Abstract
Humans develop periodontitis in response to challenge by microbial dental plaque. Inflammation begins after perturbation of gingival epithelial cells by subgingival bacteria interacting through pattern-recognition receptors, including the Toll-like receptors (TLR). Porphyromonas gingivalis is a major periodontopathogen that interacts with epithelial cells through its cell surface fimbriae (FimA), leading to colonization and/or invasion. Previous work by our group has established membrane CD14 as an essential coreceptor for TLR2-mediated activation of transfected cell lines by P. gingivalis FimA. We have shown that gingival epithelial cells express TLR2 but not CD14 on their cell surfaces. We thus speculated that P. gingivalis FimA does not readily activate epithelial innate immune responses but rather functions to promote P. gingivalis colonization in the absence of a vigorous FimA-induced response. This hypothesis was verified by the findings that primary human gingival epithelial cells responded poorly to FimA in terms of interleukin (IL)-6, IL-8, granulocyte-macrophage colony-stimulating factor, and tumor necrosis factor alpha responses, in stark contrast to the marked response to other TLR2 agonists (Pam3Cys, FSL-1) that are not strictly dependent on CD14. On the other hand, CD14-expressing human primary monocytes responded with high levels of the same cytokines to both FimA and the control TLR2 agonists. The gingival epithelial cells failed to respond to FimA even in the presence of exogenously added soluble CD14. These data indicate that the gingival epithelial cell hyporesponsiveness to FimA is attributable to the lack of membrane-expressed but not soluble CD14. In conclusion, P. gingivalis FimA differentially activates human monocytes and epithelial cells, perhaps reflecting different tactics used by P. gingivalis when interacting with different host cell types or a host strategy to limit inflammation.
Porphyromonas gingivalis is a gram-negative, black-pigmented bacterium associated with periodontal disease, a condition which results in destruction of the tooth-supporting tissues and loss of teeth (16). P. gingivalis expresses virulence factors involved in host tissue destruction and immune perturbation (10). Adhesion molecules of P. gingivalis include fimbriae (FimA) and hemagglutinins; however, P. gingivalis FimA is considered a critical determinant for the attachment of this microorganism in the oral cavity (16). In this regard, there is strong evidence supporting a crucial role for FimA in P. gingivalis adhesion to several mammalian cells types; indeed, nonfimbriated mutants of P. gingivalis, constructed by inactivation of the FimA gene, display reduced adhesion and invasion of epithelial cells compared to wild-type P. gingivalis (29). Moreover, nonfimbriated mutants display a reduced ability to induce periodontal disease in mice following oral inoculation (19). In this context, P. gingivalis FimA has been shown to induce proinflammatory cytokine release in macrophages (22) as well as inflammatory bone resorption (8, 13).
Epithelial cells function as a physical barrier and in immune surveillance through their ability to elicit an innate immune response. Human gingival epithelial cells (HGECs) express pattern recognition receptors (PRRs) including Toll-like receptor 1 (TLR1), TLR2, TLR4, and TLR6 and respond to P. gingivalis with proinflammatory cytokines, including interleukin (IL)-6, IL-8, tumor necrosis factor alpha (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) (14). Microbe-associated molecular patterns expressed by P. gingivalis such as lipopolysaccharide and FimA are recognizable by PRRs and have been implicated in the initiation and progression of periodontal disease (20). P. gingivalis FimA is detected by PRRs, such as CD14 and TLR2, resulting in activation of monocytes/macrophages (6, 21). It was furthermore reported that FimA upregulates TLR2-dependent IL-8 production in immortalized epithelial cell lines (2). Our group has previously demonstrated a strong requirement for membrane-expressed CD14 in P. gingivalis FimA-induced activation of transfected cell lines (6). Moreover, soluble CD14 could not effectively substitute for membrane CD14 and failed to support FimA-induced cell activation (6). We have thus hypothesized that FimA may be restricted with regard to the cell types it can efficiently activate. Therefore, although monocytes/macrophages may be potently activated by FimA, other cell types, such as epithelial cells, which do not generally express membrane CD14 (3, 27), may be activated relatively weakly by FimA.
