 |
| grxA | b0849 | Glutaredoxin 1 redox coenzyme for glutathione-dependent ribonucleotide reductase | 4,675 | 21.1 |
| cspG | b0990 | Homolog of Salmonella cold shock protein | 4,659 | 80.7 |
| rpmH | b3703 | 50S ribosomal subunit protein L34 | 4,648 | 1.8 |
| bfr | b3336 | Bacterioferrin, an iron storage homoprotein | 4,644 | 4.5 |
| yhhW | c4228 | Protein YhhW, putative pirin protein | 4,644 | 12.3 |
| pdhR | b0113 | Transcriptional regulator for pyruvate dehydrogenase complex | 4,590 | 5.5 |
| rplK | b3983 | 50S ribosomal subunit protein L11 | 4,576 | −1.2 |
| iscR | b2531 | Fe-S cluster-containing transcription factor | 4,564 | 2.3 |
| rseA | b2572 | Sigma E (sigma 24) factor, negative regulator | 4,530 | 1.9 |
| yceD | b1088 | Hypothetical protein | 4,522 | −1.0 |
| ryhB | b4451 | Regulatory RNA mediating Fur regulon | 4,509 | 2.4 |
| Z5401 | Z5401 | Hypothetical protein | 4,485 | 1.3 |
Signal intensity was calculated from all six biofilm arrays treated as one single sample group.
Fold change (FC) in biofilm (BF) was calculated from all 6 biofilm arrays using all 12 planktonic arrays as the baseline. Boldface indicates significant change.
Up-regulation of genes involved in stress response.
The most up-regulated genes in biofilm included genes encoding both cold shock and heat shock proteins (Table 2). The three cold shock genes cspG, cspH, and cspA were up-regulated 81-, 57-, and 16-fold, respectively, and the heat shock genes ibpB and ibpA were up-regulated 34- and 8.7-fold, respectively. The pphA gene, involved in induction of the heat shock response, was up-regulated 6.7-fold during biofilm growth. The gene encoding the transcriptional activator SoxS, which participates in controlling several genes involved in the response to oxidative stress and the transcription of which has been seen induced upon biofilm formation (39), was up-regulated 3.4-fold in VR50 biofilm while it was repressed in 83972. The transcriptions of fldA, fur, fpr, fumC, inaA, and marRAB are all activated by SoxS and were all up-regulated during biofilm growth by 2.0- to 31-fold in both 83972 and VR50. Transcription of SoxS is activated by SoxR, which also was up-regulated 6.8-fold in biofilm growth in both strains. Transcription of hha, encoding a hemolysin expression-modulating protein involved in stress response, is induced upon biofilm formation (39), and the Hha protein of virulent E. coli strains affects production of some virulence factors (17). hha was up-regulated 3.3-fold during biofilm growth in the two ABU strains. Furthermore, the stress-induced YfiD protein involved in stress resistance was also up-regulated 7.0-fold during biofilm growth in both strains. Taken together, many genes involved in stress response were highly up-regulated in biofilm of both strains, which is in agreement with previous E. coli biofilm array studies (5, 39, 44).
Up-regulation of virulence-associated genes.
E. coli 83972 carries the virulence-associated fyuA gene, encoding Yersiniabactin receptor protein (15). The c2436 gene, encoding a putative pesticin receptor protein homologous to fyuA, was up-regulated in the urinary tract in vivo (40) and was one of the most up-regulated genes in biofilm of the two ABU strains (Table 2). Interestingly, all the genes flanking the fyuA homologue, c2418 to c2440 (altogether 22 genes), were significantly up-regulated in VR50 biofilm (2.2- to 63.4-fold), and nine of them were up-regulated in 83972 (2.8- to 13.1-fold). This cluster was by far the largest contiguous up-regulated gene cluster in VR50 biofilm; several of the genes in this cluster were among the most up-regulated in both VR50 and 83972 biofilms (Table 2). The cluster is part of the pathogenicity island PAI IV536 of UPEC strain 536, which is a high-pathogenicity island encoding a siderophore system initially identified in pathogenic Yersinia spp. (16). This result indicates that both ABU strains contain PAI IV536 and that genes in this pathogenicity island may play a role during biofilm growth.
