Abstract
Most members of the genus Brucella show strong urease activity. However, the role of this enzyme in the pathogenesis of Brucella infections is poorly understood. We isolated several Tn5 insertion mutants deficient in urease activity from Brucella abortus strain 2308. The mutations of most of these mutants mapped to a 5.7-kbp DNA region essential for urease activity. Sequencing of this region, designated ure1, revealed the presence of seven open reading frames corresponding to the urease structural proteins (UreA, UreB, and UreC) and the accessory proteins (UreD, UreE, UreF, and UreG). In addition to the urease genes, another gene (cobT) was identified, and inactivation of this gene affected urease activity in Brucella. Subsequent analysis of the previously described sequences of the genomes of Brucella spp. revealed the presence of a second urease cluster, ure2, in all them. The ure2 locus was apparently inactive in B. abortus 2308. Urease-deficient mutants were used to evaluate the role of urease in Brucella pathogenesis. The urease-producing strains were found to be resistant in vitro to strong acid conditions in the presence of urea, while urease-negative mutants were susceptible to acid treatment. Similarly, the urease-negative mutants were killed more efficiently than the urease-producing strains during transit through the stomach. These results suggested that urease protects brucellae during their passage through the stomach when the bacteria are acquired by the oral route, which is the major route of infection in human brucellosis.
Brucellosis is the most common zoonosis in humans. Transmission of this disease occurs mainly by inhalation of infected aerosols, by animal contact, and by conjunctival and gastrointestinal routes. The gastrointestinal route is the most common portal of entry of Brucella in humans through ingestion of raw milk or its products and raw liver or meat (8). Transmission of Brucella melitensis, B. abortus, B. suis, and B. canis in animals also occurs by ingestion of contaminated abortions, discharge materials, or contaminated pasture plants. In contrast, gastrointestinal transmission is not important under natural conditions for B. ovis, where the sexual route seems to be the most probable route of infection (21). Most isolates of B. ovis are urease negative (10).
Urease is a multisubunit, nickel-containing enzyme that catalyzes the hydrolysis of urea, yielding ammonia and carbon dioxide. The released ammonia is used by many bacteria as a source of nitrogen, and even for the generation of ATP from a strong ammonia gradient in the case of Ureaplasma urealyticum (38). Moreover, urease is a virulence factor for several human pathogens, and it plays a major role in both urinary and gastrointestinal tract infections, although through different mechanisms (9). In urinary tract infections caused by Proteus mirabilis, urease promotes direct toxicity to renal epithelium cells and kidney stone formation (16, 24). In gastrointestinal tract infections, urease allows Helicobacter pylori colonization of the acidic environment of the stomach (23) and also allows pathogens such as Klebsiella and Yersinia enterocolitica to survive passage through the stomach (22, 43). The acidic environment of the stomach caused by the hydrochloric acid present in gastric secretions is known to form a barrier to intestinal infection. Ingested bacteria that are able to tolerate the acidic pH typical of the stomach have a better chance to survive and colonize the host. The low oral infectious dose of Shigella, for example, has been related to its natural resistance to low pH (13).
In addition, urease has been identified as an immunogenic modulator in several pathogen-induced inflammatory reactions (3, 37), suggesting that it has a role in pathogenesis independent of its intrinsic enzymatic activity.
Bacterial ureases are usually encoded in a single operon containing the genes for the three urease structural subunits (ureABC) linked to accessory genes (ureDEFG) encoding proteins involved in the assembly of native urease. Genes coding for transcription regulators and urea transporters may also be part of bacterial urease operons (25). Incorporation of nickel into urease is a complex process involving several proteins. At the same time, the metal transporters and chelating proteins involved in nickel homeostasis may affect urease activity. As a consequence, mutations in many genes outside the urease operon may result in urease-deficient phenotypes (17, 18).
Bacteria belonging to the genus Brucella (except B. ovis) usually exhibit a characteristic strong urease activity (8). The properties of this enzyme, as well as its significance in Brucella metabolism or pathogenicity, remain basically unexplored. In this work, we obtained urease-deficient mutants of B. abortus, cloned the urease genes, constructed urease-negative mutants, and analyzed the role of urease in B. abortus infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. B. abortus strains were grown in brucella broth (BB) or on brucella agar (BA) plates (Pronadisa, Spain). Escherichia coli strains were grown in Luria-Bertani broth or on Luria-Bertani medium plates. When required, media were supplemented with the following antibiotics: kanamycin (50 μg/ml), ampicillin (100 μg/ml), and nalidixic acid (15 μg/ml). Intestinal homogenates were plated on plates containing BA made selective with Brucella Selectavial (BAF) (MAST Diagnostics, United Kingdom).