This study was designed to determine whether FimA differentially activates TLR2-mediated cytokine responses in human primary monocytes and gingival epithelial cells. Our results indicate that P. gingivalis FimA is essentially inert in inducing proinflammatory cytokines, including IL-6, IL-8, TNF-α, and GM-CSF, in human primary gingival epithelial cells, despite its potent cytokine-inducing capacity in monocytes. Other TLR2 agonists, which do not strictly depend on membrane CD14 for cell activation, could equally well induce cytokine responses in epithelial cells and monocytes. Whole cells of wild-type or nonfimbriated P. gingivalis strains induced comparable and moderate levels of cytokine responses in epithelial cells, thus providing further evidence that FimA does not contribute to P. gingivalis-induced inflammation in gingival epithelial cells. Our view is that the role of FimA in P. gingivalis-epithelial cell interactions is predominantly to promote colonization and/or invasion (for a review, see reference 17) without at the same time provoking a vigorous innate immune response, as shown in the present study. The relative hyporesponsiveness of gingival epithelial cells to FimA may facilitate P. gingivalis persistence in the periodontium.
MATERIALS AND METHODS
Cell isolation and culture.
HGECs, following approval by the University of Louisville Institutional Review Board, were obtained from healthy patients after third-molar extraction. The gingiva was treated with 0.025% trypsin and 0.01% EDTA overnight at 4°C, and HGECs were isolated as previously described (24). The cell suspension was centrifuged at 120 × g for 5 min, and the pellet was suspended in K-SFM medium (Invitrogen, Carlsbad, CA) containing 10 μg/ml of insulin, 5 μg/ml of transferrin, 10 μM of 2-mercaptoethanol, 10 μM of 2-aminoethanol, 10 nM of sodium selenite, 50 μg/ml of bovine pituitary extract, 100 units/ml of penicillin/streptomycin, and 50 ng/ml of fungizone (complete medium). The cells were seeded in 60-mm-diameter plastic tissue culture dishes coated with type I collagen and incubated in 5% CO2 and 95% air at 37°C. When the cells reached subconfluence, they were harvested and subcultured. Monocytes were purified from the peripheral blood of healthy human volunteers. Briefly, monocytes were separated from lymphocytes upon centrifugation of peripheral blood over NycoPrep 1.068 (Axis-Shield, Oslo, Norway). Incidental nonmonocytes were removed by magnetic depletion using a mixture of biotin-conjugated monoclonal antibodies and magnetic microbeads coupled to antibiotin monoclonal antibody (monocyte isolation kit II; Miltenyi Biotec, Auburn, CA). Briefly, human monocytes were purified from whole human blood. Nonmonocytes (i.e., T cells, NK cells, B cells, dendritic cells, and basophils) were indirectly magnetically labeled using a cocktail of biotin-conjugated antibodies against CD3, CD7, CD16, CD19, CD56, CD123, and CD235a (glycophorin A) as well as antibiotin microbeads. Highly pure unlabeled monocytes were obtained by depletion of the magnetically labeled cells. Purified monocytes were cultured in 96-well polystyrene culture plates at 37°C under 5% CO2 in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 10 mM HEPES, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 0.05 mM of 2-mercaptoethanol.
Bacteria and FimA.
P. gingivalis 33277 and an isogenic FimA-inactivated mutant (strain JI-1; kindly provided by Fuminobu Yoshimura, Aichi Gakuin University, Nagoya, Japan) were grown at 37°C in Trypticase soy broth supplemented with 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione per liter in an anaerobic atmosphere of 85% N2, 10% H2, and 5% CO2 for 2 days. After cultivation, the bacteria were harvested by centrifugation, washed three times in phosphate-buffered saline, and heat inactivated as previously described (14, 24). FimA was purified from P. gingivalis 33277 as described previously (18).
Cytokine induction assay.
Primary epithelial cultures and cell lines at the fourth or fifth passage were harvested, seeded at a density of 0.5 × 105 cells/six-well culture plate coated with type I collagen, and maintained in 2 ml of medium. When they reached confluence, the cells were washed two times with fresh medium, and 1 ml of complete medium was added either with or without 5% FBS. The cells were incubated at 37°C under 5% CO2 for 4 h with heat-killed P. gingivalis (multiplicity of infection [MOI], 100:1), FimA (10 μg/ml), Pam3Cys and FSL-1 (both at 1 μg/ml; InvivoGen, San Diego, CA), human recombinant CD14 (0.5 μg/ml; R&D, Minneapolis, MN), and/or lipopolysaccharide binding protein (LBP) (0.05 μg/ml; R&D) suspended in 500 μl of the medium. Cell culture supernatants were then separated by centrifugation and stored at −80°C prior to a cytokine protein assay by use of Luminex 100 technology using a multiplex for four cytokines: IL-6, IL-8, GM-CSF, and TNF-α (Upstate Cell Signaling Solutions, NY). Human monocytes (1.5 × 105/well) were stimulated as described previously (9). Briefly, monocytes were incubated at 37°C under 5% CO2 with heat-killed P. gingivalis (MOI, 100:1), Pam3Cys and FSL-1, (both at 1 μg/ml), and FimA (1 or 10 μg/ml). Culture supernatants were collected at the end of the experiment and stored at −80°C until being assayed for TNF-α, IL-6 (eBioscience, San Diego, CA), and IL-8 (Cell Sciences, Canton, MA) responses using enzyme-linked immunosorbent assay kits. None of the bacterial stimuli affected cell viability as determined by trypan blue exclusion.
