Abstract
We tested 413 water buffalo cows (142 cases and 271 controls) for the presence of anti-Brucella abortus antibodies (by the skin test, the agglutination test, and the complement fixation test) and the Nramp1 genotype (by capillary electrophoresis). Four alleles (Nramp1A, -B, -C, and -D) were detected in the 3′ untranslated region of the Nramp1 gene. The BB genotype was represented among only controls, providing evidence that this genotype confers resistance to Brucella abortus. The monocytes from the BB (resistant) subjects displayed a higher basal level of Nramp1 mRNA and a lower number of viable intracellular bacteria than did the monocytes from AA (susceptible) subjects. The higher basal level of the antibacterial protein Nramp1 most probably provides the BB animals with the possibility of controlling bacteria immediately after their entry inside the cell.
The mouse gene Nramp1 (natural resistance-associated macrophage protein 1), also known as Slc11a1 (solute carrier family 11 member a1), determines the resistance or susceptibility of the host to the intracellular pathogens Mycobacterium bovis bacillus Calmette-Guérin, Salmonella enterica serovar Typhimurium, and Leishmania donovani (19). The bovine homolog of the mouse Nramp1 gene determines the resistance or susceptibility of cattle to Brucella abortus, also an intracellular pathogen (5). The transfection of the resistance-associated murine or bovine Nramp1 allele into the susceptible RAW 264.7 macrophage cell line inhibits the intracellular replication of serovar Typhimurium (19) or B. abortus (5), respectively. These results assign a critical role to the Nramp1 gene in the innate defense against intracellular infections. The product of Nramp1 functions as a transporter of Fe2+ and other divalent cations. The direction of transport of the cations remains however controversial: it is not clear whether the Nramp1 protein elevates the concentration of Fe2+ in the phagosome to favor the production of the antibacterial hydroxyl radical or, on the contrary, deprives the intraphagosomal bacterium of Fe2+ needed by the invading pathogen to survive within the phagosome (2). The Nramp1 gene is conserved in mammals, plants, insects, worms, and bacteria (11). Its presence in bacteria suggests that the intracellular pathogen and host may compete for the same nutrient (21).
Recently, to investigate the possible role of the Nramp1 gene in the resistance of water buffaloes to B. abortus infection, the 3′ untranslated region (UTR) of the gene was analyzed for polymorphism by denaturing gradient gel electrophoresis. Homozygosity for the Nramp1B allele was found to be associated with resistance to B. abortus infection (7). Given the concern for the false-positive results characterizing association studies in general (4, 9, 28), a replication of the original report seemed crucial. The present study, carried out on a larger and independent group of animals and using an independent technique (capillary electrophoresis), confirms the initial data. The study also provides biological support for the association between Nramp1 gene activity and resistance to the disease. The interest of the study goes beyond water buffalo brucellosis. The ubiquitous Nramp1 gene can be used to select goats and sheep resistant to B. melitensis, the agent responsible for most cases of human brucellosis (14).
MATERIALS AND METHODS
Study design.
Allele segregation (at the Nramp1 locus and eight microsatellite marker loci) was studied on 166 water buffalo triads (father, mother, and offspring). The animals forming the triads were not included in the association study as they belonged to an experimental herd free from brucellosis. Cases were subjects positive for brucellosis by the skin test, the agglutination test, and the complement fixation test. Controls were animals negative by the same tests. Cases and controls (142 and 271 subjects, respectively) were randomly drawn from a list of about 1,000 lactating cows distributed in three herds located in the province of Caserta (Italy). Cows were all unvaccinated and ear tagged. Herds were characterized by a high incidence of brucellosis (up to 40% of the subjects were positive by the agglutination and complement fixation tests). Cases and controls were therefore homogeneous in terms of environmental exposure and sex. Genotype analysis was carried out without knowing the results of the brucellosis tests. To avoid stratification (33), cases of brucellosis and controls were drawn in equal proportion (47 cases and 90 controls) from each herd.
Identification of Nramp1 alleles.
Capillary electrophoresis was carried out using the PE-Applied Biosystems ABI PRISM 310 analyzer equipped with a 47-cm-long and 50-μm-wide capillary. The separation medium was the POP-4 polymer. The 3′ untranslated region, nucleotide positions 1745 to 1955, of the water buffalo Nramp1 gene was amplified using the forward primer 5′-GTGGAATGAGTGGGCACAGT-3′ and the reverse primer 5′-CTCTCCGTCTTGCTGTGCAT-3′ (24). The forward primer was labeled with the fluorescent dye 6-carboxyfluorescein. PCR was carried out in 25 μl containing 1× GeneAmp PCR Gold buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 0.4 μM of each primer, 1 U of AmpliTaq Gold DNA polymerase, and 5 μl of DNA solution (0.5 to 2 ng/μl). PCRs were run with the following program: an initial step of 10 min at 95°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, and a final extension step of 7 min at 72°C. The PCR product (1 μl) was added to 11.5 μl of deionized formamide (Applied Biosystems, Foster City, CA) and 0.5 μl of GeneScan 6-carboxy-X-rhodamine 500 size standard. The samples were incubated at 94°C for 3 min, cooled at 4°C, and then loaded on the ABI PRISM 310. Electrophoresis data were acquired with the ABI PRISM 310 collection software (Applied Biosystems). The size of alleles was determined using 310 GeneScan 3.1.2 and Genotyper 2.5.2 software (Applied Biosystems).
Determination of Nramp1 alleles nucleotide sequence.
PCR products from three subjects homozygous for the identified alleles, Nramp1A, -B, -C, and -D, were sequenced in both directions. The nucleotide sequence was determined using version 2.0 of the Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and the ABI 310 PRISM genetic analyzer (Applied Biosystems). The length of the capillary was 47 cm, and the section was 50 μm. The separation medium was the POP-6 polymer (Applied Biosystems). The sequence data were analyzed using GeneScan 3.1.2 and Sequencing Analysis 3.4.1 software (Applied Biosystems).
Detection of microsatellite markers.
