Abstract
In vitro studies of obligate intracellular chlamydia biology and pathogenesis are highly dependent on the use of experimental models and growth conditions that mimic the mucosal architecture and environment these pathogens encounter during natural infections. In this study, the growth of Chlamydia trachomatis genital serovar E was monitored in mouse fibroblast McCoy cells and compared to more relevant host human epithelial endometrium-derived HEC-1B and cervix-derived HeLa cells, seeded and polarized on collagen-coated microcarrier beads, using a three-dimensional culture system. Microscopy analysis of these cell lines prior to infection revealed morphological differences reminiscent of their in vivo architecture. Upon infection, early chlamydial inclusion distribution was uniform in McCoy cells but patchy in both epithelial cell lines. Although no difference in chlamydial attachment to or entry into the two genital epithelial cell lines was noted, active bacterial genome replication and transcription, as well as initial transformation of elementary bodies to reticulate bodies, were detected earlier in HEC-1B than in HeLa cells, suggesting a faster growth, which led to higher progeny counts and titers in HEC-1B cells upon completion of the developmental cycle. Chlamydial development in the less relevant McCoy cells was very similar to that in HeLa cells, although higher progeny counts were obtained. In conclusion, this three-dimensional bead culture system represents an improved model for harvesting large quantities of infectious chlamydia progeny from their more natural polarized epithelial host cells.
Chlamydia trachomatis serovars D to K are oculogenital pathogens and the leading cause of bacterial sexually transmitted diseases (41). It is estimated there are 3 to 4 million cases of chlamydial sexually transmitted diseases annually in the United States and some 90 million cases per year worldwide (7). Since the majority of infected individuals are essentially asymptomatic and do not seek medical attention, ascending migration can occur and lead to serious complications, such as prostatitis and epididymitis in men and pelvic inflammatory disease, salpingitis, ectopic pregnancy, and infertility in women (12, 14).
Chlamydiae are obligate intracellular bacteria and, as such, must be internalized into superficial epithelial cells of the genital mucosa in order to initiate the infectious process. Infection begins with attachment of the infectious elementary bodies (EB) form to the apical surface of columnar epithelial cells, followed by entry via various endocytic mechanisms. The EB-containing endosomes exit the endocytic pathway to avoid fusion with lysosomes and travel on microtubules to the nuclear hof, where they undergo homotypic fusion with one another, and then the EBs transform into metabolically active reticulate bodies (RB). Since RB divide by binary fission and the number of progeny increases, the expanding endocytic vesicle is termed an inclusion. Eventually, RB mature back into infectious EB, and this developmental cycle ends by the release of chlamydial progeny, usually after 48 to 72 h for in vitro infections (1, 54).
In the 1970s, McCoy cells were used by many chlamydiologists worldwide for the isolation and propagation of C. trachomatis from ocular, genital, and rectal specimens. The fibroblasts were hearty, easy to manipulate, and a less cumbersome alternative to the isolation of chlamydiae from yolk sacs. A decade later, Pharmacia Biotech (Uppsala, Sweden) devised the collagen-coated dextran microcarrier bead system (Cytodex 3) for growing anchorage-dependent eukaryotic cells in suspension culture (38). The method became popularized by virologists for highly improved yields of virus progeny from fibroblasts. Thus, our laboratory adopted the microcarrier bead suspension culture system for McCoy cells and found—compared to infected McCoy cell growth in flasks—(i) an increased yield of C. trachomatis serovar E EB progeny, (ii) which were more infectious on a per particle ratio basis; (iii) an accelerated developmental cycle (54 h versus 72 h) due to a more synchronous conversion of early non-metabolically active EB into metabolically active RB, as well as a more synchronous late maturation of virtually all noninfectious RB to infectious EB; and (iv) considerable cost savings in medium, serum, plastics, and time and effort (47, 52). There were also some surprising findings at high-resolution morphological levels, including a dramatic change in the appearance of glycogen in chlamydial inclusions from the typically described granular appearance to a more globular appearance, and the seeming release of EB into the host cell cytoplasm at the end of the developmental cycle rather than release of the inclusion and its contents into the extracellular environment (52).
However, C. trachomatis serovar E, one of the most common genital isolates in the United States (8), is a mucosal pathogen adapted to growth in human polarized superficial luminal epithelial cells; fibroblasts are not natural hosts, nor do they grow in a polarized orientation. Furthermore, the original McCoy A subclone, reportedly obtained from knee joint synovial fluid from a patient with degenerative arthritis, evidently became cross-contaminated with the McCoy B subline of mouse origin, based on subsequent karyological criteria and heteroploidy (43). Routinely in our laboratory, in vitro titration on coverslips of an identical inoculum of serovar E EB stock, harvested from infected McCoy cells propagated on the microcarrier beads, always yields a higher infectivity or inclusion-forming unit (IFU) count in epithelial cells (80%) than it does in McCoy cells (50%), which emphasizes the importance of the epithelial cell environment for chlamydial growth. However, chlamydial infection in various polarized epithelial cell lines also differs. C. trachomatis serovar E infectivity is greater for the endometrial carcinoma epithelial subclone HEC-1B than for the Ishikawa endometrial epithelial cell line and the two breast cancer epithelial cell lines MCF-7 and HCC-1806 (21).
The objective of the present study was to adapt two epithelial cell lines used in chlamydia research to the microcarrier bead suspension culture system and compare the growth of C. trachomatis serovar E in these polarized epithelial cells under three-dimensional (3D) conditions, using identically grown and infected McCoy cells as historical controls. The epithelial cell lines utilized were the endometrium-derived HEC-1B cells, representative of tall columnar epithelia, and the more popular, commonly used cervix-derived HeLa cells, representative of low, squamo-columnar epithelia.
MATERIALS AND METHODS
Cell lines.
The cell lines obtained from the American Type Culture Collection (ATCC) were (i) the transformed endometrial line HEC-1B (HTB-113, ATCC), (ii) the transformed epithelial cervical line HeLa (CCL-2, ATCC), and (iii) the mouse fibroblast cell line McCoy (CRL-1696, ATCC). All cultures were routinely monitored and shown to be free of mycoplasma contamination. Stock cultures of the cell lines were propagated in flasks containing minimum essential medium (MEM; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum.
Chlamydia strain.
A human urogenital isolate of C. trachomatis serovar E/UW-5/CX was originally obtained from S. P. Wang and C.-C. Kuo (University of Washington, Seattle, WA) and has been maintained in our laboratory. For these experiments, crude chlamydia stocks were grown in HEC-1B cells in flasks at 35°C in MEM supplemented with 2 mM glutamine and 10% fetal bovine serum in an atmosphere of 5% carbon dioxide for 72 h, harvested, stored at −80°C in 2× SPG (0.2 M sucrose, 0.02 M phosphate buffer, and 5 mM l-glutamine), and subsequently titers were determined as previously described by Moorman et al. (33).
Pretitration of crude chlamydial stock.
Near-confluent or confluent monolayers of the McCoy, HEC-1B, and HeLa cells on coverslips were infected with serial dilutions of C. trachomatis serovar E EB. The EB (50 μl) were resuspended in the “open system” Dulbecco modified Eagle medium and added to the coverslips for a 1-h adsorption period at 35°C. Infected monolayers at 48 h postinfection (hpi) were fixed with methanol and then stained with the Pathfinder C. trachomatis direct specimen monoclonal antibody (Bio-Rad Laboratories, Redmond, WA). The numbers of inclusions were counted in 15 random microscopic grid fields at ×400 magnification on a Zeiss Axiovert S100 microscope equipped with 495-nm excitation and 520-nm emission filters (Carl Zeiss, Inc., Göttingen, Germany). All titrations were performed in triplicate.