Real-time PCR.
Total RNA was extracted from cultured cells by using TRIzol (Invitrogen, Carlsbad, CA) and quantified by spectrometry at 260 and 280 nm. Ten micrograms from each RNA extract was used to perform first-strand cDNA synthesis using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) in a total volume of 100 μl. Real-time PCR was performed by using 50 ng of cDNA with an ABI 7500 system (Applied Biosystems). TaqMan probes and sense and antisense primers for gene expression of human TLR1, -2, and -6 were purchased from Applied Biosystems along with probes and primers for human GAPDH as an endogenous control. Using a Universal PCR Master Mix (Applied Biosystems), the reactions were carried out according to the manufacturer's protocol.
Flow cytometry.
Human gingival epithelial cells and monocytes were washed three times with phosphate-buffered saline, and 1,000,000 cells were stained with 0.5 μg of phycoerythrin-Cy7 conjugated to anti-human CD14, 1 μg of fluorescein isothiocyanate conjugated to anti-human TLR2 or to isotype controls (mouse immunoglobulin (Ig) G1 or IgG2a [eBioscience, San Diego, CA], respectively), in 100 μl total staining buffer for 20 min at 4°C. The cells were analyzed by flow cytometry using a BD FACSCalibur and CellQuest software.
RESULTS
CD14 and TLR2 protein expression in human gingival epithelial cells and monocytes.
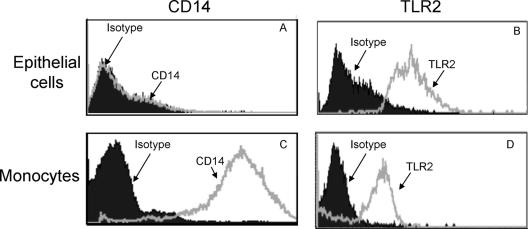
It has been shown that TLR2 and CD14 play a crucial role in recognition of FimA by monocytes (6, 9). Moreover, membrane-expressed CD14 is an essential coreceptor for TLR2-mediated cell activation by P. gingivalis FimA (6). Although epithelial cells express TLR2 (14), they may not readily respond to P. gingivalis FimA unless they coexpress CD14. We therefore examined whether TLR2 and membrane CD14 are produced by human gingival epithelial cells. Although we found that CD14 is expressed by epithelial cells at the gene level (data not shown), CD14 protein was not detectable by flow cytometry (Fig. 1A). In contrast, membrane CD14 was readily detected on human monocytes (Fig. 1C). In addition, TLR2 protein expression was detected by flow cytometry in both cell types (Fig. 1B and D). These data indicate that CD14 expression varies depending on the cell type and suggest that gingival epithelial cells may not be readily responsive to P. gingivalis FimA in terms of CD14-dependent cytokine induction.
FIG. 1.
TLR2 and CD14 protein expression by human primary monocytes and gingival epithelial cells. HGECs (A) and monocytes (C) were stained with phycoerythrin-conjugated monoclonal anti-human CD14 or its isotype control, mouse IgG1. Isolated HGECs (B) and monocytes (D) were stained with fluorescein isothiocyanate-conjugated monoclonal anti-human TLR2 or its isotype control, IgG2a, for 20 min at 4°C. The stained cells were analyzed by flow cytometry using a BD FACSCalibur and CellQuest software.