DNA from the 413 subjects included in the association study were analyzed using eight microsatellite markers. Markers were amplified by using the primer pairs listed below and the following PCR program: an initial step of 10 min at 95°C, followed by 30 cycles of 15 s at 95°C, 1 min at 57°C, 1 min at 72°C, and a final extension step of 10 min at 72°C. PCR products were separated by capillary electrophoresis on ABI PRISM 310 analyzer (Applied Biosystems, Foster City, CA). Results were analyzed with the GeneScan 3.1.2 and Genotyper 2.5.2 programs (Applied Biosystems). The primers used to amplify the microsatellite primers were forward primer 5′-TTG TCA GCA ACT TCT TGT ATC TTT-3′ and reverse primer 5′-TGT TTT AAG CCA CCC AAT TAT TTG-3′ (CSSM19); forward primer 5′-GGG AAG GTC CTA ACT ATG GTT GAG-3′ and reverse primer 5′-ACC CTC ACT TCT AAC TGC ATT GGA-3′ (CSSM42); forward primer 5′-TCT CTG TCT CTA TCA CTA TAT GGC-3′ and reverse primer 5′-CTG GGC ACC TGA AAC TAT CAT CAT-3′ (CSSM47); forward primer 5′-GGA GGG TTA CAG TCC ATG AGT TTG-3′ and reverse primer 5′-TCG CGA TCC AAC TCC TCC TGA AG-3′ (CYP21); forward primer 5′-TAC TCG TAG GGC AGG CTG CCT G-3′ and reverse primer 5′-GAG ACC TCA GGG TTG GTG ATC AG-3′ (D5S2); forward primer 5′-AGG AAT ATC TGT ATC AAC CTC AGT C-3′ and reverse primer 5′-CTG AGC TGG GGT GGG AGC TAT AAA TA-3′ (INRA006); forward primer 5′-AAA GGC CAG AGT ATG CAA TTA GGA G-3′ and reverse primer 5′-CCA CTC CTC CTG AGA ATA TAA CAT G-3′ (MAF65); and forward primer 5′-CAG CAA AAT ATC AGC AAA CCT-3′ and reverse primer 5′-CCA CCT GGG AAG GCC TTT A-3′ (RM4).
Brucella abortus transformation.
The plasmid pBBR1MCS-6Y (31) carrying the green fluorescent protein (GFP) gene constitutively expressed in B. abortus was kindly provided by M. E. Kovach (Baldwin-Wallace College, Berea, OH). The plasmid was introduced into B. abortus 2308 by electroporation, and the transformed bacteria (GFP-B. abortus) were grown in tryptose soy broth supplemented with 15 μg/ml chloramphenicol.
In vitro infection of peripheral blood mononuclear cells with GFP-B. abortus.
Peripheral blood mononuclear cells were separated by density gradient centrifugation (Lympholyte-Mammal; Cederlane, Hornby, Ontario, Canada; 1,200 × g for 20 min), distributed in 24-well plates (5 × 106 cells/well), and incubated overnight (37°C; 5% CO2) in Dulbecco's modified Eagle medium (DMEM) supplemented with water buffalo serum (4%) and penicillin and streptomycin (100 IU/ml). Wells were washed with DMEM to remove nonadherent cells. Cells were fed with DMEM medium supplemented with 10% water buffalo serum and penicillin and streptomycin (100 IU/ml) until infected. The wells were washed with DMEM to remove the antibiotics and then infected with GFP-B. abortus (106 bacteria and 106 adherent cells in 500 μl volume/well). The plates were centrifuged (750 × g for 5 min) to facilitate cell contact and then incubated for 30 min at 37°C in 5% CO2. Extracellular bacteria were killed with gentamicin (12.5 μg/well). Cells were washed with DMEM, gently scraped from the tissue culture wells, and analyzed by fluorescent microscopy (Leica DMRA; Wetzlar, Germany) or flow cytometry (Epics Elite flow cytometer; Coulter, Miami, Florida).
Viability of intracellular bacteria.
Monocytes were infected with B. abortus 2308 as described above. Following incubation (12 to 48 h), monocytes were washed with phosphate-buffered saline (PBS), lysed with 0.50% Tween 20 (40 μl/600 μl), washed again with PBS (to remove Tween 20), plated for CFU counting or stained with 100 nM SYTO9 and 15 μM propidium iodide (Molecular Probes, Eugene, Oregon) for 15 min in the dark, and analyzed by flow cytometry. For bacterial enumeration by flow cytometry, a fixed volume (75 μl) of sample was analyzed (22). The two counting methods (flow cytometry and CFU counting) displayed high correlation (y = 1.06x − 91,236; R2 = 0.99). In the assay, viable bacteria stain green, whereas dead bacteria stain red. The assay was carried out according to the manufacturer's instructions.
Influence of the Nramp1 genotype on milk yield.
Milk yield was determined from monthly sampling collected by representatives of the National Dairy Association. Individual milk yields were normalized (to the milk yield of a water buffalo cow at its fifth lactation, milked twice per day, with a lactation length of 270 days) using PUMA software (www.aia.it). Milk yield differences between genotypes were analyzed by Student's t test.
Assay of the reactive oxygen intermediates.
When the fluorochrome 2′,7′-dichlorofluorescein diacetate (DCF) crosses the cell membrane, it undergoes deacetylation by intracellular esterases and becomes nonfluorescent. Upon oxidation by reactive oxygen intermediates (ROIs), DCF becomes again fluorescent (6). To measure the production of ROIs, monocytes (106 cells suspended in 250 μl PBS) were incubated with DCF (Sigma) (final concentration, 0.4 μM) at 37°C for 15 min in humidified atmosphere in the presence of 5% CO2, washed with PBS, resuspended in the same medium (106 monocytes/250 μl), infected with B. abortus (106 CFU for 20 min), washed with PBS, and finally analyzed by flow cytometry.
Assay of the reactive nitrogen intermediates (RNIs).
The test was carried out as previously described (15). Briefly, monocytes (106/well), noninfected or infected with B. abortus (106 CFU/well for 48 h), were suspended in DMEM medium containing 10% water buffalo serum. The cell culture supernatant (100 μl) was mixed with an equal volume of Griess reagent (15) and incubated at room temperature for 10 min, and the absorbance was read at 550 nm in a spectrophotometer (Bio-Rad, Hercules, CA) using sodium nitrite (5 μM to 80 μM) as the standard.
Ultrastructural analysis of B. abortus-infected cells.
Monocytes (106) were infected with B. abortus (108 CFU suspended in 1 ml medium) for 5 min at 37°C, washed with PBS to remove nonadherent bacteria, and incubated at 37°C for 15 min, 2 h, or 24 h. Cells were then fixed with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), washed with the same buffer, and postfixed with 1% OsO4 (1 h). The pellet was then dehydrated with ethanol and embedded in Epon 812 resin. Following polymerization (70°C for 24 h), the ultrathin sections were stained on grid with uranyl acetate, followed by lead citrate, and examined with a Zeiss (Milan, Italy) type 902A electron microscope at 80 kV.
Other procedures.
The skin test was carried out using the Brucellergene OCB (Synbiotics, Lyon, France) according to the manufacturer's instructions. Agglutination and complement fixation tests were carried out as previously described (3). The Nramp1 expression level was determined as previously described (7). B. abortus DNA was identified by real-time PCR (34). The odds ratio, the confidence interval (CI) of the odds ratio, and data for Fisher's exact test and Student's t test were calculated as previously described (30). Before calculating the odds ratio, 0.5 was added to each of the four values since one of the values was 0 (30). Confidence intervals were calculated according to Woolf's method (30). Hardy-Weinberg equilibrium was calculated as previously described (10). The degrees of freedom of the χ2 test for Hardy-Weinberg equilibrium were calculated according to the formula k(k − 1)/2, where k is the number of alleles (39).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the four alleles can be found in the GenBank/DDBJ/EMBL databases under the accession numbers DQ095780, DQ095781, DQ376109, and DQ376110.