3D cell cultures.
Cytodex collagen-coated microcarrier beads (Sigma-Aldrich, Inc., St. Louis, MO) have a tendency to stick to glass surfaces, so all glassware used throughout the present study was treated with silicon to prevent bead damage and loss. The microcarrier beads were rehydrated in phosphate-buffered saline (pH 7.2) (PBS) in weight/volume ratio of 1 g of dry beads per 50 ml of PBS and then sterilized by autoclaving. The bead concentration was calculated on the basis of actual bead counts by light microscopy in 20 50-μl drops of the bead-PBS suspension; counts can range between 1.2 × 105 and 1.6 × 105 beads/ml but usually average 1.5 × 105 beads/ml; this latter count was the bead stock used throughout the present study. Regardless of the size spinner bottle to be used, the final concentration or density of beads should be 104 beads/ml.
An appropriate dilution (Table 1) of the stock bead suspension was added to 100 ml of complete MEM, supplemented with Hanks salts, in a 500-ml siliconized spinner culture bottle (Bellco Biotechnology, Inc., Vineland, NJ) and equilibrated in the medium at 37°C for 1 h prior to addition of the host eukaryotic cells. McCoy, HEC-1B, or HeLa cell confluent monolayers harvested from 150-cm2 flasks (Table 1) were resuspended in 20 ml of fresh, complete MEM and added to the beads at a cell/bead ratio of 50 to 100:1. The spinner bottle containing the cells and bead suspension was then placed in a controlled environment incubator with a rotating shaker, for slow, continuous stirring at ∼60 rpm for 1 h at 37°C. Fresh MEM was added, according to the size/volume spinner bottle (in our case up to a 500-ml volume). The spinner bottles were moved to a 37°C incubator and placed on a magnetic stir plate set at the lowest possible speed to permit rotation of the paddle blades to keep the beads in suspension (Fig. 1). Depending on the cell line, it takes anywhere from 2 to 7 days for the fibroblasts or epithelial cells to form a confluent monolayer over the bead surface. Eukaryotic host cell growth was monitored daily by phase microscopy by withdrawing a few drops of the cell-bead suspension in a Pasteur pipette, placing the drops on a slide or in a petri dish and examining the drops microscopically. For the slowly growing HEC-1B cells, one-half fresh medium was added to one-half conditioned medium every 2 to 3 days.
TABLE 1.
Estimated ratio of microcarrier beads to cells and time of incubation for establishing sufficient suspension cell cultures
| Cell line and parametera | Setup for various sizes of spinner bottles
|
||
|---|---|---|---|
| 250 ml | 500 ml | 1,000 ml | |
| McCoy | |||
| Bead stock (ml) | 25 | 50 | 100 |
| No. of flasks | 0.5 | 1 | 2 |
| Time (days) | 2 | 2 | 2-3 |
| HEC-1Bb | |||
| Bead stock (ml) | 20 | 50 | 90 |
| No. of flasks | 1 | 2-3 | 5-6 |
| Time (days) | 7-10 | 7-10 | 7-10 |
| HeLac | |||
| Bead stock (ml) | 20 | 50 | 90 |
| No. of flasks | 1 | 2 | 4 |
| Time (days) | 3-4 | 3-4 | 3-4 |
Bead stock refers to the amount (in milliliters) of Microcarrier Cytodex bead stock (estimated at 1.5 × 105 beads/ml) to add per spinner bottle. The number of flasks is the number of 150-cm2 flasks (with confluent cell monolayer) to add per spinner bottle to get the appropriate ratio of eukaryotic host cells per bead. Time refers to the eukaryotic cell growth rate on beads and depends on many conditions (passage number, medium, cell health, etc.). Daily monitoring of cell growth is necessary. Coverage of 90% of beads is desirable.
The number of beads needs to be slightly decreased to help reduce “organoid” structure formation and provide a more even cell distribution on the beads.
Because of inefficient anchoring of HeLa cells to the bead surface, the number of beads needs to be slightly decreased in order to prevent mechanical bead-to-bead interaction.
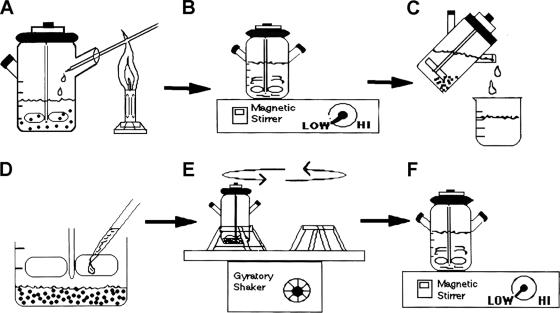
FIG. 1.
Summary diagram of the microcarrier bead culture procedure. Cytodex collagen-coated microcarrier beads were added to a spinner bottle containing MEM, at a final concentration of 104 beads/ml, and equilibrated at 37°C for 1 h. (A) McCoy, HEC-1B, or HeLa cells were aseptically added to the beads at a cell/bead ratio of 50 to 100:1 and incubated for 1 h at 37°C on slowly a rotating gyratory shaker. (B) After fresh MEM was added, the bottles were moved to a 37°C incubator, placed on a magnetic stir plate set at the lowest possible speed to keep beads in suspension, and incubated until confluent cell monolayers were obtained (2 to 7 days depending on the cell line); for the more slowly growing HEC-1B cells, one-half fresh medium was added to one-half conditioned medium every 2 to 3 days. (C) Prior to inoculation, the beads were allowed to settle down to the bottom of the spinner bottle, and four-fifths of the medium was removed. (D) Chlamydia stock was appropriately diluted in fresh MEM and aseptically added to the bead culture slurry. (E) The adsorption step consisted of a 2-h incubation at 35°C in a slow gyratory incubator shaker. (F) The volume of medium was appropriately topped back up with MEM containing cycloheximide and gentamicin, and bottles were placed on the magnetic stir plate in a 35°C incubator for various periods of time. When used for progeny harvest, the bottles were removed from the stir plate at 48 hpi but kept stationary (without stirring) in the 35°C incubator to allow completion of the chlamydial development cycle; for other uses, the bead cultures were sampled at various times postinfection. Chlamydial infection was monitored routinely throughout by phase microscopy.
Infection of host cells on the beads.
Confluent monolayers on the beads were infected by using a modified version of our adsorption inoculation method (42, 47). The bead cultures were first allowed to settle down to the bottom of the spinner bottle, and all medium but 50 ml for 250-ml bottles and 100 ml for 500-ml bottles was removed via 50-ml pipettes. Chlamydia stock, based on pretitration for each cell line, was diluted appropriately in 10 ml of fresh MEM and added to the bead culture slurry, followed by 2 h of incubation at 35°C in the slowly rotating incubator shaker. After the adsorption period, the volume of medium was topped back up to 500 ml with fresh, complete MEM, additionally supplemented with 0.5 μg of cycloheximide/ml and 10 μg of gentamicin/ml (Sigma-Aldrich, Inc.). The bottles, placed again on the magnetic stir plate set at the lowest possible speed to permit rotation of the paddle blades to keep beads in suspension, were incubated at 35°C. At 48 hpi, the bottles were removed from the stir plate but kept stationary (without stirring) in the 35°C incubator to allow completion of the C. trachomatis serovar E development cycle. This latter maneuver helps prevent detachment of the anchorage-weakened infected cells from the beads, as well as breakage of the huge, blister-like chlamydial inclusions (i.e., from mechanical shear; chlamydial development is not affected). Chlamydial infection was monitored routinely throughout by phase microscopy; fluorescence and transmission electron microscopy were also performed at selected intervals.