YliI has recently been identified as an outer membrane protein (32) and was expressed during human infection by E. coli O157:H7 (25). The yliI gene was up-regulated 8.0- and 5.9-fold during biofilm growth in 83972 and VR50, respectively. Another virulence-associated gene, ygdP, was up-regulated in biofilm growth. This gene is associated with the invasion of brain microvascular endothelial cells by E. coli K1 (2) and was significantly induced in both 83972 and VR50.
Comparison with expression in vivo.
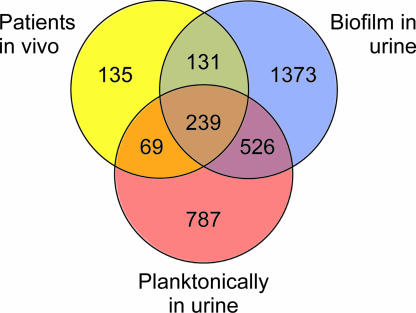
Recently, we investigated the global gene expression profile of 83972 in vivo in three patients and compared it with those in MOPS and in vitro urine (40). Normalization of the 9 patient arrays together with the 18 arrays presented in this study revealed that a majority of the genes that were significantly changed in the patients were also regulated in the same way during biofilm growth. Of a total of 574 significantly changed genes in the patients compared with expression in MOPS, 64% (370 genes) were regulated in the same way (induced or repressed) during biofilm growth of 83972, and about half of these (177 genes) showed the same regulation during biofilm growth of VR50 as well. Of the 43 genes that were up-regulated in all three patients compared with growth in both MOPS and in vitro urine (Table 1 in reference 40), 23 were up-regulated in biofilm growth of 83972. Comparison of the regulation of 83972 grown in the patients with that of grown in vitro planktonically in urine and in biofilms revealed that the gene expression in biofilm growth more closely resembled expression of 83972 genes in patients than that in planktonically grown cells (Fig. 4); of the total of 574 significantly changed genes in the patients, 131 genes showed the same regulation as that during biofilm growth but no change compared with that of planktonically grown cells in urine, while only about half as many (69 genes) showed the same regulation as planktonic cells in urine but no change during biofilm growth. Interestingly, this could indicate that strain 83972 lives a partial biofilm-like existence in the human urinary tract.
FIG. 4.
Venn diagram showing numbers of significantly changed genes in E. coli 83972 during in vivo growth in three patients, biofilm growth, and planktonic growth in urine compared with that in MOPS. The diagram reveals that gene regulation of 83972 grown in patients more closely resembles that in biofilm than that in planktonic growth in urine.
Verification of microarray results.
RT-PCR was performed to verify the transcript levels of selected genes. aaeX, yqgA, ygiD, and c2422, which all were highly up-regulated in biofilm of 83972 and VR50 (Table 2) and have not been reported to be up-regulated in other E. coli biofilm studies, were selected for RT-PCR. After 20 to 40 cycles of PCR, products of the four genes were visualized on agarose gels. The strength of the gel bands from each sample corresponded well to the actual signal intensities recorded from the corresponding microarray (unpublished data). 16S was used as a normalizing internal standard and was detected with the same intensity in all samples.
The expression levels of the fim genes reveal the sensitivity of the microarrays. The fim gene cluster of E. coli 83972 has a 4.25-kb deletion between fimB and fimD, resulting in the complete absence of fimEAIC, leaving only fimF, fimG, and fimH unaffected (26). The signal from fimEAIC was very low (absent call) and varied between 15 and 63 on the arrays hybridized with 83972, while the signals of fimBDFGH varied between 95 and 935. The signal levels (i.e., absent/present call) of the different fim genes on the arrays corresponded well to the actual presence of fim genes in E. coli 83972.