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pBluescript SK II(+) | Cloning vector | Stratagene |
| pCRIS | 35-kb EcoRI genomic fragment from 2308 ure1::Tn5 cloned in pBluescript SK II(+) | This study |
| pJQ200uc1 | Broad-host-range mobilizable suicide vector; Gmr | 31 |
| pJQ200SK | Broad-host-range mobilizable suicide vector; Gmr | 31 |
| pFJS110 | 2,556-bp fragment containing ureC1 cloned into pGEM-T Easy | This study |
| pFJS111 | pFJS110 with a 732-bp EcoRV deletion (ΔureC1) | This study |
| pFJS112 | 1,858-bp NotI fragment from pFJS111 containing ΔureC1 cloned into pJQ200uc1 | This study |
| pFJS113 | 2,131-bp fragment containing ureC2 cloned into pGEM-T Easy | |
| pFJS114 | pFJS113 with a 732-bp EcoRV deletion (ΔureC2) | |
| pFJS115 | 1,433-bp NotI fragment from pFJS111 containing ΔureC1 cloned into pJQ200SK | |
| pGEM-T Easy | PCR cloning vector | Promega |
| pBBR1MCS | Broad-host-range plasmid | 19 |
| E. coli strains | ||
| S17-1(λ-pir) | Mobilizing donor for conjugation | 36 |
| DH5α | Standard E. coli cloning strain | Gibco BRL |
| B. abortus strains | ||
| 2308 | Virulent laboratory strain | |
| BAM723 | Strain 2308 ure1::Tn5 | This study |
| 2308ΔureC1 | This study | |
| 2308ΔureC2 | This study |
To determine the effect of ammonium chloride or urea on urease production, BB was supplemented with ammonium chloride at concentrations up to 200 mM or with urea at concentrations up to 5 mM.
Random transposon mutagenesis and isolation of urease mutants.
Transposon mutagenesis was performed as previously described (34). Urease mutants were selected using urea-indole medium (Institut Pasteur Production, France). Wild-type strain 2308, used as a control, turned the medium from orange to pink in just 15 min at room temperature. Mutants that failed to change the color of the medium in 15 min were selected for further analysis.
Recombinant DNA techniques and sequencing.
Chromosomal DNA from B. abortus strains was purified by the guanidinium thiocyanate method of Pitcher et al. (29). Construction of plasmids, restriction enzyme analysis, agarose gel electrophoresis, and Southern hybridization were carried out by using standard molecular biology protocols (33).
Chromosomal DNA PCR templates were obtained from single colonies by using InstaGene matrix as described by the supplier (Bio-Rad Laboratories, United Kingdom). Two microliters of the resulting solution was used in 50-μl PCR mixtures that included 0.5 U of Taq DNA polymerase, 10 pmol of each primer, and 10 pmol of each deoxynucleoside triphosphate. The PCR conditions used were 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 2 min.
DNA sequencing data were obtained with an automatic Vistra 725 DNA sequencer. Database searches and sequence alignment were performed at the National Center for Biotechnology Information, using the BLAST network service.
Construction of mutants by homologous recombination.
In order to construct a stable urease mutant, oligonucleotides UreC#1.F (CTCAACCATCCCGAAGCCATCG) and UreC#1.R (CCGTTTTCCAGCGTGATGGC) were used to amplify a 2,556-bp region comprising the complete ureC1 gene. A PCR fragment of the expected size from B. abortus strain 2308 was gel purified and cloned into pGEM-T Easy (Promega, Madison, WI), resulting in plasmid pFJS110. An internal 732-bp EcoRV fragment from ureC1 was removed, resulting in plasmid pFJS111. The deleted region included the sequence that encodes the substrate binding site and most of the residues putatively involved in the nickel metallocenter (28). The insert in pFJS111 was sequenced to verify that no extra mutation was present and that the deletion had taken place, keeping the reading frame. A 1,858-bp NotI fragment, containing ΔureC1, was extracted from pFJS111 and subcloned into pJQ200uc1, producing pFJS112. A B. abortus mutant strain carrying the deleted ureC1 gene was obtained in a two-step process, as described previously (28). In the first step pFJS112 was mobilized into B. abortus strain 2308 from E. coli strain S17-1(λ-pir), and transconjugants were selected in BAF supplemented with gentamicin. Colonies growing in this medium represented single-crossover events. Five of these colonies were pooled and grown in BB, and 108 CFU was plated on BA containing 5% sucrose. Sucrose-resistant colonies were selected and analyzed by PCR performed with primers UreC#1.F and UreC#1.R to detect deletion of the internal EcoRV fragment in ureC1. A smooth colony with the deletion (2308ΔureC1) was selected for further work.