P. gingivalis but not purified FimA activates human gingival epithelial cells.
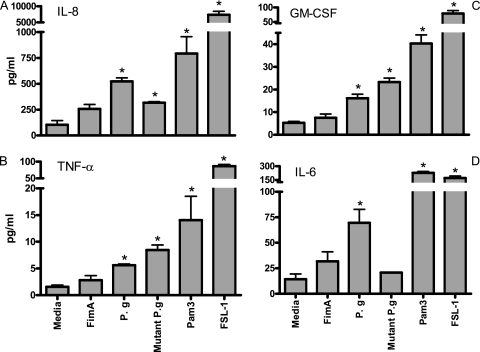
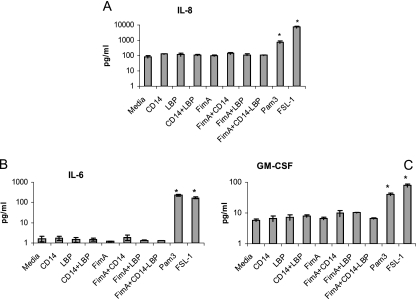
It has been shown that FimA induces cytokine release in a TLR2-dependent fashion (6, 9, 21). Therefore, to determine FimA-induced cytokine responses in gingival epithelial cells, we used TLR2 agonists as a positive control. To assess the stimulatory potential of P. gingivalis on human gingival epithelial cells, the cells were challenged with P. gingivalis, purified FimA, or the control TLR2 agonists FSL-1 (a TLR2/6 agonist) and Pam3Cys (a TLR2/1 agonist) for 4 h. Wild-type P. gingivalis and TLR2 agonists significantly upregulated IL-6, IL-8, TNF-α, and GM-CSF cytokines at the protein level compared to medium alone (P < 0.05), in contrast to FimA, which failed to induce cytokine responses (Fig. 2). Additionally, IL-8, TNF-α, and GM-CSF were upregulated in epithelial cells challenged by nonfimbriated P. gingivalis (mutant P. gingivalis) compared to medium alone (Fig. 2). Since CD14 plays an important role in TLR2-dependent cytokine induction by FimA (6), we examined whether the epithelial cells could become responsive to FimA in the presence of serum (FBS) as a source of soluble CD14. Thus, we challenged epithelial cells with P. gingivalis, FimA, or TLR2 agonists in the presence of 5% FBS (Fig. 3). Even though 5% FBS resulted in some background increase in cytokine responses compared to medium alone, the proinflammatory cytokines IL-6, IL-8, GM-CSF, and TNF-α were not significantly upregulated after challenge with FimA for 4 h (Fig. 3). Addition of exogenous human CD14 in purified form with or without LBP did not improve proinflammatory cytokine production by FimA in epithelial cells (Fig. 4). These data indicate that human epithelial cells are hyporesponsive to P. gingivalis FimA even in the presence of soluble CD14, in contrast to other TLR2 agonists or heat-killed P. gingivalis (Fig. 2).
FIG. 2.
Cytokine induction in epithelial cells challenged with P. gingivalis or TLR2 agonists (Pam3Cys or FSL-1). HGECs were treated with heat-killed P. gingivalis (P. g; MOI, 100:1), nonfimbriated P. gingivalis (Mutant P. g; MOI, 100:1), FimA (10 μg/ml), or the TLR2 agonist Pam3Cys (Pam3) or FSL-1 (both at 1 μg/ml) for 4 h. Induction of IL-8 (A), TNF-α (B), GM-CSF (C), and IL-6 (D) was determined in culture supernatant by use of Luminex 100 technology (Upstate Cell Signaling Solutions, NY). Data are presented as the means ± standard deviations of triplicate determinations, from one of three independent sets of experiments that yielded similar findings. Statistically significant (P < 0.05) induction of cytokine release is indicated by an asterisk.
FIG. 3.
Cytokine induction in the presence of 5% FBS in human gingival epithelial cells. HGECs were challenged by P. gingivalis (P. g; MOI, 100:1), FimA (10 μg/ml), or TLR2 agonists (Pam3 or FSL-1; 1 μg/ml) for 4 h at 37°C in the presence of 5% FBS. Induction of the cytokines TNF-α (A), IL-6 (B), and GM-CSF (C) was determined in culture supernatants by use of Luminex 100 technology (Upstate Cell Signaling Solutions, NY). Data are presented as the means ± standard deviations of triplicate determinations. Statistically significant (P < 0.05) induction of cytokine release compared to treatment with medium plus 5% FBS is indicated by an asterisk.
FIG. 4.