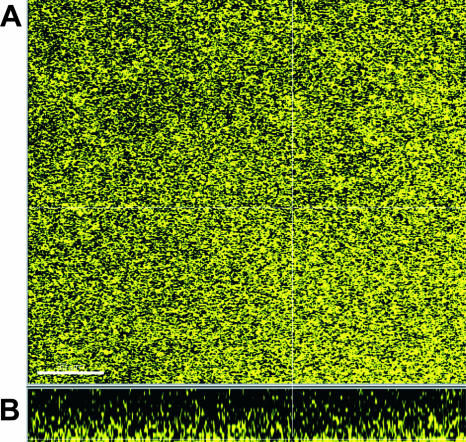
The down-regulation and low expression levels of motility genes were supported by results from motility testing on low-agar urine plates showing that 83972 and VR50 were nonmotile. The down-regulation of genes encoding extracellular components, indicating a lack of highly structured biofilm morphology, was supported by examination of the biofilm structure using scanning confocal laser microscopy. Fluorescently tagged versions of 83972 and VR50 grown in urine for 40 h revealed biofilms with a smooth and flat structure; Fig. 5 shows the biofilm formation of VR50yfp after 40 h in the flow-chamber system.
FIG. 5.
Top view (A) and side view (B) of biofilm formation by ABU strain VR50 grown in human urine in a flow chamber. VR50 tagged with yellow fluorescent protein was grown in flow chambers in human urine at 37°C for 40 h, and biofilm formation was monitored by a scanning confocal laser microscope. The strain showed a flat biofilm structure lacking a highly structured matrix. The scale bar represents 30 μm.
Functional analysis of selected up-regulated genes.
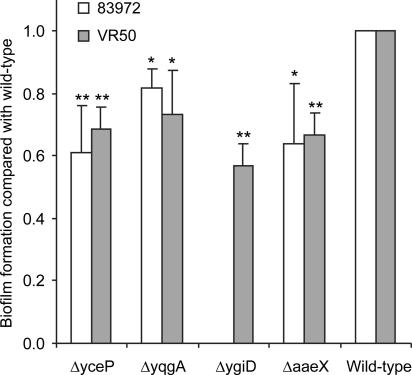
To further verify the microarray results, four genes, which were highly up-regulated during biofilm formation in both 83972 and VR50, were selected for knockout mutagenesis. Three and four positive knockout mutants of 83972 and VR50, respectively, were obtained and subsequently analyzed for biofilm formation in microtiter plates in pooled human urine (Fig. 6). All knockout mutants showed significantly reduced biofilm formation in human urine compared to the wild type (paired two-tailed t test, P < 0.05). The three mutants of 83972, i.e., 83972ΔyceP, 83972ΔyqgA, and 83972ΔaaeX, reduced biofilm formation by 18 to 39%, and the four mutants of VR50, i.e., VR50ΔyceP, VR50ΔyqgA, VR50ΔygiD, and VR50ΔaaeX, reduced biofilm formation by 26 to 43%.
FIG. 6.
Biofilm formation in polystyrene microplates in human urine of knockout mutants of the two ABU strains, 83972 and VR50. The cells were incubated for 16 h at 37°C, and A590 was measured after staining with crystal violet and washing three times with PBS. Values are shown relative to those of the wild type and are means of at least three separate experiments. Error bars indicate standard deviation (δn − 1). All mutants formed significantly less biofilm than the corresponding wild-type strain (paired two-tailed t test; *, P < 0.05; **, P < 0.01).
DISCUSSION
More than 50% of all microbial infections have been associated with the formation of biofilms (10). Notably, many persistent and chronic bacterial infections are now believed to be linked to the formation of biofilms. Moreover, virtually all medical implants are prone to bacterial colonization and biofilm formation (19). Urinary tract infections often tend to become chronic, with many recurrent episodes. Moreover, patients with indwelling urinary catheters invariably get urinary tract infections within a few weeks (20). Arguably, biofilm formation could well be associated with the infectivity of UT-colonizing E. coli strains. Indeed, previous studies have advocated the importance of bacterial biofilm formation in urinary tract infections, notably in chronic cystitis and infections associated with catheters (33, 37).