Similarly, oligonucleotides UreC#2.F (CGACATTCCCGCCAATACCG) and UreC#2.R (GGTCGAGCACAAGCCAATCG) were used to clone a 2,131-bp fragment including the ureC2 gene, producing plasmid pFJS113. The resulting insert was completely sequenced to verify the construction as described above. An internal NcoI fragment was deleted to produce plasmid pFJS114. The mutated ureC2 fragment was cloned as a PstI/XhoI insert into pJQ200SK, producing pFJS115, which was used to mutate the gene ureC2 as described above.
Measurement and characterization of urease activity.
Urease activity was determined by measuring the amount of ammonia released from urea per unit of time.
Crude extracts containing urease were prepared as follows. Exponential cultures of bacteria were recovered by centrifugation, washed, and resuspended in phosphate-buffered saline (PBS) to a concentration of 108 CFU/ml. The preparations were then lysed using three 10-s cycles with a FastPrep system (Bio 101, Vista, CA) at the maximum setting, cooled on ice, and centrifuged for 5 min at 14,000 rpm at 4°C to remove the cell debris. Crude extracts were aliquoted and stored at −80°C until they were used. For standard urease reactions 5 to 10 μl of extract was added to a tube containing 200 μl of 50 mM urea in PBS and incubated for 5 min at 37°C. The amount of ammonia released from urea hydrolysis was determined colorimetrically by the modified Berthelot reaction (35), and the optical densities at 595 nm of the samples were determined. The amount of ammonia present was then inferred from a standard NH4Cl concentration curve. The total protein concentration was measured by a Bradford assay (6).
The urea Km of B. abortus urease was calculated from a Lineweaver-Burk plot of the initial reaction rates obtained in standard assays with urea concentrations ranging from 0.5 to 50 mM. To determine the optimal pH for urease, phosphate buffer at pHs ranging from 5.5 to 8.0 were used. Urease activity was expressed in micromoles of urea hydrolyzed per minute per milligram of protein.
In vitro susceptibility of Brucella to acid pH.
B. abortus strains were grown in BB until the end of the exponential phase, washed in sterile water, and resuspended at a concentration of 109 CFU/ml in citrate buffer (pH 2.0) or phosphate buffer (pH 4.0) for 30 min in the presence or absence of different concentrations of urea. Bacteria were washed three times in normal PBS, and the survivors were counted after dilution and plating (20).
Competitive mouse infection assay.
BALB/c mice (CRIFA, Spain) were infected with 1:1 mixtures of B. abortus 2308 (wild type) and mutants in order to minimize animal-to-animal variation. Two hundred microliters of a suspension containing approximately 1010 bacteria was administered orally to groups (n = 5) of 5- to 6-week-old female BALB/c mice that previously were held without food for 6 h. Mice were sacrificed 90 min after inoculation, and the terminal ileum (around 15 cm) was removed aseptically and homogenized with 5 ml of BB containing 20% glycerol. Samples were serially diluted and plated in triplicate on BAF plates supplemented and not supplemented with the appropriate antibiotic.
Statistical analysis.
A statistical analysis was performed using Prism3, version 3.0 (GraphPad Software, San Diego, CA). Statistical significance was calculated using either a nonparametric Mann-Whitney test or an unpaired t test. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited in the GenBank database under accession number AF361941.
RESULTS
Characterization and regulation of urease activity.
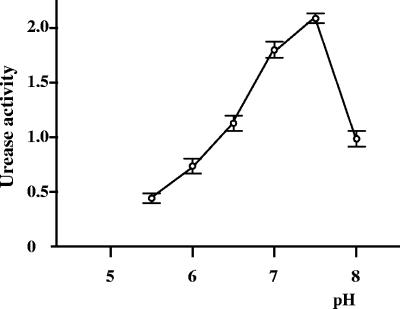
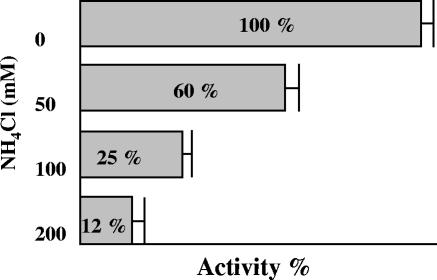
Crude cell extracts of B. abortus 2308 were used to determine the urease kinetic parameters. The Km for urea was found to be 13.44 ± 0.8 mM (mean ± standard deviation of five determinations), and the activity was found to be maximal at pH 7.3 (Fig. 1). To determine the factors that control urease expression in B. abortus, we analyzed the effect of nitrogen availability on urease production by adding increasing concentrations of ammonium chloride to bacteria growing in BB. The urease activity decreased as the concentration of added ammonium chloride increased (Fig. 2). Furthermore, urease production was found to be independent of the presence of urea in the culture medium.
FIG. 1.