RESULTS
Allele identification at the Nramp1 locus.
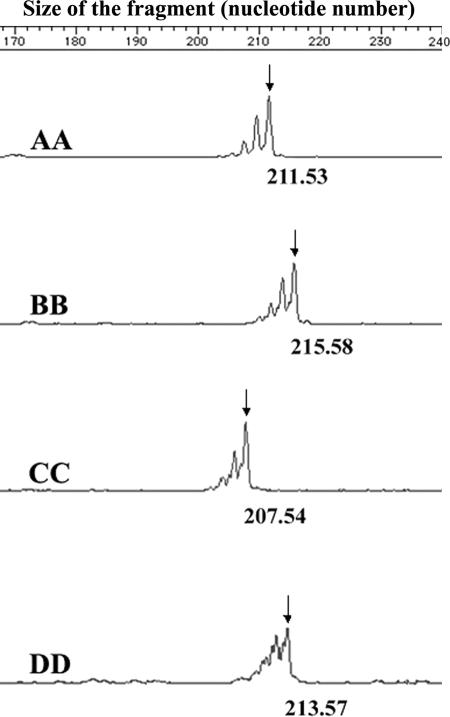
A preliminary study established an association between the Nramp1 BB genotype and the absence of anti-B. abortus antibodies in water buffaloes. The same study reported the identification of 6 out of 64 water buffaloes that were BB and positive for the B. abortus tests at the same time (7). As a limit in the analytical power of the technique used in the preliminary study (denaturing gradient gel electrophoresis) was suspected, 166 triads (father, mother, and offspring) and the 6 exceptional animals were tested by capillary electrophoresis. This technique displayed the presence of four alleles, Nramp1A, -B, -C, and -D, at the Nramp1 locus. Figure 1 shows the profile of subjects homozygous for each allele. Family data (the triads) indicated that the four alleles behave as codominant (data not shown). As for the six exceptional animals, four displayed the CD genotype and two displayed the BD genotype.
FIG. 1.
Graphic representation of the Nramp1A, -B, -C, and -D alleles obtained from an ABI PRISM 310 analyzer. The amplification reaction yields DNA fragments of slightly different sizes. The size of each allele is given by the highest (most representative) peak, which is marked with an arrow.
The BB genotype confers resistance to B. abortus infection.
A sample of 413 water buffaloes, independent of the sample included in the preliminary study (7), was analyzed by capillary electrophoresis. Genotype analysis showed that the BB homozygous subjects were all found among the 271 controls (the animals exposed to B. abortus but negative by the B. abortus tests) (Table 1). The data reported in Table 1 were used to calculate the odds ratio (the ratio of the odds of being positive to the B. abortus tests for the BB and the non-BB subjects). The odds ratio was 0.07, and its 95% CI was 0.004 to 1.139. Thus, BB animals are 7% as likely as non-BB animals to be positive to B. abortus tests. The same data were compared by Fisher's exact test. The two-sided P value was <0.0060, indicating that there would be a less than 0.6% chance of randomly picking animals with so much association if the BB genotype and the B. abortus-negative status were not associated.
TABLE 1.
Association between the BB genotype and resistance to B. abortus infectiona
| Genotype | No. of:
|
Total | |
|---|---|---|---|
| Cases | Controls | ||
| BB | 0 | 13 | 13 |
| Non-BB | 142 | 258 | 400 |
| Total | 142 | 271 | 413 |
The odds ratio was 0.07; the CI (calculated according to Woolf's method) was 0.004 to 1.139.
As determined by Fisher's exact test, the P value was <0.0060.
The Nramp1 alleles are not in Hardy-Weinberg equilibrium in seropositive water buffaloes.
The Hardy-Weinberg law states that, under certain assumptions (such as the absence of selection, stratification, or genetic drift), allele frequencies can be used to calculate the expected genotype frequencies (10). In the present study, this fundamental principle of population genetics was exploited to exclude the presence of stratification in the sample being studied, a condition which could vitiate the interpretation of the results (33), and to gain further supportive evidence for the association between the Nramp1 BB genotype and the B. abortus-negative status. The 142 brucellosis cases and 271 controls included in the association study were therefore screened separately for the deviation of genotype distribution from the Hardy-Weinberg law at eight polymorphic microsatellite marker loci and the Nramp1 locus. Genotype frequencies at the marker loci were in Hardy-Weinberg equilibrium among brucellosis cases as well as among controls (P = 0.30 to 0.50), excluding the presence of stratification in the population sample being studied (data not shown). Genotype frequencies at the Nramp1 locus were in Hardy-Weinberg equilibrium among controls (P = 0.20) (Table 2), but not among brucellosis cases (P < 0.001) (Table 3). Since a disequilibrium generated by the failure of any of the assumptions of the Hardy-Weinberg law was expected to influence both cases and controls, the evidence that only the cases do not fulfill the Hardy-Weinberg law, supports the results from the case-control study.
TABLE 2.
Nramp1 genotype distribution among controlsa
| Genotype | Frequencies of seronegative animals
|
|
|---|---|---|
| Observed | Expected (formula for expected frequency) | |
| AA | 68 | 68 (n × p2) |
| AB | 73 | 68 (n × 2pq) |
| AC | 21 | 27 (n × 2pr) |
| AD | 42 | 41 (n × 2ps) |
| BB | 13 | 17 (n × q2) |
| BC | 11 | 13 (n × 2qr) |
| BD | 24 | 20 (n × 2qs) |
| CC | 5 | 3 (n × r2) |
| CD | 11 | 8 (n × 2rs) |
| DD | 3 | 6 (n × s2) |
| Total | 271 | 271 |
p, q, r, and s indicate the observed frequencies of the Nramp1A, -B, -C, and -D alleles, respectively. p = (AA + AB/2 + AC/2 + AD/2)/n = 0.50; q = (BB + AB/2 + BC/2 + BD/2)/n = 0.25; r = (CC + AC/2 + BC/2 + DC/2)/n = 0.10; s = (DD + AD/2 + BD/2 + CD/2)/n = 0.15. n (number of observed animals) = 271. Degrees of freedom (DF) were calculated according to the formula k(k-1)/2, where k is the number of alleles (39). χ2 = {∑[(observed frequency − expected frequency)2/expected frequency]} = 8.9; DF = 6; P = 0.20.
TABLE 3.