The bacterial transcriptome is a dynamic entity that reflects the organism's immediate, ongoing response to its environment. DNA microarray-assisted functional genomics provides the global expression profile of the genome. Microarray expression profiling therefore provides a comprehensive snapshot of the transcriptome. The expression profile discloses how the bacterium adapts to an environmental niche. Adaptation to a given host environment is an extremely important parameter and underlies the capacity of an infectious agent to persist in a host. In nature, biofilms usually comprise multiple species; however, UTI in humans by E. coli are almost always caused by single strains (clones). We believe that our experimental approach, with surface-associated biofilm formation in human urine, mimics in vivo scenarios considerably, notably that of catheterized patients. Both our E. coli ABU strains, 83972 and VR50, were good biofilm formers under these conditions and provided ample material for global gene expression profiling. The microarray data revealed that the gene expression profiles of the two ABU E. coli strains, 83972 and VR50, during biofilm formation differ significantly from those observed during planktonic growth. Thus, expression of 815 genes was significantly changed in both strains during biofilm growth in urine compared with the gene profile in planktonic growth, and the large majority of these genes were affected in the same way (Fig. 3). The down-regulated genes were mainly involved in translation, metabolism, and energy production, while transcription was the largest up-regulated group, suggesting that the two strains responded in much the same way to biofilm conditions.
Comparison of previous studies on gene expression profiles in bacterial biofilms has suggested that there may not be biofilm-specific genes. Instead, biofilms have a unique pattern of gene expression, where subsets of genes that are expressed in biofilms are also expressed under various planktonic conditions (28). A few other studies have addressed global gene expression in E. coli biofilms (5, 39, 44). However, these studies employed laboratory strains (E. coli K-12) and were performed in artificial growth media like minimal medium. Also, in all cases cells were harvested prior to stabilization of RNA. As expected, relatively few genes were affected in the same manner when comparing the individual studies. Nevertheless, in spite of the very different strains and experimental conditions, some of the biofilm-associated genes from previous studies were also affected in our ABU strains during biofilm formation in urine. For example, 8 out of 45 genes found to be up-regulated in E. coli K-12 strain MG1655 during biofilm formation in flow cells in minimal medium were also induced in both 83972 and VR50 during biofilm growth in urine, i.e., b2146, cydA, frmR, hyaA, hybO, hypA, ndh, and ybfA (44). Of the 24 genes up-regulated in E. coli K-12 strain TG1 during biofilm formation in minimal medium compared with both exponential and stationary growth phases (5), 7 and 13 were up-regulated in 83972 and VR50 biofilm, respectively, i.e., rpoE, rseA, sixA, tatE, yceP, yebE, and yqcC were induced in both strains, whereas cpxP, pspABCD, and yjbO were induced in VR50. Interestingly, the biofilm-related yceP and yliH genes (18) were highly up-regulated in our two ABU strains and have been reported to be up-regulated in all three preceding E. coli biofilm array studies (5, 39, 44). The exact roles of these two genes have not been fully revealed, and mutants show different biofilm-forming capacity depending on growth medium; they appear to be global regulators of several genes involved in catabolite repression and stress response and regulation of the uptake and export of signaling pathways (18). In our study, ΔyceP mutants of 83972 and VR50 reduced biofilm formation in microplates in human urine by 39% and 31%, respectively.
It is interesting to note that some genes previously reported to be important for biofilm formation in E. coli were not affected in our ABU strains. Expression of fimbriae, notably type 1 fimbriae, has been observed to enhance biofilm formation in K-12 strains and is thought to promote attachment in the initial stages of biofilm formation (35). Our two ABU strains differ in their abilities to express fimbriae; strain VR50 can express functional type 1, whereas strain 83972 cannot (26, 41, 42). Yet they show similar growth characteristics and ability to form biofilm (Fig. 1), and they have similar gene regulation during biofilm growth (Fig. 3). Interestingly, in both strains neither intact nor residual fimbria-encoding gene clusters were activated during biofilm formation in urine. Human urine does not seem to promote expression of type 1 fimbriae; the fim gene cluster has not been reported up-regulated in human urine, neither in the UPEC isolate CFT073 (45) nor in 83972 grown in vitro or in vivo (40), and ABU strains expressing functional type 1 fimbriae in Luria-Bertani medium failed to do so in human urine (41). Our results suggest that type 1 fimbriae are not crucial for establishment of biofilm by UTI isolates in urine. Despite strain 83972's inability to express type 1 fimbriae, it forms significantly more biofilm than UPEC isolates CFT073 and 536 (Fig. 1), and even though VR50 is capable of expressing these surface elements, the fim genes are not expressed in urine biofilm. Several other genes encoding reportedly biofilm-associated products, such as Ag43 and extracellular matrix components, were not up-regulated either. Meanwhile, several virulence-associated genes, such as the fyuA gene, encoding Yersiniabactin receptor protein, and its adjacent genes belonging to the PAI IV536 as well as yliI and ygdP, associated with virulence of E. coli O157:H7 and K1, respectively, were found to be induced in our strains during biofilm growth. This suggests that many virulence-associated genes are expressed during biofilm growth.