Cytokine induction in human gingival epithelial cells in the presence of soluble CD14 and/or LBP. HGECs were challenged with TLR2 agonists (Pam3 or FSL-1; 1 μg/ml) or FimA (10 μg/ml) in the presence of soluble human recombinant CD14 (0.5 μg/ml) and/or LBP (0.05 μg/ml) for 4 h at 37°C. Cell culture supernatants were assayed for IL-8 (A), IL-6 (B), and GM-CSF (C) by use of Luminex 100 technology (Upstate Cell Signaling Solutions, NY). Data are presented as the means ± standard deviations of triplicate determinations. Statistically significant (P < 0.05) induction of cytokine release is indicated by an asterisk.
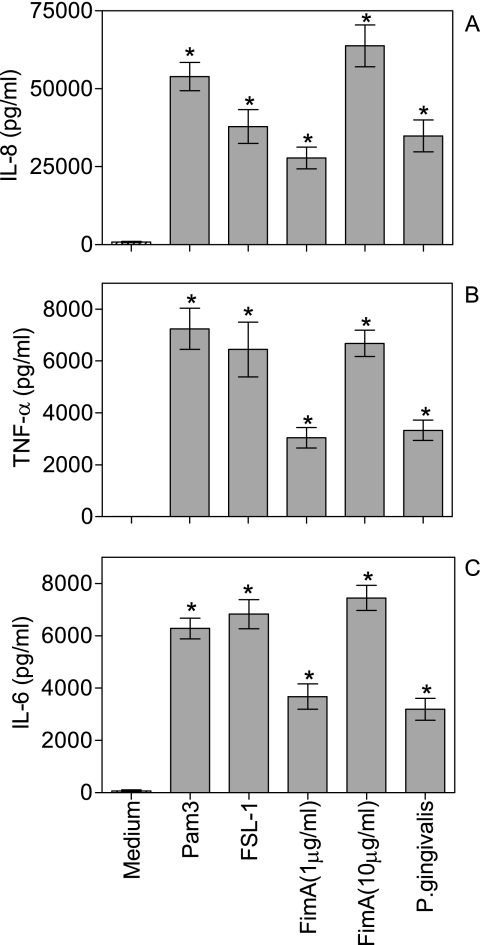
P. gingivalis and FimA activate human monocytes.
In contrast to gingival epithelial cells, human monocytes readily responded with IL-6, IL-8, and TNF-α expression at the protein level when challenged with purified FimA (Fig. 5). In addition to the 10-μg/ml concentration (also used for epithelial cell activation), a 1-μg/ml concentration of FimA was also proinflammatory (Fig. 5). The highest FimA-induced cytokine responses were comparable to those induced by the TLR2 agonists Pam3Cys and FSL-1 (both used at 1 μg/ml) and were higher than the responses induced by P. gingivalis at an MOI of 100:1 (Fig. 5). Therefore, P. gingivalis FimA is not inherently noninflammatory but may activate or not a particular cell type, depending on its repertoire of expressed PRRs.
FIG. 5.
Cytokine induction in human primary monocytes. Purified human primary monocytes were challenged by TLR2 agonists (Pam3 or FSL-1; 1 μg/ml), FimA (1 or 10 μg/ml) or P. gingivalis (MOI, 100:1) for 16 h at 37°C. Culture supernatants were assayed for IL-8 (A), TNF-α (B), and IL-6 (C) responses using enzyme-linked immunosorbent assay kits. Data are presented as the means ± standard deviations of triplicate determinations. Statistically significant (P < 0.05) induction of cytokine release is indicated by an asterisk.
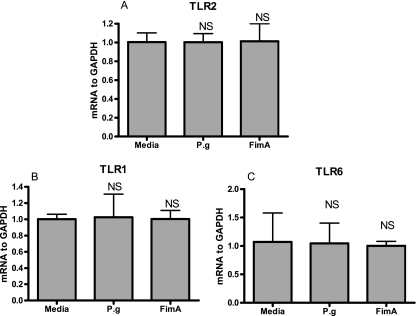
TLR expression in human gingival epithelial cells.
To rule out the possibility that the lack of stimulatory effect on epithelial cells by P. gingivalis FimA is due to possible downregulation of expression of TLR2 and/or its signaling partners (TLR1 and TLR6), we investigated the expression of these PRRs in epithelial cells upon exposure to FimA. Expression of TLR1, -2, and -6 at the mRNA level was not influenced following incubation of epithelial cells exposed to FimA for 4 h (Fig. 6) and 12 h (data not shown).
FIG. 6.