Nramp1 genotype distribution among brucellosis casesa
| Genotype | Frequencies of seropositive animals
|
|
|---|---|---|
| Observed | Expected (formula for expected frequency) | |
| AA | 71 | 56 (n × p2) |
| AB | 24 | 23 (n × 2pq) |
| AC | 2 | 18 (n × 2pr) |
| AD | 10 | 27 (n × 2ps) |
| BB | 0 | 2 (n × q2) |
| BC | 2 | 3 (n × 2qr) |
| BD | 10 | 5 (n × 2qs) |
| CC | 3 | 1 (n × r2) |
| CD | 17 | 4 (n × 2rs) |
| DD | 3 | 3 (n × s2) |
| Total | 142 | 142 |
p, q, r, and s indicate the observed frequencies of the Nramp1A, -B, -C, and -D alleles, respectively. p = (AA + AB/2 + AC/2 + AD/2)/n = 0.63; q = (BB + AB/2 + BC/2 + BD/2)/n = 0.13; r = (CC + AC/2 + BC/2 + DC/2)/n = 0.10; s = (DD + AD/2 + BD/2 + CD/2)/n = 0.15. n (number of observed animals) = 142. Degrees of freedom (DF) were calculated according to the formula k(k-1)/2, where k is the number of alleles (39). χ2 = {∑[(observed frequency − expected frequency)2/expected frequency]} = 79.8; DF = 6; P < 0.001.
Culling of seropositive animals increases the frequency of the BB genotype.
The infection of the BB water buffaloes with a virulent B. abortus strain, an experiment which could provide direct proof of the protection conferred by the BB genotype, was vetoed by the sanitary authority (7). Field studies, however, produced dividends in another direction, providing the possibility of testing a herd where (thanks to the goodwill of the owner) a control program against brucellosis had been in operation for several years. The approach consisted of the rapid and systematic culling of the subjects who were positive for brucellosis by the serological tests (agglutination and complement fixation). In this herd, now brucellosis free, about 17% (36 out of 215) of the lactating cows are BB, a considerably higher percentage than that (4.8%, or 13 out of 271) found among the seronegative lactating cows included in the present study (Table 2). Although the frequency of the BB genotype before the control program was started is not known, the result, as it stands, suggests that the BB genotype may well have played a central role in the outcome of B. abortus infection, at least in this herd.
Nramp1 expression level in AA and BB monocytes.
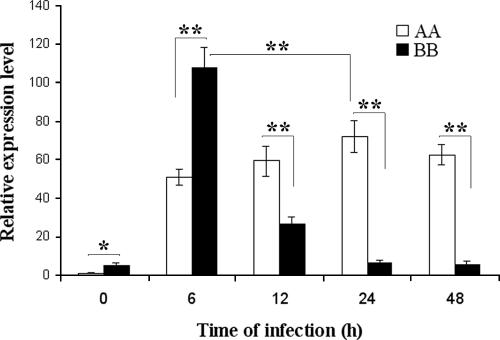
The level of the Nramp1 messenger expressed by the AA and BB monocytes was measured by real time PCR before and after in vitro infection with B. abortus 2308 (106 CFU/well). The experiment was carried out on 10 susceptible (AA) and 10 resistant (BB) animals, all negative by the B. abortus tests. Each animal was tested in two independent experiments, each time in triplicate. The two blood samples were collected at 1-month intervals. Noninfected BB monocytes displayed basal levels of Nramp1 messenger approximately fivefold higher than those of the noninfected AA monocytes. Upon infection, the level of the Nramp1 messenger in the BB monocytes peaked in 6 h and then declined to the basal level in approximately 24 h; in the AA monocytes, the level of the Nramp1 messenger peaked in 24 h and remained up-regulated (20 to 40 times the basal level) for the following 24 h. The peak level of the Nramp1 messenger was significantly higher in BB than in AA monocytes (Fig. 2).
FIG. 2.
Nramp1 mRNA levels in monocytes from AA or BB animals. The expression level of Nramp1 was measured before infection (time zero), and 6, 12, 24, and 48 h after B. abortus 2308 infection. Data are means ± standard deviations (error bars) of 10 AA and 10 BB subjects. Each subject was tested in triplicate in two independent experiments. Differences marked with one asterisk are significant (P ≤ 0.05); differences marked with two asterisks are highly significant (P ≤ 0.01).
Number of intracellular bacteria in AA and BB monocytes.
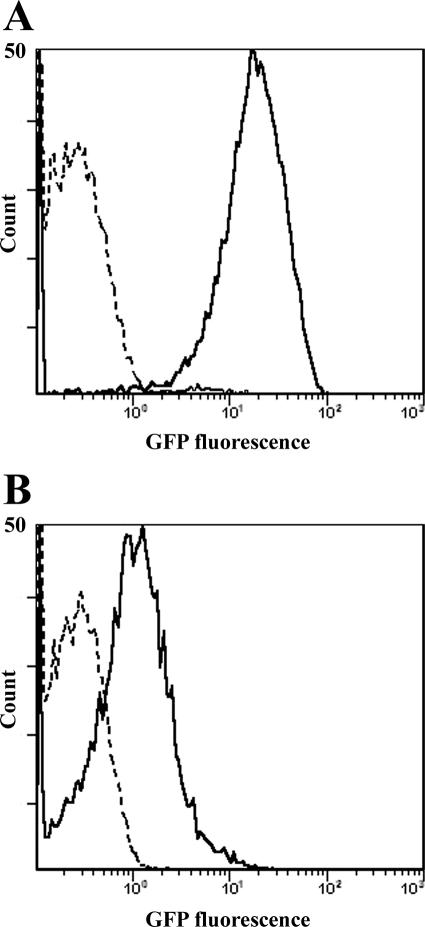
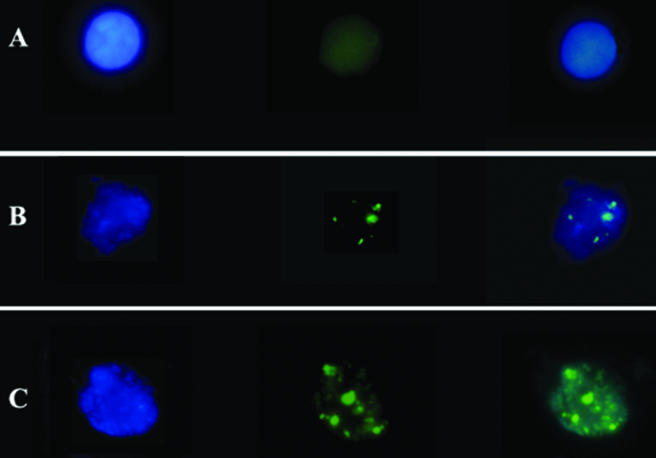
Real-time PCR experiments are often criticized for not being consistently reproducible (8, 25, 36). The biological activity of the AA and BB monocytes was therefore studied by additional approaches. The monocytes from AA and BB animals (the same used in the experiment described above) were infected in vitro with GFP-B. abortus (106 CFU/well) and then analyzed by flow cytometry and fluorescence microscopy. Both techniques showed that at 24 h after infection, BB monocytes harbored a reduced number of intracellular bacteria compared with that by AA monocytes. Representative images of the results obtained testing the monocytes from 10 AA and 10 BB subjects are shown in Fig. 3 and 4.