In a recent study (40), we investigated the global gene expression profile of 83972 in vivo in three patients. Comparison of the regulation of 83972 grown in the patients with those grown in vitro planktonically in urine and in biofilms revealed that the gene regulation during biofilm growth more closely resembled the regulation in patients than that in planktonically grown cells did (Fig. 4). Notably, about half of the genes up-regulated in 83972 in all three patients (Table 1 in reference 40) were induced in biofilm growth of both 83972 and VR50; the exceptions (i.e., genes up-regulated in patients but not in biofilm) were genes involved in nitrate/nitrite metabolism, the regulation of which probably reflects the specific in vivo environment in the patients. Interestingly, the fact that nearly all genes up-regulated in vivo, with exceptions for the nitrate/nitrite-related genes, were up-regulated in biofilm as well could indicate that strain 83972 partially lives a biofilm-like life in the human urinary tract.
There seems to be multiple mechanisms by which bacteria can form biofilms, employing combinations of various extracellular matrix molecules, such as surface proteins and sugar polymers and even DNA, for the establishment and build-up of the biofilm, combined with biofilm-associated changes in metabolism (6, 28, 48). The identification of key factors in biofilm formation has proven a difficult task, and the existence of a universal biofilm gene expression pattern has been questioned (3). Genetic and environmental factors greatly influence the biofilm formation; it has been demonstrated that diverse E. coli isolates respond very differently to changing environmental conditions (38), and many reports studying the influence of a specific “biofilm gene” have seen different, sometimes even reverse, effects on biofilm formation depending on growth media and strains used (4, 18, 23, 36, 55). In the present study, we used two genetically different clinical ABU isolates and studied these in their natural growth medium. From the present study it is not possible to identify the exact mechanism underlying the biofilm-forming capacity of the two ABU strains. However, the gene expression data indicate that the biofilms formed by these two strains lack a highly structured matrix, which was supported by microscopic analysis of the strains in the flow-cell system. Also, the results suggest that there must be other important determinants in biofilm formation that need to be discovered in order to explain why these two ABU strains are good biofilm formers and are superior to the UPEC isolates CFT073 and 536. The gene expression profiling also reveals that the biofilm life represents a very different growth mode, with many genes similarly expressed in the two strains. Down-regulation of translational genes and up-regulation of genes encoding transport systems for drugs and metabolites, together with induction of many transcriptional and stress-related genes, indicate that the biofilm growth mode exerts great pressure on the cells, forcing them to slow down their growth rate and leading to a metabolic upset due to an intracellular stress condition.
Acknowledgments
We thank Birthe Jul Jørgensen for expert technical assistance and Lionel Ferrières for providing the laser confocal microscopy picture.