Optimum pH for urease activity. The urease activities of Brucella protein extracts were assayed in different buffer conditions with pHs ranging from 5.5 to 8. The experiment was performed three times in duplicate. Each point is the arithmetic mean of six independent experimental values. The error bars indicate standard deviations. Urease activity is expressed in micromoles of urea hydrolyzed per minute per milligram of protein.
FIG. 2.
Urease activity in the presence of ammonium chloride. B. abortus 2308 cultures were grown in BB in the presence of different amounts of added ammonium chloride, and their urease activities were measured. The activity is expressed as a percentage of the maximum activity that was obtained when no ammonium chloride was added to the medium. The experiment was performed in triplicate with three technical measurements per replicate. The bars indicate means, and the error bars indicate standard deviations.
Production of B. abortus 2308 transposon mutants.
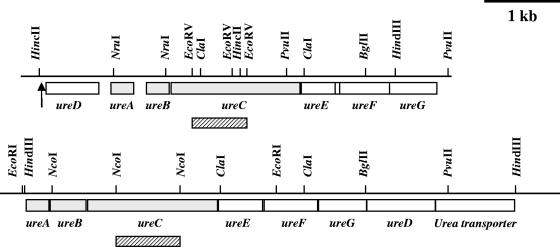
In the initial screening of B. abortus 2308 carrying transposon insertions we identified seven mutants with different levels of urease activity. Six mutants were negative after 1 week of incubation, and one mutant showed some activity after 1 week (control strain 2308 was positive in 15 min at room temperature). Southern hybridization of the genomic DNA of the mutants indicated that they contained a single copy of Tn5 and that all but one mutation clustered in the same chromosomal region (Fig. 3).
FIG. 3.
Map of the ure1 and ure2 cluster region. The boxes represent the ORFs. The shaded boxes represent the structural genes, and the open boxes represent the accessory genes. The striped bars under ureC indicate the sequences deleted in mutants 2308ΔureC1 and 2308ΔureC2. The location of the Tn5 insertion in mutant BAM723 is indicated by an arrow.
One of the mutants that remained negative after 1 week of incubation, BAM723, was selected in order to clone and sequence the affected genes.
Subcloning and sequencing of a B. abortus urease gene cluster.
The kanamycin resistance encoded by Tn5 was used to clone the transposon and flanking DNA from urease-negative mutant BAM723 into plasmid pBluescript SKII. A large EcoRI fragment that was around 35 kb long containing Tn5 was obtained in recombinant plasmid pCRIS, which was subcloned further to facilitate DNA sequence analysis.
A total of 5.7 kbp of DNA was sequenced, which revealed seven potential open reading frames (ORFs) after translation of both strands. These ORFs corresponded to the conserved structural proteins UreA, UreB, and UreC, as well as the accessory proteins UreD, UreE, UreF, and UreG (Fig. 3). The genes encoding these proteins were all transcribed in the same direction, and they were each preceded by sites similar to the E. coli consensus ribosome-binding (Shine-Dalgarno) sequence. Furthermore, a sequence (TGTCGGGACTCTCGTTGCAGCG; nucleotides 220 to 241) similar to the consensus σ54 promoter sequence (NRYTGGCACGN4TTGCWNNW) (2) was found upstream of the ureD ATG start codon. This could be the promoter for the urease operon. A putative transcriptional terminator was found between ureA and ureB (nucleotides 1,508 to 1,537), although no sequence resembling a promoter could be found in this region. This structure indicates that there are two possible transcriptional units, ureDA and ureBCEFG, although this possibility requires further analysis. Shortly after the stop codon for ureG there is a sequence corresponding to a half-copy of the Brucella palindromic repeated DNA element Bru-RS2 (14) and a putative Arg tRNAACG gene (nucleotides 5,631 to 5,700). Both of these features result in a strong secondary structure in the mRNA that could serve as a transcriptional termination signal.
The nucleotide sequence of the cloned genome segment containing Tn5 from mutant BAM723 showed that the transposon was inserted at nucleotide position 280 of the sequence determined (Fig. 3). This position corresponded to the 5′ untranslated sequence upstream of ureD. This insertion completely abolished the formation of active urease in mutant BAM723.
The only Tn5 insertion with the urease activity affected that mapped out of this ure1 region was localized in the cobT gene by sequencing the neighboring DNA. Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) plays a central role in the synthesis of α-ribazole-5′-phosphate, an intermediate for the lower ligand of cobalamin (40). Cobalamin is the largest and most complex cofactor found in biological systems.
Construction of urease deletion mutants of B. abortus 2308 by gene replacement.
A stable mutant having a deletion in the ureC1 gene was constructed as described in Materials and Methods. This mutant, 2308ΔureC1, had an in-frame deletion affecting 244 amino acids of the UreC1 polypeptide.