FIG. 3.
Representative flow cytometry profiles of monocytes from AA and BB water buffaloes, noninfected and infected in vitro with GFP-B. abortus. (A) Noninfected (dashed line) and infected (solid line) AA monocytes. (B) Noninfected (dashed line) and infected (solid line) BB monocytes. For each sample, at least 2 × 104 events were analyzed. The mean channels (calculated on 10 AA and 10 BB subjects, each tested twice in independent experiments) were 4 to 16 and 40 to 66, respectively. The mean channel of noninfected monocytes (AA or BB) was 2 to 3.
FIG. 4.
Representative fluorescent microscopy profiles of AA and BB monocytes, noninfected or infected in vitro with GFP-B. abortus for 24 h and then stained with 4′,6-diamino-2-phenylindole (DAPI). (A) Noninfected monocytes (AA and BB monocytes were indistinguishable). (B) Infected BB monocytes. (C) Infected AA monocytes. Left panel, cells analyzed with the 340-to-380-nm filter (DAPI). Central panel, cells analyzed with the 450-to-490-nm filter (GFP). Right panel, overlay; ×1,000 magnification and oil immersion. The experiment included 10 AA and 10 BB subjects, each tested twice in independent experiments.
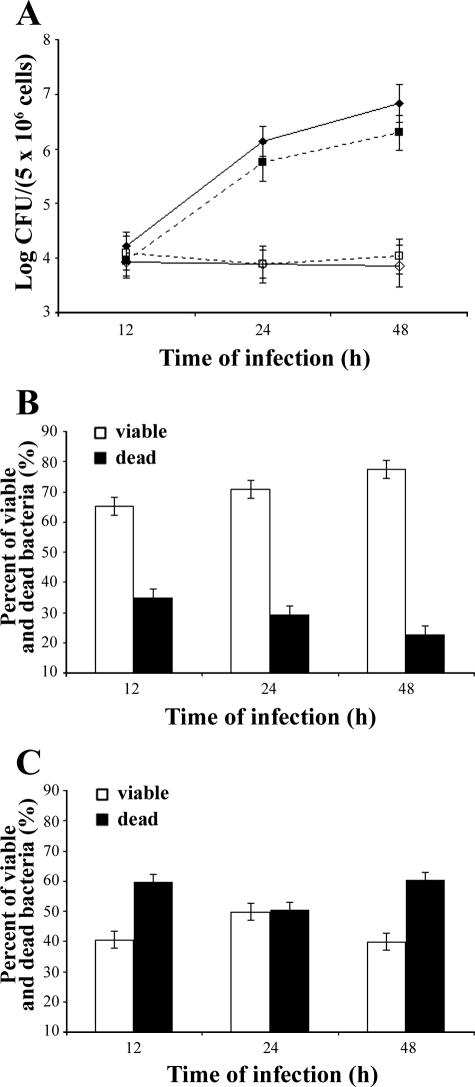
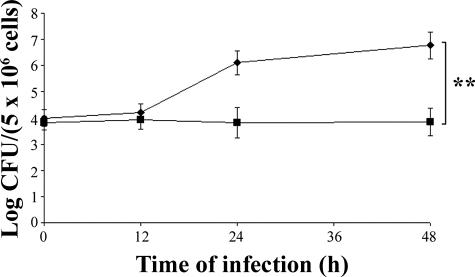
Next, the study focused on establishing whether BB monocytes killed intracellular bacteria more efficiently than did AA monocytes. To answer this question, the monocytes from AA and BB animals were infected with B. abortus 2308 and the percentage of viable (SYTO9 stained) intracellular bacteria was determined by flow cytometry at 12, 24, and 48 h after infection. Within this time frame, the percentage of viable intracellular bacteria rose from 62% ± 2.95 to 80% ± 4.06 in the AA monocytes, while it remained almost constant (about 40% ± 2.80) in the BB monocytes (Fig. 5). The viable bacteria present inside the AA and BB monocytes were also counted by the CFU method. The results (Fig. 6) confirm the capacity of the BB monocytes to control the replication of intracellular B. abortus more efficiently than AA monocytes do. Figures 5 and 6 show results from 10 AA and 10 BB subjects. Taken together, these results provide biological support for the association between the BB genotype and resistance to B. abortus infection, thus confirming epidemiological data.
FIG. 5.
Viable and dead intracellular bacteria recovered from AA or BB monocytes. Cells were infected in vitro with B. abortus 2308, incubated for 12 to 48 h, and then lysed. Intracellular bacteria were stained (with SYTO9 and propidium iodide) and analyzed by flow cytometry. SYTO9 stains viable cells, and propidium iodide stains dead cells. The experiment included 10 AA and 10 BB subjects, each tested twice in independent experiments. (A) Viable (⧫) and dead (▪) bacteria recovered from AA monocytes and viable (⋄) and dead (□) bacteria recovered from BB monocytes. (B) Percentages of viable and dead bacteria present in the AA monocytes. (C) Percentages of viable and dead bacteria present in the BB monocytes. Error bars indicate standard deviations.
FIG. 6.
Viable intracellular bacteria recovered from AA (⧫) or BB (▪) monocytes at 12 to 48 h after infection with B. abortus 2308. Bacteria were counted by the CFU method. The double asterisk marks a highly significant difference (P ≤ 0.01). The experiment included 10 AA and 10 BB subjects, each tested twice in independent experiments. Error bars indicate standard deviations.
Production of reactive oxygen intermediates and reactive nitrogen intermediates in AA and BB monocytes.
Upon activation by bacteria, macrophages exhibit an increased production of ROIs and RNIs, which are both the primary mediators of macrophage antibacterial activity (12). The monocytes from 10 BB subjects, noninfected as well as infected with B. abortus, displayed significantly higher ROI generation than the monocytes from 10 AA subjects (Table 4). The BB monocytes, when infected with B. abortus, also displayed a significantly higher production of RNIs. Noninfected AA and BB monocytes were instead indistinguishable (Table 5). Nonactivated monocytes in fact do not express nitric oxide synthase enzyme and therefore cannot produce measurable levels of RNIs (12).
TABLE 4.
Reactive oxygen intermediates production in AA and BB subjects
| Samples | Mean fluorescence channel for indicated genotypea
|
Pb | |
|---|---|---|---|
| AA | BB | ||
| Controlc | 1.10 ± 0.20 | 1.03 ± 0.15 | NS |
| Noninfected | 6.93 ± 1.20 | 17.77 ± 2.36 | 0.0209 |
| Infected | 28.67 ± 4.04 | 83.67 ± 6.66 | 0.0024 |
Values are the means ± standard deviations of 10 AA and 10 BB subjects, each tested twice in independent experiments.
P was calculated by Student's t test. NS, nonsignificant difference.
Control refers to cells not stained with DCF.
TABLE 5.