Fluorescence and transmission electron microscopy.
For microscopy studies, the host eukaryotic cells were grown in 3D bead cultures in 250-ml spinner bottles, infected with crude chlamydia stock, and incubated at 35°C for 70 h. Samples (5-ml aliquots per sample) of uninfected and infected cells were obtained at 0, 6, 12, 24, 36, 48, 50, 52, 54, 56, 62, and 70 hpi. After the beads were allowed to settle out in 15-ml tubes, the medium was removed, and the samples were washed twice in PBS.
For fluorescence microscopy, the bead cultures were fixed with 1 ml of ice-cold 100% methanol for 30 min at 25°C, washed, and transferred to 1.5-ml microfuge tubes for staining with the Pathfinder Chlamydia trachomatis Direct Specimen Monoclonal Antibody (100 μl/pellet) for 30 min at 37°C. Excess stain was gently removed, and the bead cultures were resuspended in 100 μl of DakoCytomation fluorescence mounting medium (Dako North America, Inc., Carpinteria, CA), stored at 4°C until all samples were collected, and finally examined on a Zeiss Axiovert S100 microscope equipped with an AxioCam Color digital charge-coupled device camera. Images were captured by using Zeiss AxioCam Photoshop software, adjusted in Adobe Photoshop, and analyzed.
For electron microscopy analysis, the bead cultures were fixed with a solution of 2% glutaraldehyde-0.5% paraformaldehyde, prepared in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h at 25°C, agar enrobed, and postfixed in 1% osmium tetroxide, followed by alcoholic uranyl acetate, as previously described (50). After dehydration in increasing concentrations of ethanol, infiltration and embedding in Epon-Araldite 812 resin, and curing at 60°C for 48 h, silver-gold thin sections were cut on a Reichert Ultracut S microtome (Leica Microsystems, Inc., Bannockburn, IL), counterstained, and examined in a Phillips Tecnai-10 electron microscope (FEI) operated at 60 kV.
Harvest of chlamydia progeny EB.
Progeny EB were collected from each infected cell line grown on the bead cultures at 52 hpi. The beads were allowed to settle to the bottom of the spinner bottles, and all but 100 ml of medium (culture supernatants) was transferred to sterile centrifuge bottles and placed on ice until further use. The bead pellets were resuspended in the residual medium and distributed into three 50-ml tubes; 10 ml of glass beads (ca. 3 mm in diameter) were added to each tube. The tubes were briefly vortexed to remove as many eukaryotic cells as possible from the beads and then sonicated for 15 min in a water bath sonifier to lyse the infected cells and release chlamydia progeny. Intact beads were removed by pouring the lysate through a nylon (100-μm pore size) cell strainer (BD Biosciences Discovery Labware, Bedford, MA). Lysates, split between the centrifuge bottles containing culture supernatants, were centrifuged according to routine protocol to separate the EB from host cell debris, the EB were purified in self-generating Percoll gradients, and EB particle counts were determined via our spectrophotometric standard curve, diluted to 1.0 × 109 EB per ml in 2× SPG and stored at −80°C for subsequent titration in each cell line.
Real-time PCR analysis.
HEC-1B and HeLa cells were grown in 500-ml spinner bottles and then infected with C. trachomatis serovar E with a high multiplicity of infection (ca. 20 EB per cell). For real-time PCR, samples (5 ml of bead suspension per sample) were collected at 3, 12, 24, 36, and 48 hpi and used for total RNA and genomic DNA (gDNA) isolation. After a brief wash with PBS, cells were lysed and homogenized in 400 μl of the RLT buffer provided in the RNeasy minikit (QIAGEN, Valencia, CA), supplemented with β-mercaptoethanol and with proteinase K (200 μg/ml; QIAGEN). Lysates were then incubated for 15 min at 55°C to allow digestion of the bead-coating collagen; this step was added to prevent clogging of the spin columns by collagen. After removal of the beads by centrifugation, a 150-μl aliquot of each lysate was used for gDNA isolation using the QIAamp blood minikit (QIAGEN); the rest of the lysate was used for total RNA isolation with the RNeasy minikit, including an on-column DNase step. All samples from a same time course experiment were processed in parallel, using the same protocol and the same elution volumes. In addition, genomic DNA from Percoll-purified serovar E EB (chlamydial gDNA) or from uninfected HeLa cells (host cell gDNA), used as standards in the quantitative PCR (qPCR) assays, was isolated with the QIAamp blood minikit, using the recommended optimal lysis buffer and protocol. All genomic DNA and total RNA samples were quantitated by an optical density reading and either visualized after agarose gel electrophoresis stained with ethidium bromide (gDNA) or analyzed for integrity on a BioAnalyzer 2100 (RNA; Agilent Technologies, Inc., Palo Alto, CA). Two micrograms of total RNA from all bead culture samples was then reverse transcribed using the SuperScript II kit (Invitrogen, Carlsbad, CA) in the presence of 200 ng of random hexamers (Integrated DNA Technologies, Coralville, IA) under the conditions recommended by the manufacturer. Prior to qPCR, the quality of all cDNA was checked by conventional PCR with primers specific for human aldolase as previously described (21). In addition, for each RNA sample, an “RT-minus” control, in which the reverse transcriptase (RT) enzyme was omitted, was also included.
Real-time PCR assays were carried out in a 96-well plate format; reaction plates were set up using the laboratory robotic workstation Biomek 2000 (Beckman Coulter, Inc., Fullerton, CA), and amplifications were performed in a Bio-Rad iCycler (Bio-Rad Laboratories, Hercules, CA). The following primer sets were used: (i) C. trachomatis CT446/euo- and CT443/omcB-specific primers, kindly provided by R. J. Belland, and (ii) primers specific for two human genes located on different chromosomes, i.e., myeloperoxidase (MPO; chromosome 17; from Linzmeier and Ganz [29]) and coagulation factor VIII (F8; chromosome X; modified from Boeckman et al. [4]). Reaction mixtures contained 1× Platinum Quantitative PCR Supermix-UDG (Invitrogen), 0.8× SYBR green (Cambrex BioScience, Inc., Rockland, ME), 5 mM MgCl2, 300 nM concentrations of each primer, and 1 μl of standard or experimental sample in a total reaction volume of 50 μl. The amplification conditions were as follows: 1 cycle of 2 min at 50°C and 2 min at 95°C, followed by 50 cycles of either 15 s at 95°C, 15 s at 55°C, and 15 s at 72°C for the chlamydia-specific primer sets or 18 s at 95°C, 18 s at 55°C, and 20 s at 72°C for the MPO and F8 host-specific primer sets; a melting curve analysis was also included in each run to ensure that a single PCR product was amplified. All samples were tested in triplicate, and all qPCR assays used had similar high efficiencies of at least 97%.
qPCR controls included blanks (no DNA or cDNA template), gDNA, or cDNA from uninfected HEC-1B and HeLa cells when testing chlamydia-specific primer sets to ensure that no PCR amplification was detected, and RT-minus controls for each RNA sample to assess the contribution of gDNA to the overall euo and omcB qPCR signal detected in the corresponding RT reaction.