P. gingivalis FimA does not influence TLR1, -2, and -6 expression in human gingival epithelial cells. HGECs were challenged with either P. gingivalis (P.g; MOI, 100:1) or FimA (10 μg/ml) for 4 h at 37°C. Real-time PCR was performed with an ABI 7500 system (Applied Biosystems). TaqMan probes and sense and antisense primers for gene expression of human TLR1, -2, and -6 were purchased from Applied Biosystems along with probes and primers for the human endogenous control, GAPDH. Using a Universal PCR Master Mix (Applied Biosystems), the reactions were carried out according to the manufacturer's protocol. The ratio of TLR2 (A), TLR1 (B), and TLR6 (C) mRNAs was normalized to GAPDH mRNA. Data are presented as the means ± standard deviations of triplicate determinations. NS, not statistically significant.
DISCUSSION
Periodontal disease is an infectious chronic inflammatory condition initiated by a multitude of microorganisms, among which P. gingivalis is considered to be a major pathogen (25, 26). Electron microscopy studies have revealed that FimA constitutes a prominent surface molecule of P. gingivalis (11), perhaps being the first molecule of this pathogen coming into contact with epithelial cells. In this regard, oral gingival epithelial cells provide a physical barrier against invading bacteria and play an important role in the host's innate defense (1). However, P. gingivalis has the ability to both adhere to and invade epithelial cells via a FimA-dependent process (17). Epithelial cells challenged with P. gingivalis secrete proinflammatory cytokines, including IL-6, IL-8, TNF-α, IL-1β, and GM-CSF (14, 23). Our current findings suggest that FimA does not significantly contribute to the P. gingivalis-induced proinflammatory response in epithelial cells. The lack of epithelial cell responsiveness to FimA was established using purified protein, and moreover, the ability of P. gingivalis to induce modest cytokine responses in epithelial cells was not tightly correlated with its fimbriation status, in contrast to the case with monocytes/macrophages (5, 6). The observation that FimA is essentially noninflammatory in gingival epithelial cells may represent a bacterial strategy for utilizing an adhesin for colonization and/or invasion without the necessary contact of FimA and epithelial cells resulting in a robust innate response, which could eliminate the invading pathogen. An alternative consideration is that the lack of response to FimA is a tactic employed by the host to prevent continual activation and thus chronic inflammation, although our working hypothesis is that reduced perturbation of epithelial cells is a bacterial strategy to enhance colonization. We have confirmed that the cytokine induction in human gingival epithelial cells challenged by the FimA-deficient mutant of P. gingivalis was upregulated except for IL-6. IL-6 is not as strictly dependent on NF-κB as the other three cytokines examined, since the IL-6 gene in epithelial cells contains cyclic AMP-responsive elements that are important for its transcriptional regulation (15). Thus, it is possible that although FimA itself did not induce substantial IL-6 levels (the apparent increase compared to basal levels is not statistically significant), it helped P. gingivalis (the wild type but not of course the FimA-deficient mutant) to come into closer contact with GECs by virtue of its adhesin function. Under these conditions, the interaction of another surface molecule with the GECs may have been enhanced, resulting in activation of a cyclic AMP-dependent pathway for inducing an IL-6 response.
The ability of P. gingivalis FimA to activate monocytes/macrophages is strongly dependent on the presence of CD14 and TLR2 (6, 9). Mouse macrophages deficient in either CD14 or TLR2 fail to respond to FimA (4, 6). We have previously shown that the human colonic epithelial cell line SW620 does not respond to P. gingivalis FimA even when TLR2 is cotransfected with TLR1 or TLR6 as signaling partners (6). These transfectants fail to respond to FimA even in the presence of serum, which is a source of soluble CD14 (6). We have now shown that the same concept applies to a more physiologically relevant epithelial cell type, namely primary gingival epithelial cells. Indeed, even the addition of purified soluble CD14, with or without LBP, was not sufficient for supporting FimA-induced gingival epithelial cell activation. The TLR2-transfected SW620 cells became highly responsive to P. gingivalis FimA only upon cotransfection with CD14, indicating a strong requirement for membrane-expressed CD14 in TLR2-dependent cell activation by FimA. In stark contrast, the presence of membrane CD14 was not essential for the abilities of Pam3Cys and MALP-2 (FSL-1) to activate TLR2/1- and TLR2/6-dependent cell activation, respectively, in transfected SW620 cells (6). The failure of FimA to activate an inflammatory response in HGECs would be expected to dramatically affect the proinflammatory potential of P. gingivalis, without this, however, resulting in complete abrogation of the host response (being activated by other bacterial surface molecules) as shown by our findings.