2308ΔureC1 had a stable urease-negative phenotype. The mutant and the parental strain had identical growth patterns, indicating that the disrupted gene was not required for normal cell functions.
During this study the complete sequence of B. melitensis strain 16 M was published (12), and this was followed by publication of the sequences of B. suis (27) and B. abortus strains 9-941 (15) and 2308 (7). The published sequences confirmed our findings and showed that a second urease locus (ure2) is present in the large chromosomes of the three species. To determine the functionality of ure2, we constructed a strain 2308 mutant having a deletion in the ureC2 gene. 2308ΔureC2 had the same urease activity as the parental strain. This result, together with the complete lack of urease activity in the ureC1 mutants, clearly indicated that the ure2 locus did not produce any measurable phenotype under the experimental conditions used.
In vitro susceptibility of the urease mutants to acid.
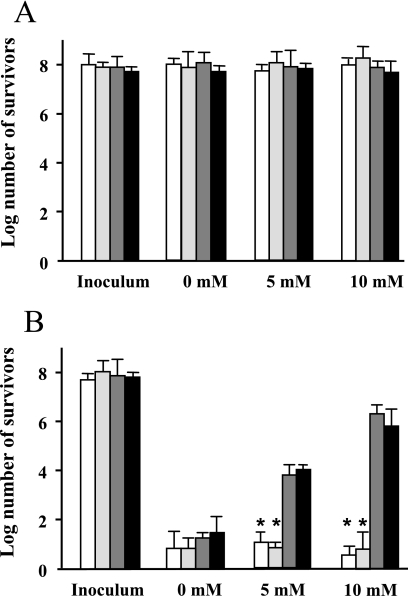
Since during gastrointestinal infection Brucella spp. must be able to survive passage through the stomach before invading the host, the ability of B. abortus to survive exposure to acid was investigated in the presence and absence of urea. B. abortus 2308 (urease-positive strain) and the urease mutants BAM723, 2308ΔureC1, and 2308ΔureC2 were exposed to different acid conditions in the presence and absence of urea. As shown in Fig. 4, B. abortus strain 2308 and all the mutants tested survived exposure to pH 4.0 for 30 min equally well, independent of the presence of urea. However, there was a marked decrease in survival when they were exposed to pH 2.0. When urea was incorporated into the assay buffer, the wild-type strain and the 2308ΔureC2 mutant exhibited dose-dependent increases in survival compared to the urease-negative strains. At the end of the assays, buffer pH values were determined, and in all cases they were very similar to the initial values (data not shown).
FIG. 4.
Survival of B. abortus 2308 and urease mutants after acid exposure. Bacteria were resuspended in buffer at pH 4.0 (A) or in buffer at pH 2.0 (B) in the presence of different concentrations of urea (indicated at the bottom). After 30 min of incubation at 37°C, bacteria were diluted and plated to count the survivors. The data are the arithmetic means and standard deviations from three separate experiments. An unpaired t test was performed to determine if the survival of each strain was significantly different than the survival of the corresponding wild-type control. An asterisk indicates that the P value is <0.05. Light gray bars, BAM723; open bars, 2308ΔureC1; dark gray bars, 2308; solid bars, 2308ΔureC2.
Virulence analysis.
The virulence of each of the urease mutants was compared to the virulence of the parental strain in a macrophage survival assay and by intraperitoneal injection of the bacteria into mice. The urease mutants exhibited the same behavior as the parental strain in the two models (data not shown). Based on these data, we concluded that urease does not play a role in these models of infection.
To investigate if urease could play a role in establishment of a B. abortus infection when the bacteria were administered by the oral route, we determined the level of survival after transit though the stomach. The assay was performed with the BAM723 mutant and the 2308ΔureC1 and 2308ΔureC2 mutants. In the case of BAM723, we used kanamycin for selection. In order to have a selectable marker for the deletion mutants, we first introduced plasmid pBBR1MCS (19) by conjugation from S17-1(pBBR1MCS). The transconjugants were selected on BAF plates containing chloramphenicol. A 1:1 mixture of B. abortus 2308 and each urease mutant was administered orally to mice.
In order to allow bacterial passage through the stomach, mice were sacrificed 90 min after infection, and bacteria from the terminal ileum were plated on media with and without an antibiotic to compare the numbers of urease-negative mutants (antibiotic resistant) and parental bacteria (antibiotic susceptible).