Reactive nitrogen intermediates production in AA and BB subjectsa
| Samples | Reactive nitrogen intermediates (μM) for indicated genotype
|
Pb | |
|---|---|---|---|
| AA | BB | ||
| Noninfected | 2.64 ± 0.13 | 3.14 ± 0.06 | NS |
| Infected | 25.86 ± 5.08 | 46.09 ± 7.80 | 0.0363 |
Figures are the means ± standard deviations of 10 AA and 10 BB subjects, each tested twice in independent experiments.
P was calculated by Student's t test. NS, nonsignificant difference.
Ultrastructural studies of AA and BB monocytes infected with B. abortus.
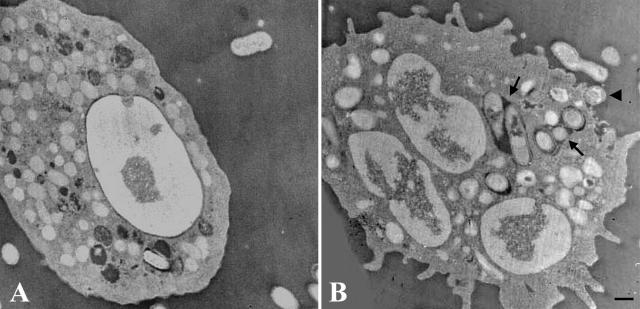
The intracellular traffic of B. abortus in AA and BB monocytes was also explored. At 2 h postinfection, individual phagosomes (characterized by walls tightly apposed to the bacteria) prevail in the AA monocytes, while phagolysosomes (spacious vesicles bearing one or more bacteria) prevail in the BB monocytes. In addition, BB monocytes show evident cell activation processes (Fig. 7). Thus, the Nramp1 gene apparently controls the intracellular bacterial replication by several means, such as the production of ROIs and RNIs and the activation of phagocytic cells.
FIG. 7.
Micrographs of AA and BB monocytes 2 h postinfection with B. abortus 2308. (A) AA monocytes contain almost exclusively phagosomes with apposed walls. (B) BB monocytes contain numerous phagolysosomes (arrows) and display evident cell activation processes. Note the phagosome formation around a bacterium (arrowhead). Images are representative of differences observed between three AA and three BB subjects. Bar, 1 μm.
Resistance to B. abortus is polygenic.
Upon incubation with GFP-B. abortus, the monocytes from 2 AA animals (2 out of 10) displayed flow cytometric profiles very close to those of the BB animals (a reduced number of intracellular bacteria). A posteriori, it was discovered that both these animals remained anti-B. abortus antibody negative (by the agglutination and complement fixation tests), although they were exposed to the pathogen for several years. The flow cytometric test was extended to 10 more animals (AA, AB, or CD) that shared the characteristic of remaining anti-B. abortus antibody-negative over several years of exposure to B. abortus. Again, the monocytes from these animals and those from BB animals were indistinguishable. A likely explanation is that, in addition to Nramp1, other genes might confer resistance to brucellosis. If this is the case, the infection of monocytes with GFP-B. abortus promises to become a valuable test for the identification of B. abortus-resistant animals.
Nramp1 alleles and milk production.
Host resistance to pathogen infection sometimes carries a fitness cost (40). Therefore, it seemed crucial to ascertain whether the BB genotype adversely affected milk yield, the most important production trait for water buffalo breeders. No difference was found in milk yield between BB and AC, AD, BD, or AC cows (t0.95, 0.08 to 1.3; degrees of freedom, 17 to 23).
The BB animals are Brucella DNA negative.
The persistence of Brucella over extended periods of time in individuals apparently free of disease is well documented in both ruminants (17, 35) and humans (41). Brucella DNA was sought in the blood of 10 BB animals. The Brucella DNA was examined by real-time PCR using an assay with a detection limit of 10 fg of Brucella DNA (five genome equivalents). Ten blood samples collected from each animal at an interval of approximately 2 weeks were all negative. The same assay detected the presence of Brucella DNA in the blood of 4 out of 10 anti-B. abortus antibody-positive subjects (antibody titer measured by the complement fixation test was 20 to 40 IU). The possibility that the 10 BB animals were all intermittent carriers cannot be excluded, but it seems remote. On the basis of the available evidence (negative results to the B. abortus tests and absence of Brucella DNA in the blood), the conclusion that BB water buffaloes are not carriers seems sufficiently prudent.
DISCUSSION
At present, the control of brucellosis (in water buffalo as well as in other species) is based on the serological identification and slaughter of infected (seropositive) animals. Latent infections, prolonged incubation periods of the disease, and inadequate protection provided by vaccination limit the success of this approach. We are exploring an alternative solution: the control of brucellosis by selective breeding. The results reported here demonstrate that the approach is feasible.
Case-control studies can detect associations between host genes and disease resistance very efficiently (4, 16, 27). The design of these studies is also conceptually simple: the frequency of the allele conferring resistance in a sample of cases is compared with the frequency in a sample of controls. The expectation is that the allele conferring resistance will display a higher frequency among the controls. However, well-designed association studies require the absence of stratification in the source population (33). Only in this case does the genotype distribution observed in the cases also represent the genotype distribution in the controls. Stratification occurs when the population under study contains genetically different groups (or strata) as a result of selection, inbreeding, or other forms of nonrandom mating. In the present study, the distribution of the alleles at eight distinct loci among cases and controls fulfills the Hardy-Weinberg law. Any potential bias introduced by stratification can therefore be excluded. In this context, the lack of Hardy-Weinberg equilibrium between cases and controls observed at the Nramp1 locus (Tables 2 and 3) becomes strong evidence for the correlation between the BB genotype and resistance to B. abortus infection. Actually, the test for Hardy-Weinberg disequilibrium in a gene bank of affected individuals has been proposed as a valid method of searching for disease-susceptible loci (16).
The biological plausibility of the candidate gene is also a critical requisite for an association study. Here the function of Nramp1 has been exploited in the AA (susceptible) and BB (resistant) animals by using independent approaches. The Nramp1 basal expression level was much higher in BB than in AA animals. When the monocytes were infected in vitro with B. abortus, the expression of Nramp1 lasted at a sustained level for about 24 h in the BB animals, but much longer in the AA animals (Fig. 2). These results suggest that the higher basal gene level gives the BB animals the opportunity to rapidly oppose the pathogen during the early hours of infection. The increased production of ROIs and RNIs (Tables 4 and 5) and earlier activation of the BB monocytes (Fig. 7) concur with the above interpretation and with the notion that innate immunity acts in the early hours after exposure to microorganisms (23). The evidence that the BB monocytes rapidly destroy the pathogen (Fig. 5 and 6) perhaps also suggests why BB animals do not form anti-B. abortus antibodies upon contact with the pathogen: the ingested bacteria are rapidly destroyed by phagocytes, and the inflammatory signaling is consequently too short to induce a systemic response (antibody production). What becomes striking here is the similarity in innate immune response between BB water buffaloes (resistant to B. abortus infection and specific antibody production) and that of CKR5Δ32/CKR5Δ32 individuals (resistant to human immunodeficiency virus infection and specific antibody production) (13).