Real-time PCR assays using the euo and omcB primer sets and chlamydial gDNA standards (0.11 pg to 11 ng per well, corresponding to 102 to 107 genome copies per well) were used to determine the levels of chlamydial transcripts (RT − RT-minus) and of chlamydial genomes (gDNA) in bead culture samples harvested at various times postinfection. Similarly, MPO- and F8-specific qPCR assays using serial dilutions of uninfected HeLa cell gDNA as standards (150 pg to 150 ng per well) were used to evaluate the relative amounts of host cell genomic material in all bead culture aliquots (gDNA); the amounts of host gDNA determined from these assays that targeted genes located on two independent chromosomes were then averaged for each bead culture sample and used to normalize chlamydial euo and omcB transcript levels, as well as chlamydial genome copy numbers.
One consideration during assay design was possible ploidy differences between the HEC-1B and HeLa cell lines, which, if confirmed for chromosomes 17 (MPO) and X (F8), would introduce a bias in the normalized data. Preliminary karyotyping of a small sample (20 to 30 cells) of our epithelial cell line subclones showed that, although most of the cells analyzed in both cell lines were aneuploid, more heterogeneity was found in HEC-1B compared to HeLa cells regarding total numbers of chromosomes per cell (mixture of diploid- and triploid-like cells versus mostly triploid-like cells, respectively). Some intra- and inter-cell line variation was also observed in the numbers of chromosome 17 (MPO) and chromosome X (F8) per cell but, importantly, the average numbers of these chromosomes/cell were not significantly different between the two cell populations (data not shown), therefore validating our normalization strategy.
RESULTS
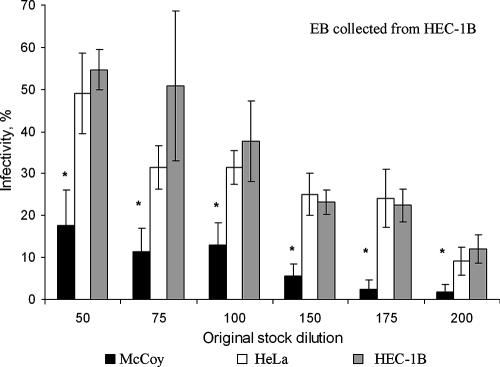
The C. trachomatis serovar E progeny stock, harvested from infected HEC-1B cells in flask culture and used for all subsequent experiments reported here, was titrated for infectivity units (IFU) on coverslips of McCoy, HEC-1B, and HeLa cells. At the same multiplicity of infection, the percentage of chlamydial infection was consistently higher in HEC-1B cells (55% at a 1:50 dilution; P < 0.01) than in McCoy cells (17%). HEC-1B and HeLa cells had essentially the same sensitivity to chlamydial infection (Fig. 2).
FIG. 2.
The infectivity of C. trachomatis serovar E is greater for HEC-1B and HeLa cells than for McCoy cells. Confluent cell monolayers grown on glass coverslips were inoculated with various dilutions (1:50 to 1:200) of a chlamydial stock originally grown in HEC-1B cells; initial titration showed that a 1:50 dilution of this stock resulted in 50% infectivity in HEC-1B cell monolayers. At 48 hpi, the percentage of infected cells for each cell line and each dilution of inoculum was determined after staining with fluorescent anti-chlamydia-specific antibodies. The mean percent infectivity (± the standard deviation) in McCoy, HEC-1B, and HeLa cells was calculated from three independent titration experiments; each experiment included triplicate coverslips. Statistical analysis was performed by using the Student t test (*, significantly different from other cell lines [P < 0.01]).
Comparative growth of HEC-1B and HeLa cells with McCoy cells in the 3D bead suspension cell culture system monitored by light microscopy.
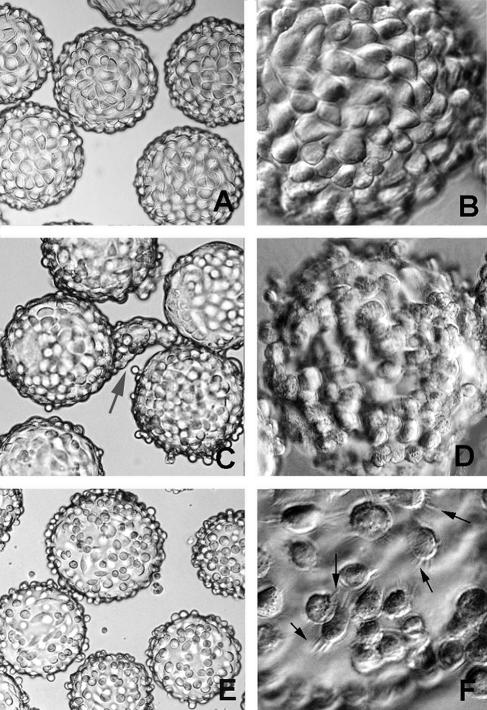
Within 2 to 3 days of seeding, the faster- growing McCoy cells form even monolayers on the collagen-coated beads (Fig. 3A and B). HEC-1B cells are the slowest-growing cell line of the three and require a prolonged period of cultivation before monolayers are formed on the beads—usually by 7 to 10 days (Fig. 3C). Once the HEC-1B monolayers become confluent and tight junctions and junctional complexes are formed, the endometrial epithelial cells assume their tall columnar orientation. As in polarized cell culture, these epithelial cells have a tendency to form “organoids” (31), which resemble the glandular structures from which they were derived (Fig. 3C). With time, the organoids will detach, leaving a more normal monolayer. Alternatively, the fetal bovine serum concentration can be reduced temporarily from 10 to 3% to reduce organoid formation. Eventually, lush monolayers remain (Fig. 3D). In contrast, HeLa cells form uneven monolayers on the beads (Fig. 3E and F). Extensions emanate from the HeLa cells as if trying to anchor the cells to the collagen surface but spaces remain (Fig. 3F). Where confluent patches of cells do occur, the low columnar architecture of cervical cells is apparent.
FIG. 3.
Light microscopy analysis of McCoy, HEC-1B, and HeLa cell monolayer formation on microcarrier beads. (A and B) Even distribution of McCoy cells over the bead surface, 2 days after initial seeding. (C and D) Confluent HEC-1B monolayers were obtained 7 to 10 days after seeding. Polarized endometrial cells tend to form small and large “organoid”-like structures (gray arrow in panel C); with time, the organoids detach, leaving a lush monolayer (D). (E) Uneven distribution of HeLa cells over the bead surface, with the presence of free-floating cells at day 3 after seeding. (F) Extensions emanate from the HeLa cells (black arrows) as if trying to anchor to the collagen surface, but spaces remain. Magnifications: ×150 (A, C, and E) and ×450 (B, D, and F).
C. trachomatis infection of host cell monolayers on the 3D culture system monitored by immunofluorescence and electron microscopy.
It should be noted that a 10-fold-higher inoculum was used for ease of localizing EB in the host cells at 6 and 12 hpi but, thereafter, the inoculum was reduced to the standard amount—a dilution of stock inoculum to infect 60 to 80% of host cells—to prevent host cell cytotoxicity prior to the completion of the developmental cycle.