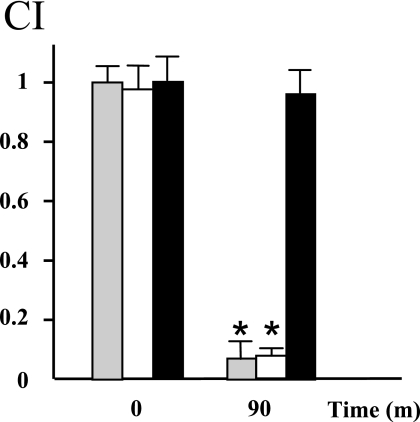
In vitro competitive growth experiments were carried out first to verify that strains did not have growth defects under normal laboratory conditions. The mutants did not exhibit any deficiency when they were grown in vitro in the presence of the wild-type strain. The competitive index was calculated by dividing the output ratio of mutant to wild-type bacteria by the input ratio of mutant to wild-type bacteria. In general, the bacterial mixtures recovered after passage through the stomach were enriched around 10-fold for urease-producing bacteria (Fig. 5). In other words, more than 90% of the Brucella colonies which survived transit through the stomach were identified as B. abortus 2308.
FIG. 5.
Survival of the urease mutants after transit through the mouse stomach. A 1:1 mixture of 2308 and a urease mutant was administered orally to groups of five mice, and the experiment was performed twice. The bars indicate the competitive indexes (CI) for the three mutants with wild-type strain 2308 from one of the experiments. The error bars indicate standard deviations. The results of the mouse competitive index assay were analyzed with a nonparametric Mann-Whitney test using log-transformed competitive indexes to determine if the indexes were significantly different than 1. An asterisk indicates that the P value is <0.05. Gray bars, BAM723/2308; open bars, 2308ΔureC1/2308; solid bars, 2308ΔureC2/2308.
Southern blot analysis of DNA of bacteria surviving transit through the stomach provided further confirmation of the correct identification of the mutants (data not shown).
DISCUSSION
Most Brucella isolates exhibit potent urease activity that has been hypothesized to play a role in the pathogenesis of disease (39). However, the role of this enzyme in virulence has not been explained yet. Furthermore, some isolates, such as the well-known laboratory strain B. abortus 544, are urease negative, while they seem to retain most of their pathogenic potential. In order to obtain insight into the function of Brucella urease, we first determined the Km for urea and the optimal pH of the urease enzyme. The value observed for the urea Km (13.4 mM) was not consistent either with the low Km values observed for ureases with a role in nitrogen assimilation or with the high Km values often exhibited by uropathogens. However, urease activity in Brucella was dependent on the nitrogen (ammonium chloride) availability in the culture medium; it was maximal in the absence of added ammonium chloride and practically disappeared when the concentration of ammonium was greater than 100 mM. This downregulation of urease expression by high-quality nitrogen sources, such as ammonia, has been found in other bacteria, like Klebsiella aerogenes, and suggests that the urease has a role in the assimilation of nitrogen (25) from urea or at least some dependence on nitrogen metabolism in Brucella.
When the pH was considered, the urease activity of B. abortus was optimal at a pH near neutrality (pH 7.3). The urease produced by H. pylori also has a neutral pH optimum, and this urease activity is essential for survival at the extreme pH of the stomach. The neutral pH optimum of ureases indicates that even when the bacteria have to deal with extreme pH values, the pH of the bacterial cytoplasm does not vary, remaining at the neutral physiological values required for most bacterial processes.
The Tn5 insertion mutants whose urease activity was affected allowed us to clone and sequence a urease operon from B. abortus. Our data, later confirmed by whole-genome sequencing, revealed an operon with the organization ureDABCEFG found in many bacteria (25). We predicted the presence of a promoter of the σ54 type for the Brucella urease operon. This could indicate that urease expression in Brucella is under the control of a system analogous to the Ntr system of the enteric bacteria that responds to nitrogen availability and takes advantage of the alternative sigma factor σ54 (32). This type of regulation would also explain the observed dependence of urease expression on the ammonium concentration in the growth medium.
Whole-genome sequencing revealed the presence of a second urease operon (ure2) with all the genes potentially active in B. suis and B. melitensis. On the other hand, B. abortus had two −1 frameshift deletions in ureE2, and B. ovis had deletions in ureG2 and ureT. These data support our results indicating that ure2 did not contribute to the urease activity of B. abortus 2308. Unpublished work from our laboratory showed that ureC1 mutants of other Brucella strains were also urease negative, suggesting additional reasons for the lack of urease activity resulting from the ure2 locus in Brucella.
A sequence comparison and phylogenetic analysis of Brucella ure genes indicated that the ure2 operon clustered with the ure genes from Yersinia rather than with Brucella ure1 or ure genes from other α-proteobacteria. Not only did the differences between Brucella ure1 and ure2 affect the nucleotide sequence, but the gene order was also different and a putative urea transporter was also present in ure2. These differences argue against duplication as the origin of ure2; rather, they suggest that this locus may have been acquired via a lateral transfer event. ureC2 was expressed under normal growth conditions, as shown by real-time reverse transcription-PCR (data not shown), and although no contribution to urease activity could be measured in this study, we cannot rule out the possibility that ure2 has some function that ensures its maintenance in the Brucella genome.