No single serological test can reliably differentiate between B. abortus and other bacteria (in particular Yersinia enterocolitica O:9) that share antigenic epitopes with B. abortus (18). Here, cases and controls were diagnosed using a combination of tests. The cases were animals positive by the skin test, the complement fixation test, and the agglutination test. In particular, the skin test has been repeatedly shown to be the most specific test for brucellosis (18, 29). The controls were animals negative by the same tests. Many BB animals remained B. abortus antibody negative, though they were exposed for several years to B. abortus. This observation provides convincing evidence that BB animals are inherently resistant and not just subjects erroneously diagnosed as noninfected. At the same time, in view of the complex serological cross-reaction of B. abortus with other bacteria, we cannot formally exclude the possibility that pathogens other than B. abortus might have contributed to the higher frequency of the BB genotype observed in the herd where the culling of anti-B. abortus-positive subjects was carried out.
Several reports describing the Nramp1 polymorphisms in cattle, zebu, and the water buffalo have been published recently. The relevance of these reports to the present data deserves a comment. Paixao et al. (32) found that Holstein (Bos taurus taurus) and Indian zebu (Bos taurus indicus) subjects resistant to brucellosis display the 3′ UTR Nramp1 genotype associated with resistance to the disease. On the contrary, Kumar et al. (26) found that, in the Indian zebu and crossbred (Bos taurus indicus × Bos taurus taurus) cattle, the same genotype is not associated with resistance to brucellosis. The Holstein animals screened by Paixao et al. (32) (81 animals) and the zebu or crossbred animals tested by Kumar et al. (26) (100 animals) were all homozygous for the allele conferring resistance. In the absence of data showing that alleles at unrelated loci are in Hardy-Weinberg equilibrium in cases and controls, the excess of homozygosity at the Nramp1 locus points to either mistyping of genotypes or population stratification (33, 37). Since both of these conditions can lead to spurious conclusions (33, 37), the results from these studies (26, 32) require caution in their interpretation. Ables et al. (1) have described several Nramp1 single-nucleotide polymorphisms present in bovine and water buffalo breeds. The authors did not investigate a possible association between these polymorphisms and resistance to brucellosis. The variants, located in introns 4 and 5 and in exon 5 of the Nramp1 gene, are in any case distinct from the microsatellite polymorphism in the 3′ UTR described in this study. Finally, the evidence that the Nramp1 gene is not involved in the control of B. melitensis infection in mice (20) is not in conflict with the results reported here. First, the Nramp1 polymorphisms in water buffalo (this paper) and in mice (38) are distinct, and direct comparison is therefore questionable; second, although the pathogen persists in the macrophages of both species, the disease in ruminants is localized in the reproductive system, while in mice it is localized in the reticuloendothelial system. Phrased another way, the role of Nramp1 is very likely influenced by the host (mouse versus water buffalo).
In conclusion, this study demonstrates the feasibility of using selection to increase the frequency of genes providing resistance to infectious diseases. The approach may have a positive impact on the economics of the dairy industry and hopefully contribute to changing the culture of animal health control by slaughter.
Acknowledgments
We thank two anonymous reviewers for valuable comments on the manuscript; M. E. Kovach (Baldwin-Wallace College, Berea, OH) for the generous gift of the pBBR1MCS-6Y plasmid; Giuseppe Blaiotta (University of Naples Federico II) for help with electroporation; and Raffaele Garofalo and Giovanni Garofalo for collecting blood samples.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Ables, G. P., M. Nishibori, M. Kanemaki, and T. Watanabe. 2002. Sequence analysis of the NRAMP1 genes from different bovine and buffalo breeds. J. Vet. Med. Sci. 64:1081-1083. [DOI] [PubMed] [Google Scholar]
- 2.Alter-Kultunoff, M., S. Ehrlich, N. Dror, A. Azriel, M. Eilers, H. Hauser, H. Bowen, C. H. Barton, T. Tamura, K. Ozato, and B. Z. Levi. 2003. Nramp-1 mediated innate resistance to intraphagosomal pathogens is regulated by IRT-8, PU.1 and Miz-1. J. Biol. Chem. 278:44025-44032. [DOI] [PubMed] [Google Scholar]
- 3.Alton, G. G., W. H. Jones, and D. E. Pietz. 1975. Laboratory techniques in brucellosis, p. 64-124. WHO monograph series 55. World Health Organization, Geneva, Switzerland. [PubMed]
- 4.Anonymous. 1999. Freely associating. Nat. Genet. 22:1-2. [DOI] [PubMed] [Google Scholar]
- 5.Barthel, B., J. Feng, and J. A. Piedrahita. 2001. Stable transfection of the bovine NRAMP1 gene into murine RAW264.7 cells: effects on Brucella abortus survival. Infect. Immun. 69:3110-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass, D. A., J. W. Parce, L. R. Dechatelet, P. Szejda, M. C. Seeds, and M. Thomas. 1983. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J. Immunol. 130:1910-1917. [PubMed] [Google Scholar]
- 7.Borriello, G., R. Capparelli, M. Bianco, D. Fenizia, F. Alfano, F. Capuano, D. Ercolini, A. Parisi, S. Roperto, and D. Iannelli. 2006. Genetic resistance to Brucella abortus in water buffalo (Bubalus bubalis). Infect. Immun. 74:2115-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustin, S. A., V. Benes, and M. W. Pfaff. 2005. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 34:597-601. [DOI] [PubMed] [Google Scholar]
- 9.Cardon, L. R., and J. I. Bell. 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2:91-99. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli-Sforza, L. L., and W. F. Bodmer. 1971. The genetics of human populations, p. 39-70. W. H. Freeman and Co., San Francisco, CA.