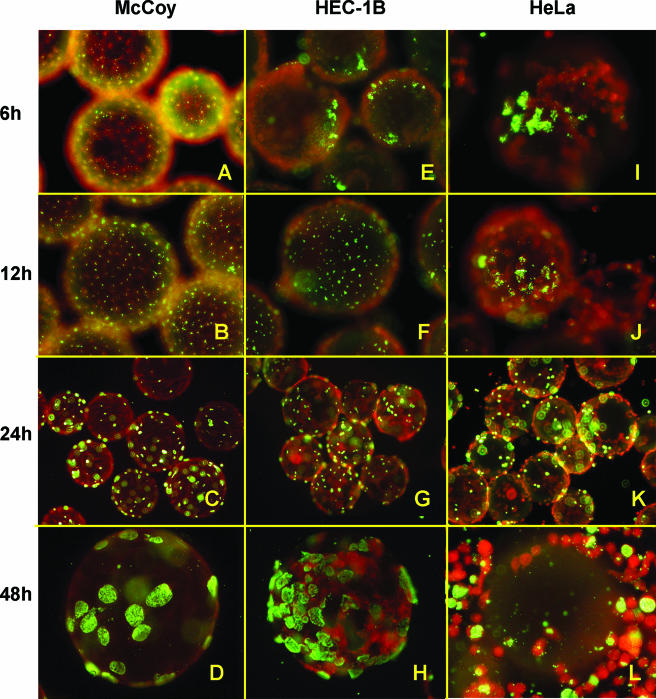
Chlamydia infection of the McCoy and epithelial cell monolayers on the beads was first monitored by fluorescence microscopy. By 6 to 12 hpi, attached/early entered EB in endocytic vesicles were evenly distributed in McCoy cell monolayers (Fig. 4A and B). Inclusions, although small, were visible by 24 hpi (Fig. 4C) and quite enlarged by 48 hpi (Fig. 4D). In contrast, attachment to and early entry of EB into the epithelial cells was “patchy” and clustered (Fig. 4E, F, I, and J). This pattern mimics early chlamydial infection previously observed in primary human and pig genital tract epithelial cell models cultured ex vivo (20, 33). Inclusions, still somewhat clustered, were easily detected by 24 hpi in the HEC-1B cells (Fig. 4G) and HeLa cells (Fig. 4K). The clustered inclusion pattern of development in HeLa cells was somewhat dictated by the patchy monolayer development. Very large inclusions containing kinetically active, bouncing chlamydiae predominate in the epithelial cells, especially in the HEC-1B cells, by 48 hpi (Fig. 4H). However, a significant number of infected and uninfected HeLa cells have come off the microcarrier beads by this time (Fig. 4L). Harvest of EB progeny from the infected cells in this 3D suspension culture system is usually performed by 52 hpi—versus 72 hpi from the same cells cultured as monolayers in flasks. Thus, the serovar E developmental cycle is definitely accelerated in the 3D bead suspension culture of epithelial cells, which was previously reported for serovar E development in McCoy cell bead cultures (52).
FIG. 4.
Fluorescence microscopy analysis of C. trachomatis serovar E inclusion formation in McCoy, HEC-1B, and HeLa cells seeded on beads. Infected 3D bead cultures were sampled at various times postinfection; the beads were then washed, fixed, and stained with fluorescein isothiocyanate-labeled anti-C. trachomatis specific antibodies and Evans blue counterstain. Representative fields of bead cultures of the three cell lines, sampled at 6 hpi (A, E, and I), 12 hpi (B, F, and J), 24 hpi (C, G, and K), and 48 hpi (D, H, and L) are shown. Note the even chlamydial EB attachment and distribution (green) throughout the McCoy cell monolayer (red) (A and B) versus the more “patchy” patterns of EB distribution in HEC-1B (E and F) and HeLa (I and J) epithelial cells at 6 and 12 hpi. Inclusions were readily visible in all cell line monolayers at mid-cycle (C, G, and K). At 48 hpi, higher chlamydial inclusion numbers were found in HEC-1B (D) than in McCoy (H) bead cultures, whereas numerous HeLa cells have come off the beads by that time (L). Magnifications: ×300 (A, B, E, F, I, and J), ×150 (C, G, and K), ×450 (D, H, and L).
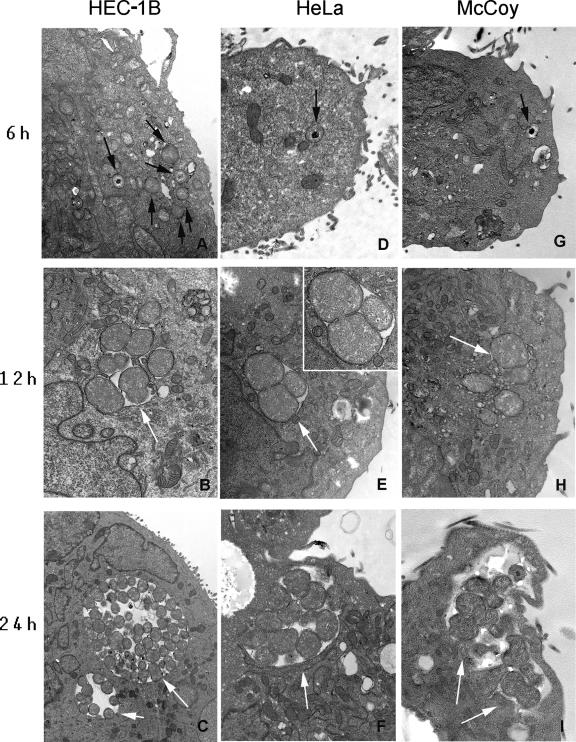
By transmission electron microscopy analysis, the entry of EB into HEC-1B cells is apparently more efficient, resulting in earlier transformation of EB into RB at 6 hpi (Fig. 5A). By comparison, only EB were detected in HeLa (Fig. 5D) and McCoy cells (Fig. 5G) at 6 hpi. Similarly, early developing inclusions at 12 hpi were larger and contained more dividing RB in HEC-1B cells (Fig. 5B) than in HeLa (Fig. 5E) and McCoy (Fig. 5H) cells. The apparent accelerated development of chlamydiae in HEC-1B cells was also evident at 24 hpi; larger inclusions containing many RB, some transitioning intermediate bodies and mature EB (Fig. 5C), were readily detected, whereas smaller inclusions with fewer RB were found in HeLa (Fig. 5F) and McCoy (Fig. 5I) cells at the same times postinfection.
FIG. 5.
Representative transmission electron photomicrographs of C. trachomatis-infected bead cultures of HEC-1B, HeLa, and McCoy cells at early stages of infection. At 6 hpi, higher numbers of intracellular chlamydial particles (black arrows) were found in HEC-B cells (A) than in HeLa (D) and McCoy (G) cells; note that the early transformation of EB into RB is only visible at that stage in HEC-1B cells (panel A versus panels D and G). The apparently accelerated chlamydial development in HEC-1B cells compared to HeLa and McCoy cells was also noticeable at 12 hpi (compare panel B versus panels E and H) and at 24 hpi (panel C versus panels F and I) since the inclusions in HEC-1B cells were larger and appeared to contain more numerous RB particles (white arrows). (E, inset) RB undergoing binary fission in HeLa cells at 12 hpi. Magnification, ×17,900.