Urease is a complex enzyme that requires an elaborate pathway for assembly in its active form. Thus, it was not surprising to find mutations producing a urease-deficient phenotype that mapped outside the ure genes. It has been reported previously that mutations in a nickel transport system in B. suis and Actinobacillus pleuropneumoniae produced a urease-deficient phenotype (4, 17). In this study, we found an additional locus, cobT, which was required for maintaining the urease-positive phenotype in B. abortus. Until now, we ignored the reason for the lack of urease activity of this mutant.
One of the proposed roles of Brucella urease in pathogenicity was thought to be inhibition of phagosome acidification by ammonia release. However, we did not observe any difference between the wild-type and urease mutant strains when they were compared in a macrophage survival assay or after intraperitoneal infection of mice. Moreover, Porte et al. (30) have shown that at the early stage of phagocytosis, neutralization of the intraphagosomal pH by addition of ammonium chloride can be detrimental for Brucella. Most of the available evidence indicates that urease does not play a significant role in the intracellular survival of Brucella.
The most common route of brucella infection in humans is through the gastrointestinal tract. The ability of a pathogen to resist killing in the acidic environment of the stomach increases the likelihood of intestinal colonization and invasion. The low pH of the gastric juices has been suggested to play a role in the prevention of Brucella infection, while proton pump inhibitors, antacids, and other drugs that decrease gastric acidity have been implicated in food-borne brucellosis (42). Urease is involved in the survival of bacteria in their transit through the acid contents of the stomach in a number of bacterial pathogens, like H. pylori and Y. enterocolitica (11). Accordingly, we decided to examine the role of urease in a model involving intragastric inoculation of mice. Since previous experiments had shown that there was noticeable variation from mouse to mouse, we inoculated mixed cultures into individual mice rather than use different groups of mice for each strain, thus reducing individual differences to a minimum. Using this model, we demonstrated that urease mutants BAM723 and 2308ΔureC1 had a competitive deficiency compared to the wild-type bacteria in terms of passage through the mouse stomach. The same pattern of acid sensitivity in vitro and attenuation in the mouse oral model has been observed for ureC1 mutants of B. abortus S19, B. melitensis Rev1, and B. suis 1330 (unpublished results). The transit time through the stomach of a liquid-fed mouse can be estimated to be 15 min (26). Thus, the acid exposure conditions used in this work were milder than the acid stress conditions experienced by Brucella during passage through the stomach of a healthy human, which include a pH of less than 3 and a gastric transit time of 2 h (11), or through the abomasum of a ruminant (41). This difference reinforces the importance of urease in the establishment of Brucella alimentary infections in humans.
B. ovis is the only urease-negative Brucella species. Transmission of B. ovis between animals occurs mainly by the sexual route. The fact that oral transmission of B. ovis is not very important could be a consequence of the lack of an active urease in this organism, stressing the role of this enzyme in the oral transmission of Brucella.
Recently, it has been shown that urease can also contribute to the establishment of A. pleuropneumoniae infections in pigs through the respiratory tract (5). If this mechanism is also operative in Brucella, then it is possible that urease-negative mutants are deficient in establishing infections not only by the digestive route but also through aerosol inhalation, the two main routes of Brucella infection in humans. In addition, the B. melitensis vaccine strain Rev.1 is known to be secreted into the milk of vaccinated animals, which is a major route of transmission to humans (1). Based on this, the introduction of mutations abolishing urease activity in vaccine strains could be an additional procedure to improve the safety of Brucella vaccines by minimizing the possibility of human infection.
Acknowledgments
This work was supported by grant BIO2004-06117 from the Spanish “Ministerio de Educación y Ciencia” and by grant PI052499 from the “Instituto de Salud Carlos III.”
M.C.R. was the recipient of a scholarship from the Fundación Marqués de Valdecilla-IFIMAV.