This work was supported by grants from the Danish Medical Research Council (22-03-0462) and the Danish Research Agency (2052-03-0013).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Andersson, P., I. Engberg, G. Lidin-Janson, K. Lincoln, R. Hull, S. Hull, and C. Svanborg. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed] [Google Scholar]
- 3.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16-19. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J. M. Ghigo, and U. Dobrindt. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 6.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revised. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Buckles, E. L., F. K. Bahrani-Mougeot, A. Molina, C. V. Lockatell, D. E. Johnson, C. B. Drachenberg, V. Burland, F. R. Blattner, and M. S. Donnenberg. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72:3890-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darouiche, R. O., W. H. Donovan, M. Del Terzo, J. I. Thornby, D. C. Rudy, and R. A. Hull. 2001. Pilot trial of bacterial interference for preventing urinary tract infection. Urology 58:339-344. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.deLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt, U., L. Emody, I. Gentschev, W. Goebel, and J. Hacker. 2002. Efficient expression of the alpha-haemolysin determinant in the uropathogenic Escherichia coli strain 536 requires the leuX-encoded tRNA(5)(Leu). Mol. Genet. Genomics 267:370-379. [DOI] [PubMed] [Google Scholar]
- 18.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donlan, R. M., and B. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, V., L. Ferrières, and P. Klemm. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 22.Hedlund, M., R. D. Duan, A. Nilsson, M. Svensson, D. Karpman, and C. Svanborg. 2001. Fimbriae, transmembrane signaling, and cell activation. J. Infect. Dis. 183(Suppl. 1):S47-S50. [DOI] [PubMed] [Google Scholar]
- 23.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hull, R., D. Rudy, W. Donovan, C. Svanborg, I. Wieser, C. Stewart, and R. Darouiche. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J. Urol. 163:872-877. [PubMed] [Google Scholar]
- 25.John, M., I. T. Kudva, R. W. Griffin, A. W. Dodson, B. McManus, B. Krastins, D. Sarracino, A. Progulske-Fox, J. D. Hillman, M. Handfield, P. I. Tarr, and S. B. Calderwood. 2005. Use of in vivo-induced antigen technology for identification of Escherichia coli O157:H7 proteins expressed during human infection. Infect. Immun. 73:2665-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klemm, P., V. Roos, G. C. Ulett, C. Svanborg, and M. A. Schembri. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 28.Lazazzera, B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8:222-227. [DOI] [PubMed] [Google Scholar]
- 29.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindberg, U., L. A. Hanson, U. Jodal, G. Lidin-Janson, K. Lincoln, and S. Olling. 1975. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr. Scand. 64:432-436. [DOI] [PubMed] [Google Scholar]
- 32.Marani, P., S. Wagner, L. Baars, P. Genevaux, J. W. de Gier, I. Nilsson, R. Casadio, and G. von Heijne. 2006. New Escherichia coli outer membrane proteins identified through prediction and experimental verification. Protein Sci. 15:884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris, N. S., D. J. Stickler, and R. J. C. McLean. 1999. The development of bacterial biofilms on indwelling urethral catheters. World J. Urol. 17:345-350. [DOI] [PubMed] [Google Scholar]
- 34.Oelschlaeger, T. A., U. Dobrindt, and J. Hacker. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33-38. [DOI] [PubMed] [Google Scholar]
- 35.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 36.Re, S. D., and J. M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation of Escherichia coli. J. Bacteriol. 188:3073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid, G., J. D. Denstedt, Y. S. Kang, D. Lam, and C. Nause. 1992. Microbial adhesion and biofilm formation on ureteral stents in vitro and in vivo. J. Urol. 148:1592-1594. [DOI] [PubMed] [Google Scholar]
- 38.Reisner, A., K. A. Krogfelt, B. M. Klein, E. L. Zechner, and S. Molin. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 40.Roos, V., and P. Klemm. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 74:3565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roos, V., E. M. Nielsen, and P. Klemm. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262:22-30. [DOI] [PubMed] [Google Scholar]
- 42.Roos, V., M. A. Schembri, G. C. Ulett, and P. Klemm. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799-1806. [DOI] [PubMed] [Google Scholar]
- 43.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 out-competes UPEC strains in human urine. Infect. Immun. 74:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 45.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 49.Stamm, W. E., and S. R. Norrby. 2001. Urinary tract infections: disease panorama and challenges. J. Infect. Dis. 183(Suppl. 1):S1-S4. [DOI] [PubMed] [Google Scholar]
- 50.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svanborg, C., and G. Godaly. 1997. Bacterial virulence in urinary tract infection. Infect. Dis. Clin. N. Am. 11:513-529. [DOI] [PubMed] [Google Scholar]
- 52.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 54.van Dyk, T. K., L. J. Templeton, K. A. Cantera, P. L. Sharpe, and F. S. Sariaslani. 2004. Characterization of the Escherichia coli AeaAB efflux pump: a metabolic relief valve? J. Bacteriol. 186:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L., J. Li, J. C. March, J. J. Valdes, and W. E. Bentley. 2005. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 187:8350-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warren, J. W. 2001. Catheter-associated urinary tract infections. Int. J. Antimicrob. Agents 17:299-303. [DOI] [PubMed] [Google Scholar]
- 57.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood, T. K., A. F. Gonzalez Barrios, M. Herzberg, and J. Lee. 2006. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 72:361-367. [DOI] [PubMed] [Google Scholar]