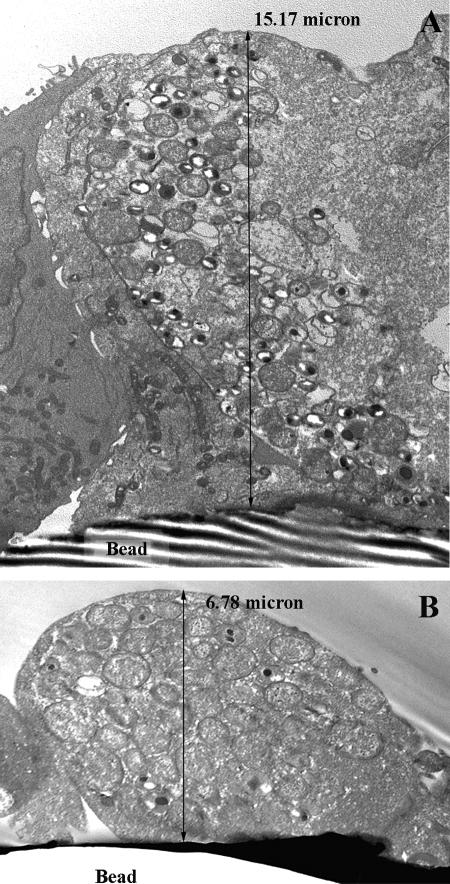
Ultrastructural analysis of sagittal views of the infected epithelial cells on bead cultures at 36 hpi confirmed the chlamydial inclusions to be larger and more mature in HEC-1B cells (Fig. 6A) than in HeLa cells (Fig. 6B). These data also illustrated the preservation of the tall columnar architecture of endometrial cells versus the low columnar stature of HeLa cells in polarized 3D culture. By 50 to 52 hpi, mature inclusions containing predominantly EB were present in both epithelial cell lines. Since at later incubation times postinfection, i.e., 54 to 60 hpi, most inclusions in the host cells were empty or contained only envelope ghosts and residual debris, 52 hpi was chosen as the time for harvesting progeny EB from all cell lines from the bead cultures.
FIG. 6.
Transmission electron photomicrograph sagittal views of HEC-1B and HeLa epithelial cells from bead cultures infected with C. trachomatis serovar E for 36 hpi. Epithelial cells grown on beads established monolayers with architectural characteristics similar to the tissue from which they originated; the tall columnar endometrium-derived HEC-1B cells on beads (A) were, on average, two- to threefold taller than the low columnar cervical-derived HeLa cells grown in the same conditions (B). Consistent with other data, chlamydial inclusions were larger and more mature in HEC-1B (A) than in HeLa (B) cells. Magnification: ×7,530.
Progeny numbers and infectivity.
Total EB particle counts were performed on progeny harvested from 500-ml bead cultures of each of the three cell lines. The number of EB recovered from infected McCoy cells (6.5 × 1010/ml) and HEC-1B cells (5.6 × 1010/ml) were similar and routinely ≥4-fold higher than the number of EB recovered from infected HeLa cells (1.4 × 1010/ml).
All purified EB stocks were then adjusted to the same concentration (1.0 × 109/ml), and titers were determined for infectivity on monolayers of the cell lines on coverslips. The pattern was similar to that illustrated in Fig. 2, i.e., the infectivity of serovar E EB was greater for epithelial cells than for fibroblasts, although not significant in this case. Even though EB infectivity for the two epithelial cell lines was similar, it is reproducibly better in HEC-1B cells than in HeLa cells (data not shown).
Comparative growth of C. trachomatis in HEC-1B and HeLa cells monitored by real-time PCR.
In order to confirm the ultrastructural data, chlamydial development in HEC-1B and HeLa cells grown on beads was evaluated at the nucleic acid level by monitoring cell-associated chlamydial genome copy numbers and chlamydial transcript levels throughout the developmental cycle (3 to 48 hpi). Of note, the primer sets specific for human genes (MPO and F8) used to normalize chlamydial growth in HEC-1B and HeLa cells did not amplify any PCR product when tested on murine fibroblasts McCoy cells; therefore, qPCR analysis was only performed on the two more relevant human genital epithelial cell lines.
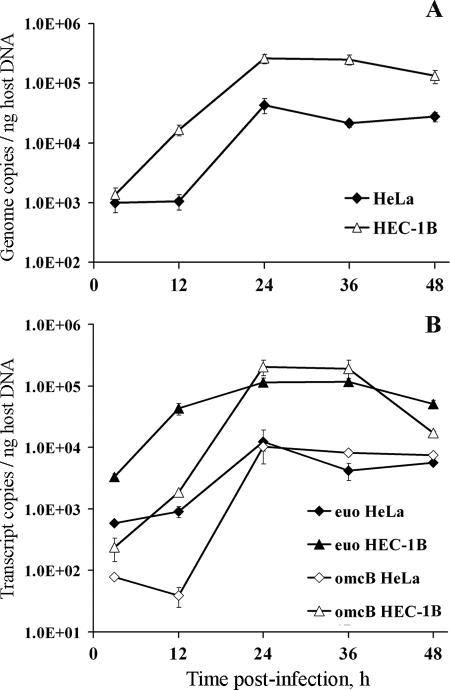
Interestingly, whereas chlamydial genome numbers (normalized to nanograms of host gDNA) were not significantly different at 3 hpi in HEC-1B and HeLa cell cultures as determined by qPCR, a 12.3-fold increase in bacterial genomes was detected in cultures of HEC-1B cells between 3 and 12 hpi; no change in the genome content was found in infected HeLa cells during that time interval (Fig. 7A). Between 12 and 24 hpi, chlamydial genome replication was detected in both cell lines, with a more dramatic increase in HeLa compared to HEC-1B cells, i.e., 40.5-fold versus 15.6-fold, respectively. No increase was found in bacterial genome numbers at later times in either cell line; however, the overall chlamydial genome accumulation throughout the developmental cycle was higher in HEC-1B compared to HeLa cells, resulting in 2 to 3.5 times more genomes in the former cell line by 48 hpi, depending on the experiment (Fig. 7A).
FIG. 7.
Real-time PCR analyses of C. trachomatis serovar E genome numbers and transcript levels in HEC-1B and HeLa cells grown in bead cultures. Aliquots of cultures were sampled at 3, 12, 24, 36, and 48 hpi, lysed, and used for concomitant genomic DNA (gDNA) and total RNA isolation. (A) Chlamydial genome copy numbers in each bead sample were determined by qPCR using euo- and omcB-specific primer sets and chlamydial gDNA standards; copy numbers obtained with the two primer sets were nearly identical and were therefore averaged. Chlamydial genomes were then normalized to the relative amount of host cell gDNA in the same sample, as determined by MPO- and F8-specific qPCR assays (averaged values). Normalized chlamydial genome copy numbers detected in both cell lines are shown on the y axis; sampling times are indicated on the x axis. (B) The levels of specific chlamydial transcripts in bead samples were determined after 2 μg of total RNA of each sample was reverse transcribed, along with an RT-minus (no enzyme) control, and analyzed by qPCR using the same euo- and omcB-specific assays and chlamydial gDNA standards described above. For each qPCR assay and each sample, the adjusted transcript copy numbers (i.e., RT − RT-minus) were then normalized to host cell gDNA as indicated above. Normalized chlamydial transcript levels are shown on the y axis; sampling times are indicated on the x axis. All time course samples were tested in triplicate, and all qPCR assays used had similar high efficiencies (≥97%). The figure shows average data from triplicate wells (± the standard deviation) from a representative time course experiment. Two independent experiments were performed and showed similar patterns.