The inability of FimA to utilize soluble CD14 for epithelial cell activation stands in sharp contrast to the ability of lipopolysaccharide to readily activate CD14-negative cells in the presence of soluble CD14 (6, 28). In this regard, FimA behaves similarly to peptidoglycan, which fails to activate CD14-negative cells even in the presence of soluble CD14, although the same ligand becomes proinflammatory when interacting with CD14-expressing cells (12). The biochemical basis for the inability of FimA to utilize soluble CD14 for TLR2 activation is currently uncertain. However, membrane CD14-mediated TLR2 activation by FimA requires formation of FimA-CD14-TLR2 complexes in lipid rafts, as suggested by fluorescence resonance energy transfer studies (6, 7). A plausible explanation, therefore, is that the binding of FimA to soluble CD14 blocks the ability of either CD14 or FimA itself to efficiently interact with TLR2. In this respect, it has been argued that the critical regions of CD14 for ligand binding may differ between the soluble and membrane forms of this receptor (12, 28). It was previously shown that an immortalized gingival epithelial cell line that expresses TLR2 responds to P. gingivalis FimA with IL-8 induction in the presence of soluble CD14 and LBP (2). These findings appear to be in conflict with our present findings. It is possible, however, that certain immortalized cell lines may behave differently than primary cells, although the precise mechanism involved is uncertain.
Our results further show that the lack of FimA stimulatory effect on gingival epithelial cells cannot be attributed to downregulation of TLR2 or its signaling partners (TLR1 and TLR6). In conclusion, CD14-nonexpressing human primary gingival epithelial cells are relatively hyporesponsive to P. gingivalis FimA, in contrast to human primary monocytes. On the other hand, other TLR2 agonists without a strict requirement for membrane CD14 (Pam3Cys and FSL-1) are proinflammatory in both cell types. It appears possible that P. gingivalis may come in close association with gingival epithelial cells through contact with its FimA without activating a robust FimA-mediated innate defense response, as would be expected to occur in monocytes/macrophages.
Acknowledgments
This study was supported by grants HRSAC76, HF01199-01, and CDC-PA04189 (to D.F.K.), and U.S. Public Health Service grant DE015254 (to G.H.).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 21 November 2006.
REFERENCES
- 1.Andrian, E., D. Grenier, and M. Rouabhia. 2006. Porphyromonas gingivalis-epithelial cell interactions in periodontitis. J. Dent. Res. 85:392-403. [DOI] [PubMed] [Google Scholar]
- 2.Asai, Y., Y. Ohyama, K. Gen, and T. Ogawa. 2001. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect. Immun. 69:7387-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillot, L., S. Medjane, K. Le-Barillec, V. Balloy, C. Danel, M. Chignard, and M. Si-Tahar. 2004. Response of human pulmonary epithelial cells to lipopolysaccharide involves Toll-like receptor 4 (TLR4)-dependent signaling pathways: evidence for an intracellular compartmentalization of TLR4. J. Biol. Chem. 279:2712-2718. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis, G., P. Ratti, and E. Harokopakis. 2005. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. J. Biol. Chem. 280:38902-38913. [DOI] [PubMed] [Google Scholar]
- 5.Hajishengallis, G., H. Sojar, R. J. Genco, and E. DeNardin. 2004. Intracellular signaling and cytokine induction upon interactions of Porphyromonas gingivalis fimbriae with pattern-recognition receptors. Immun. Investig. 33:157-172. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis, G., R. I. Tapping, E. Harokopakis, S.-I. Nishiyama, P. Ratti, R. E. Schifferle, E. A. Lyle, M. Triantafilou, K. Triantafilou, and F. Yoshimura. 2006. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell. Microbiol. 8:1557-1570. [DOI] [PubMed] [Google Scholar]