Editor: A. Camilli
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Banai, M. 2002. Control of small ruminant brucellosis by use of Brucella melitensis Rev. 1 vaccine: laboratory aspects and field observations. Vet. Microbiol. 90:497-519. [DOI] [PubMed] [Google Scholar]
- 2.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beswick, E. J., I. V. Pinchuk, K. Minch, G. Suarez, J. C. Sierra, Y. Yamaoka, and V. E. Reyes. 2006. The Helicobacter pylori urease B subunit binds to CD74 on gastric epithelial cells and induces NF-κB activation and interleukin-8 production. Infect. Immun. 74:1148-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosse, J. T., H. D. Gilmour, and J. I. MacInnes. 2001. Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosse, J. T., and J. I. MacInnes. 2000. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can. J. Vet. Res. 64:145-150. [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chain, P. S. G., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 73:8353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomel, B. B., E. E. DeBess, D. M. Mangiamele, K. F. Reilly, T. B. Farver, R. K. Sun, and L. R. Barrett. 1994. Changing trends in the epidemiology of human brucellosis in California from 1973 to 1992: a shift toward foodborne transmission. J. Infect. Dis. 170:1216-1223. [DOI] [PubMed] [Google Scholar]
- 9.Collins, C. M., and S. E. D'Orazio. 1993. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol. Microbiol. 9:907-913. [DOI] [PubMed] [Google Scholar]
- 10.Corbel, M. J., and D. M. Hendry. 1985. Urease activity of Brucella species. Res. Vet. Sci. 38:252-253. [PubMed] [Google Scholar]
- 11.De Koning-Ward, T. F., and R. M. Robins-Browne. 1995. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect. Immun. 63:3790-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorden, J., and P. L. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halling, S. M., and B. J. Bricker. 1994. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol. Microbiol. 14:681-689. [DOI] [PubMed] [Google Scholar]
- 15.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187:2715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, D. E., R. G. Russell, C. V. Lockatell, J. C. Zulty, J. W. Warren, and H. L. Mobley. 1993. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect. Immun. 61:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jubier-Maurin, V., A. Rodrigue, S. Ouahrani-Bettache, M. Layssac, M. A. Mandrand-Berthelot, S. Kohler, and J. P. Liautard. 2001. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. K., S. B. Mulrooney, and R. P. Hausinger. 2005. Biosynthesis of active Bacillus subtilis urease in the absence of known urease accessory proteins. J. Bacteriol. 187:7150-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 20.Krishnamurthy, P., M. Parlow, J. B. Zitzer, N. B. Vakil, H. L. Mobley, M. Levy, S. H. Phadnis, and B. E. Dunn. 1998. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect. Immun. 66:5060-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madkour, M. M. 1989. Brucellosis. Butterworths, London. United Kingdom.
- 22.Maroncle, N., C. Rich, and C. Forestier. 2006. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res. Microbiol. 157:184-193. [DOI] [PubMed] [Google Scholar]
- 23.Marshall, B. J., L. J. Barrett, C. Prakash, R. W. McCallum, and R. L. Guerrant. 1990. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology 99:697-702. [DOI] [PubMed] [Google Scholar]
- 24.Mobley, H. L., G. R. Chippendale, K. G. Swihart, and R. A. Welch. 1991. Cytotoxicity of the HpmA hemolysin and urease of Proteus mirabilis and Proteus vulgaris against cultured human renal proximal tubular epithelial cells. Infect. Immun. 59:2036-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreto, M., L. Guzman, and A. Diez. 1982. A pattern for gastric emptying in mice. Am. J. Physiol. 242:G333-G336. [DOI] [PubMed] [Google Scholar]
- 27.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 29.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of genomic DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 30.Porte, F., J. P. Liautard, and S. Kohler. 1999. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 32.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Sangari, F., and J. Agüero. 1991. Mutagenesis of Brucella abortus: comparative efficiency of three transposon delivery systems. Microb. Pathog. 11:443-446. [DOI] [PubMed] [Google Scholar]
- 35.Senior, B. W., N. C. Bradford, and D. S. Simpson. 1980. The ureases of Proteus strains in relation to virulence for the urinary tract. J. Med. Microbiol. 13:507-512. [DOI] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering, transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 37.Skurnik, M., S. Batsford, A. Mertz, E. Schiltz, and P. Toivanen. 1993. The putative arthritogenic cationic 19-kilodalton antigen of Yersinia enterocolitica is a urease beta-subunit. Infect. Immun. 61:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, D. G., W. C. Russell, W. J. Ingledew, and D. Thirkell. 1993. Hydrolysis of urea by Ureaplasma urealyticum generates a transmembrane potential with resultant ATP synthesis. J. Bacteriol. 175:3253-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, L. D., and T. A. Ficht. 1990. Pathogenesis of Brucella. Crit. Rev. Microbiol. 17:209-230. [DOI] [PubMed] [Google Scholar]
- 40.Trzebiatowski, J. R., G. A. O'Toole, and J. C. Escalante-Semerena. 1994. The cobT gene of Salmonella typhimurium encodes the NaMN:5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-alpha-d-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 176:3568-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittek, T., K. Schreiber, M. Furll, and P. D. Constable. 2005. Use of the d-xylose absorption test to measure abomasal emptying rate in healthy lactating Holstein-Friesian cows and in cows with left displaced abomasum or abomasal volvulus. J. Vet. Intern. Med. 19:905-913. [DOI] [PubMed] [Google Scholar]
- 42.Young, E. J. 2000. Brucella species, p. 2053-2060. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 4th ed. Churchill Livingstone, Philadelphia, PA.
- 43.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]