Expression of euo and omcB transcripts, previously shown to occur early and late in the chlamydial developmental cycle, respectively (3, 44), was also monitored in the present study. At 3 hpi, approximately 5.5 times more euo transcripts were detected in HEC-1B cell bead cultures than in HeLa cell bead cultures (after normalization to the nanogram amounts of host gDNA; Fig. 7B), but whether this difference was the result of an “earlier start” in euo gene expression or of a “more active” transcriptional activity in HEC-1B cells was not determined. Indeed, because of the experimental setup and “in-suspension” inoculation procedure, the first bead samples were not collected any earlier than 3 hpi, the time by which the peak in euo expression may already have occurred (3, 44, 56). Notably, some increase in euo transcript levels was detected between 3 and 12 hpi, particularly in HEC-1B cells for which a 13.0-fold change was found, compared to a 1.6-fold increase in HeLa cells during the same time interval; a reverse trend was observed between 12 and 24 hpi. At later times (>24 hpi), steady or decreased levels in euo transcripts were detected in both HEC-1B and HeLa cells, suggesting a reduction in transcriptional activity for this early gene.
Real-time PCR analysis of late gene omcB expression showed a decline in transcript levels in serovar E-infected HeLa cells grown on beads during the 3- to 12-hpi time interval (Fig. 7B), which was consistent with previous findings in nonpolarized HeLa cells (3), and was likely to result from the degradation of “carryover” transcripts present in EB from the inoculum. In contrast, omcB transcript levels increased by ∼7.9-fold in HEC-1B cultures during the same time period, suggesting that omcB transcription was initiated earlier in this cell line compared to HeLa cells. Then, between 12 and 24 hpi, a large increase in omcB transcript levels was found in both cell lines but was more dramatic in HeLa cells, with a 260-fold increase versus a 108-fold increase in HEC-1B cells; nevertheless, overall omcB transcript levels were about 20-fold higher in the HEC-1B cell line at 24 hpi. At later times, while in HEC-1B cultures no change (24 to 36 hpi) and a decline (36 to 48 hpi) in omcB transcript levels were found, these transcripts were detected at steady levels in HeLa cells up to the last sampling time (48 hpi). It is noteworthy that very similar patterns were found when euo and omcB transcript levels were normalized to host cell β2-microglobulin transcripts (data not shown).
The more active growth of C. trachomatis serovar E in endometrial HEC-1B cells compared to endocervical HeLa cells was confirmed by Western blot analysis. Indeed, the production of chlamydial OmcB and HSP-60 copy-1 proteins, expressed at mid-to-late cycle, was first detected at 24 hpi in both cell lines, but the intensity of the signal was more intense in HEC-1B cell lysates than in HeLa cell lysates (data not shown).
DISCUSSION
It is well documented that the topography and cytoskeletal organization of nonpolarized epithelial cells cultured in a 2D fashion in vitro on impermeable plastic or glass surfaces resembles that of fibroblasts: plasma membrane proteins are evenly distributed circumferentially, the perinuclear region is broad with the Golgi complex juxtaposed to the nucleus, and microtubules extend horizontally out to the periphery in an orientation parallel to the basement membrane. In contrast, epithelial cells cultured in vitro in a more relevant 3D polarized orientation on suitable collagen or natural extracellular matrices are three to five times taller and have a different topography: plasma membrane proteins are separated, by tight junctions and junctional complexes, into distinct apical and basolateral membranes and are functionally compartmentalized therein; the Golgi complex assumes a supranuclear position in the apical cytoplasm domain, where chlamydial inclusions are localized, and microtubules are arranged vertically in an apical-to-basal axis parallel to the lateral membrane (55). New technology, such as nanotechnology engineering, reaffirms that the 3D in vitro cultivation of epithelial cells is crucial for such cells to sense, feel, and respond properly (18, 34, 36) to receptor complex presentation, cytoskeletal orientation or function, and signaling, i.e., tyrosine phosphorylation and Rho/Ras/Rac activation, all of which have been implicated as important in chlamydial infection.
In vitro culture methodologies to mimic to some degree the polarized organization of epithelial cells in vivo have existed for several years and have been used for a variety of experimental scenarios. Example methodologies include the application of collagen-coated polycarbonate Transwell filter chamber inserts (Costar) and/or extracellular matrix-coated filter Invasion chambers (BioCoat Matrigel; Becton Dickinson) to examine the mechanisms of pathogen entry and intracellular fate and the reactivity to antibiotics and innate inflammatory response cells (11, 17, 22, 23, 25-28, 30, 40). However, large-scale culture for harvesting many progeny microorganisms for stock quantities to pursue experimental questions using the methods described above is impractical in terms of both effort and cost. Exploiting the microcarrier bead culture system allows us to routinely recover 1010 infectious C. trachomatis serovar E organisms/ml, which is ∼10-fold greater than the number of progeny EB that can be harvested from equivalent infected host cell numbers in flasks. Furthermore, EB from the flask 2D system are less infectious. The nuances of how signaling and trafficking conditions in the 3D epithelial cell environment permit an accelerated and more synchronous chlamydial developmental cycle are not yet understood, but such in vivo-like relevant growth has the potential to affect experimental data interpretation.
C. trachomatis infection or biological function parameters have, indeed, been reported to differ between 3D versus 2D culture of epithelial cells, as well as differ between 3D culture of HEC-1B versus that of HeLa cells. For example, cumulative interleukin-8 (IL-8) concentrations were two- to threefold greater in nonpolarized HeLa cells cultured in vitro than in polarized HeLa cells in response to serovar E infection for 24 to 48 h. Further, IL-8 accumulation in polarized HeLa cells was twice that of IL-8 accumulation in polarized HEC-1B cells (16, 17). The latter results are also likely a reflection of the fundamentally different physiological functions between endometrial versus endocervical epithelial cells. The endometrial-cell-specific chemokine message for ENA-78 and GCP-2 was upregulated in polarized HEC-1B cells in vitro in response to serovar E infection (53). Although IL-8 can be expressed by the endometrium in vivo at specific stages of gestation and menstruation (2), it is probable that lower genital tract infection induces an ascending cascade of signals that programs endometrial epithelial cells to produce a cytokine signature such that these cells can notify the immune system to distinguish between the subtleties of physiological inflammation versus infection-induced inflammation. In a similar vein, polarized culture of epithelial cells in vitro can discriminate between the polarized infection-directed spread of serovar E and serovar L2 pathogens. Luminal serovar E EB escape from polarized endometrial epithelial cells via their apical domains to spread canalicularly to the upper genital tract, whereas the invasive, lymphotrophic serovar L2 EB exit via the basal domains (50). This latter result may be due to microtubule orientation, which in polarized cells is parallel to the lateral membrane with an apical-to-basal axis, on which serovar L2 are known to traffic (9). In addition, polarized HeLa cells, cocultured with human neutrophils and monocyte-derived macrophages, stimulate the release of 100-fold-higher amounts of tumor necrosis factor alpha from serovar L2 infection compared to serovar E infection (17). Finally, another significant biological function difference involves the pharmacokinetics of antibiotic uptake. Polarized HEC-1B cells internalize and concentrate threefold more [14C]azithromycin than nonpolarized HEC-1B cells (39, 51), which is reflected in lower 90% minimal bactericidal concentrations for inhibiting serovar E infection (37).