- 7.Hajishengallis, G., M. Wang, E. Harokopakis, M. Triantafilou, and K. Triantafilou. 2006. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect. Immun. 74:5658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanazawa, S., Y. Kawata, Y. Murakami, K. Naganuma, S. Amano, Y. Miyata, and S. Kitano. 1995. Porphyromonas gingivalis fimbria-stimulated bone resorption in vitro is inhibited by a tyrosine kinase inhibitor. Infect. Immun. 63:2374-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harokopakis, E., and G. Hajishengallis. 2005. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur. J. Immunol. 35:1201-1210. [DOI] [PubMed] [Google Scholar]
- 10.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 11.Hongo, H., H. Takano, and M. Morita. 6 September 2006. Dense fimbrial meshwork enhances Porphyromonas gingivalis adhesiveness: a scanning electron microscopic study. J. Periodont. Res. doi: 10.1111/j.1600-0765.2006.00922.x. [DOI] [PubMed]
- 12.Jin, Y., D. Gupta, and R. Dziarski. 1998. Endothelial and epithelial cells do not respond to complexes of peptidoglycan with soluble CD14 but are activated indirectly by peptidoglycan-induced tumor necrosis factor-alpha and interleukin-1 from monocytes. J. Infect. Dis. 177:1629-1638. [DOI] [PubMed] [Google Scholar]
- 13.Kesavalu, L., B. Chandrasekar, and J. L. Ebersole. 2002. In vivo induction of proinflammatory cytokines in mouse tissue by Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 17:177-180. [DOI] [PubMed] [Google Scholar]
- 14.Kinane, D. F., H. Shiba, P. G. Stathopoulou, H. Zhao, D. F. Lappin, A. Singh, M. A. Eskan, S. Beckers, S. Waigel, B. Alpert, and T. B. Knudsen. 2006. Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun. 7:190-200. [DOI] [PubMed] [Google Scholar]
- 15.Krueger, J., A. Ray, I. Tamm, and P. B. Sehgal. 1991. Expression and function of interleukin-6 in epithelial cells. J. Cell. Biochem. 45:327-334. [DOI] [PubMed] [Google Scholar]
- 16.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont, R. J., and O. Yilmaz. 2002. In or out: the invasiveness of oral bacteria. Periodontol. 2000 30:61-69. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J. Y., H. T. Sojar, A. Amano, and R. J. Genco. 1995. Purification of major fimbrial proteins of Porphyromonas gingivalis. Protein Expr. Purif. 6:496-500. [DOI] [PubMed] [Google Scholar]
- 19.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami, Y., S. Hanazawa, A. Watanabe, K. Naganuma, H. Iwasaka, K. Kawakami, and S. Kitano. 1994. Porphyromonas gingivalis fimbriae induce a 68-kilodalton phosphorylated protein in macrophages. Infect. Immun. 62:5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa, T., Y. Asai, M. Hashimoto, and H. Uchida. 2002. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 32:2543-2550. [DOI] [PubMed] [Google Scholar]
- 22.Saito, A., H. T. Sojar, and R. J. Genco. 1997. Interleukin-1 gene expression in macrophages induced by surface protein components of Porphyromonas gingivalis: role of tyrosine kinases in signal transduction. Oral Microbiol. Immunol. 12:135-140. [DOI] [PubMed] [Google Scholar]
- 23.Sandros, J., C. Karlsson, D. F. Lappin, P. N. Madianos, D. F. Kinane, and P. N. Papapanou. 2000. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J. Dent. Res. 79:1808-1814. [DOI] [PubMed] [Google Scholar]
- 24.Shiba, H., S. G. Venkatesh, S. U. Gorr, G. Barbieri, H. Kurihara, and D. F. Kinane. 2005. Parotid secretory protein is expressed and inducible in human gingival keratinocytes. J. Periodontal. Res. 40:153-157. [DOI] [PubMed] [Google Scholar]
- 25.Slots, J., L. Bragd, M. Wikstrom, and G. Dahlen. 1986. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J. Clin. Periodontol. 13:570-577. [DOI] [PubMed] [Google Scholar]
- 26.Socransky, S. S., A. D. Haffajee, J. M. Goodson, and J. Lindhe. 1984. New concepts of destructive periodontal disease. J. Clin. Periodontol. 11:21-32. [DOI] [PubMed] [Google Scholar]
- 27.Uehara, A., S. Sugawara, K. Watanabe, S. Echigo, M. Sato, T. Yamaguchi, and H. Takada. 2003. Constitutive expression of a bacterial pattern recognition receptor, CD14, in human salivary glands and secretion as a soluble form in saliva. Clin. Diagn. Lab. Immunol. 10:286-292. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Viriyakosol, S., J. C. Mathison, P. S. Tobias, and T. N. Kirkland. 2000. Structure-function analysis of CD14 as a soluble receptor for lipopolysaccharide. J. Biol. Chem. 275:3144-3149. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]