- 11.Cellier, M., G. Prive, A. Belouchi, T. Kwan, V. Rodriguez, W. Chia, and P. Gros. 1995. The natural resistance associated macrophage protein (Nramp) defines a new family of membrane proteins conserved throughout evolution. Proc. Natl. Acad. Sci. USA 92:10089-10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darrah, P. A., M. K. Hondalus, Q. Chen, H. Ischiropoulos, and D. M. Mosser. 2000. Cooperation between reactive oxygen and nitrogen intermediates in killing of Rhodococcus equi by activated macrophages. Infect. Immun. 68:3587-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, L. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, and R. Detels. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 14.De Massis, F., A. Di Girolamo, A. Petrini, E. Pizzigallo, and A. Giovannini. 2005. Correlation between animal and human brucellosis in Italy. Clin. Microbiol. Infect. 11:632-636. [DOI] [PubMed] [Google Scholar]
- 15.Ding, A. H., C. F. Nathan, and D. J. Stuehr. 1988. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. J. Immunol. 141:2407-2412. [PubMed] [Google Scholar]
- 16.Feder, J. N., A. Gnirke, W. Thomas, Z. Tsuchihashi, D. A. Ruddy, A. Basava, F. Dormishian, R. Domingo, Jr., M. Ellis, A. Fullan, L. Hinton, L. Jones, B. Kimmel, G. Kronmal, P. Lauer, V. Lee, D. Loeb, F. Mapa, E. McClelland, N. Meyer, G. Mintier, N. Moeller, T. Moore, E. Morikang, C. Prass, L. Quintana, S. Starnes, K. Schatzman, K. Brunke, D. Drayna, N. Risch, R. Bacon, and R. Wolff. 1996. A novel class I like gene is mutated in patients with hereditary haemochromatosis. Nat. Genet. 13:399-408. [DOI] [PubMed] [Google Scholar]
- 17.Fitch, T. A. 2003. Intracellular survival of Brucella: defining the link with persistence. Vet. Microbiol. 92:213-223. [DOI] [PubMed] [Google Scholar]
- 18.Godfroid, J., C. Saegerman, V. Wellemans, K. Walraven, J. Letesson, A. Tibor, A. McMillan, S. Spencer, M. Sanna, D. Bakker, R. Puillot, and B. Garin-Bastuji. 2002. How to substantiate eradication of bovine brucellosis when aspecific serological reactions occur in the course of brucellosis testing. Vet. Microbiol. 90:461-477. [DOI] [PubMed] [Google Scholar]
- 19.Govoni, G., and P. Gros. 1998. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47:277-284. [DOI] [PubMed] [Google Scholar]
- 20.Guilloteau, L. A., J. Dornand, A. Gross, M. Olivier, F. Cortade, Y. Le Vern, and D. Kerbueuf. 2003. Nramp1 is not a major determinant in the control of Brucella melitensis infection in mice. Infect. Immun. 71:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruenheid, S., and P. Gros. 2000. Genetic susceptibility to intracellular infections: Nramp1, macrophage function and divalent cations transport. Curr. Opin. Microbiol. 3:43-48. [DOI] [PubMed] [Google Scholar]
- 22.Hoefel, D., W. L. Grooby, P. T. Monisa, S. Andrews, and C. P. Sainta. 2003. Enumeration of water-borne bacteria using viability assays and flow cytometry: a comparison to culture-based techniques. J. Microbiol. Methods 55:585-597. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, Jr., and R. A. Esekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 24.Horĭn, P., I. Rychlik, J. W. Templeton, and L. G. Adams. 1999. A complex pattern of microsatellite polymorphism within the bovine NRAMP1 gene. Eur. J. Immunogenet. 26:311-313. [DOI] [PubMed] [Google Scholar]
- 25.Hugget, J., K. Dheda, S. Bustin, and A. Zumla. 2005. Real-time RT-PCR normalization; strategies and considerations. Genes Immun. 6:279-284. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, N., A. Mitra, I. Ganguly, R. Singh, S. M. Deb, S. K. Srivastava, and A. Sharma. 2005. Lack of association of brucellosis resistance with (GT)13 microsatellite allele at 3′ UTR of Nramp1 gene in Indian zebu (Bos indicus) and crossbred (Bos indicus × Bos taurus) cattle. Vet. Microbiol. 111:139-143. [DOI] [PubMed] [Google Scholar]
- 27.Lander, E. S., and N. J. Schork. 1994. Genetic dissection of complex traits. Science 265:2037-2047. [DOI] [PubMed] [Google Scholar]
- 28.Lohmueller, K. E., C. L. Pearce, M. Pike, E. S. Lander, and J. N. Hirschorn. 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33:177-182. [DOI] [PubMed] [Google Scholar]
- 29.MacDiarmid, S. C. 1987. A theoretical basis for the use of a skin test for brucellosis surveillance in extensively managed cattle herds. Rev. Sci. Tech. Off. Int. Epizoot. 6:1029-1035. [DOI] [PubMed] [Google Scholar]
- 30.Motulski, H. 1995. Intuitive biostatistics, p. 80-105. Oxford University Press, New York, NY.
- 31.Murphy, E., G. T. Robertson, M. Parent, S. D. Hagius, R. M. Roop, I. I. P. H. Elzer, and C. L. Baldwin. 2002. Major histocompatibility complex class I and II expression on macrophages containing a virulent strain of Brucella abortus measured using green fluorescent protein-expressing brucellae and flow cytometry. FEMS Immunol. Med. Microbiol. 33:191-200. [DOI] [PubMed] [Google Scholar]
- 32.Paixao, T. A., C. Ferreira, A. M. Borges, D. A. Oliveira, A. P. Lage, and R. L. Santos. 2006. Frequency of bovine Nramp1 (Slc11a1) alleles in Holstein and zebu breeds. Vet. Immun. Immunopathol. 109:37-42. [DOI] [PubMed] [Google Scholar]
- 33.Pritchard, J. K., and N. A. Rosenberg. 1999. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 65:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Queipo-Ortuno, M. I., J. D. Colmenero, J. M. Reguera, M. A. Garcia-Ordonez, M. E. Pachon, M. Gonzalez, and P. Morata. 2005. Rapid diagnosis of human brucellosis by SYBR green I-based real-time PCR assay and melting curve analysis in serum samples. Clin. Microbiol. Infect. Dis. 11:713-718. [DOI] [PubMed] [Google Scholar]
- 35.Ray, W. C., R. R. Brown, D. A. Stringfellow, P. R. Schunrrenberger, C. M. Scanlan, and A. I. Swan. 1988. Bovine brucellosis: an investigation of latency in progeny of culture-positive cows. J. Am. Vet. Med. Assoc. 192:182-186. [PubMed] [Google Scholar]
- 36.Sundberg, R. 2005. Statistical modelling in case-control real-time RT-PCR assays, for identification of differently expressed genes in schizophrenia. Biostatistics 1:1-22. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, D., J. S. Witte, M. Slattery, and D. Goldgar. 2004. Increased power for case-control studies of single nucleotide polymorphisms through incorporation of family history and genetic constraints. Genet. Epidemiol. 27:215-224. [DOI] [PubMed] [Google Scholar]
- 38.Vidal, S. M., D. Malo, K. Vogan, E. Skamene, and P. Gros. 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73:469-485. [DOI] [PubMed] [Google Scholar]
- 39.Weir, B. S. 1996. Genetic data analysis II: methods for discrete population genetic data, p. 91-139. Sinauer Associates, Sunderland, MA.
- 40.Woolhouse, M. E. J., J. P. Webster, E. Domingo, B. Charlesworth, and B. R. Levin. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32:569-577. [DOI] [PubMed] [Google Scholar]
- 41.Young, E. J. 1995. An overview of human brucellosis. Clin. Infect. Dis. 21:283-290. [DOI] [PubMed] [Google Scholar]