Another important finding from the present study using the original microcarrier bead system suggests that tall columnar epithelia, represented by the endometrial HEC-1B cell line, polarized better than low-columnar epithelia, represented by the cervical HeLa cell line. Our laboratory has experienced the same results when trying to culture polarized HeLa cells in vitro on the commercial filter-coated chamber inserts. Several hypotheses could be proposed to explain why HeLa cells do not anchor strongly to these matrices, which compromises cell-cell confluence and strong tight junction formation. Perhaps the adhesive and polarizing capabilities of HeLa cells were affected during immortalization. It is also possible that the natural matrix for correct cervical epithelial behavior was not mimicked in the available commercial microcarrier beads or filter chamber inserts; thus, polarized culture of HeLa cells might be able to be improved or achieved by slight changes in the matrix support (18, 45).
Chlamydial progeny yield from HEC-1B cells was, and reproducibly is, greater (2- to 3.5-fold and ≥4-fold as determined by qPCR and titration, respectively) than from HeLa cells, which is not entirely due to the premature loss of HeLa cells from the beads. These data have been confirmed by Deka et al. (15), who routinely recover 10-fold-greater C. trachomatis serovar E infectious progeny from HEC-1B cells (42,833 ± 1,167 IFU/ml) than from HeLa cells (4,767 ± 623 IFU/ml). Recently, Miyairi et al. (32) showed that chlamydial growth rate is influenced by the tropic cell type used. For instance, oculogenital strains of C. trachomatis grew faster in endocervical epithelial HeLa cells than in conjunctival epithelial cells and, reciprocally, prototype ocular strain serovar A growth was faster in the conjunctiva-derived cell line than in HeLa cells; the differences in chlamydial growth were, in fact, correlated to differences in the length of the lag phase (i.e., attachment, endocytosis, and primary differentiation of EB into RB) and of the generation time (i.e., binary fission of RB). Therefore, one could argue that polarized tall columnar epithelial cells (HEC-1B) and low squamo-columnar cervical cells (HeLa) may provide different growth environments for chlamydiae, even for the same strain, leading to the differences in growth rates seen in the present study. Although no difference in C. trachomatis serovar E attachment to and/or entry into the two genital epithelial cell lines was noted (Fig. 2), active bacterial genome replication and transcription (Fig. 7), as well as EB-to-RB initial conversion (Fig. 5), were detected earlier in HEC-1B cells than in HeLa cells, which suggests a shorter lag phase for this strain in the endometrial HEC-1B cell line, leading to higher progeny counts and titers in these cells upon completion of the developmental cycle. Therefore, the greater infectivity of chlamydia progeny EB harvested from HEC-1B cells is a reflection of the faster growth of the serovar E strain in this cell line than in HeLa cells.
A particularly interesting and curious finding, also of biological importance, was that EB attachment to both HEC-1B and HeLa cells in the microcarrier bead system is “patchy” and clustered. Since this finding is quite similar to our previous findings of patchy EB attachment to polarized primary human endometrial gland epithelial cells (33) and to polarized swine genital organ tissue cultured ex vivo (20), in contrast to an even distribution EB attachment pattern in the same cells cultured on glass coverslips, the pattern of attachment appears to be a 3D culture phenomenon, at least in part. The significance of this to receptor complex presentation is not yet clear given the varying reports of EB attachment to nonpolarized epithelia in compartmentalized domains such as caveolae (13, 24, 35, 49), lipid rafts (19, 46), and TARP-enriched pedestals (5, 10).
Since our laboratory started using a bead culture model for growing chlamydiae, advances in 3D suspension culture systems have been described, including (i) growing epithelial cell lines in rotating wall vessels under low shear microgravity conditions that are thought to better mirror in vivo conditions at various anatomic sites such as the uterus (34, 48) and (ii) substituting acellular collagen-rich small submucosa pieces for beads as bioscaffolds (6) for greater tissue differentiation and organoid formation. Such systems may, indeed, represent improved in vivo-like culture models for studying chlamydial biology and pathogenesis in epithelial cells.
Acknowledgments
We thank R. J. Belland and T. P. Hatch (Department of Molecular Sciences, University of Tennessee Health Science Center) who, respectively, generously provided the chlamydia-specific real-time PCR primers and antibodies used in this study. We also gratefully acknowledge J. M. Rary (Department of Pediatrics) and R. Dykes (Department of Microbiology) for their help with karyotyping our epithelial cell lines. We acknowledge the use of the transmission electron microscope in the Electron Microscopy Core Facility and the use of the robotic workstation Beckman Biomek 2000 and Bio-Rad iCycler in the Molecular Biology Core Facility.
This study was supported by Public Health Service grant AI13446 from the National Institute for Allergy and Infectious Diseases (P.B.W.).
Editor: D. L. Burns
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Abdelrahman, Y. M., and R. J. Belland. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Arici, A., E. Seli, L. M. Senturk, L. S. Gutierrez, E. Oral, and H. S. Taylor. 1998. Interleukin-8 in the human endometrium. J. Clin. Endocrinol. Metab. 83:1783-1787. [DOI] [PubMed] [Google Scholar]
- 3.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeckman, F., L. Tan, M. Brisson, R. Park, and K. Hamby. 2001. Real-time multiplex PCR from genomic DNA using the iCycler iQ detection system. Bulletin 2679. Bio-Rad Laboratories, Richmond, CA.
- 5.Carabeo, R. A., S. S. Grieshaber, E. Fischer, and T. Hackstadt. 2002. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 70:3793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho, H. M., L. D. Teel, G. Goping, and A. D. O'Brien. 2005. A three-dimensional tissue culture model for the study of attach and efface lesion formation by enteropathogenic and enterohaemorrhagic Escherichia coli. Cell Microbiol. 7:1771-1781. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2003. Sexually transmitted disease surveillance 2002 supplement: Chlamydial Prevalence Monitoring Project annual report 2002. [Online.] http://www.cdc.gov/std/chlamydia2002/chlamydia2002.pdf.
- 8.Centers for Disease Control and Prevention. 2005. sexually transmitted disease surveillance 2004 supplement: Chlamydia Prevalence Monitoring Project annual report 2004, p. 24. [Online.] http://www.cdc.gov/std/Chlamydia2004/ctsupplement_2004FINAL.pdf.
- 9.Clausen, J. D., G. Christiansen, H. U. Holst, and S. Birkelund. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 25:441-449. [DOI] [PubMed] [Google Scholar]
- 10.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart, P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darville, T. L. 2000. Chlamydia spp., p. 229-261. In J. P. Nataro (ed.), Persistent bacterial infections. ASM Press, Washington, DC.
- 13.Dautry-Varsat, A., A. Subtil, and T. Hackstadt. 2005. Recent insights into the mechanisms of Chlamydia entry. Cell. Microbiol. 7:1714-1722. [DOI] [PubMed] [Google Scholar]
- 14.Dean, D., R. J. Suchland, and W. E. Stamm. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 182:909-916. [DOI] [PubMed] [Google Scholar]
- 15.Deka, S., J. Vanover, J. Sun, J. Kintner, J. Whittimore, and R. Schoborg. An early event in herpes simplex virus type-2 replication cycle is sufficient to induce Chlamydia trachomatis persistence. Cell. Microbiol., in press. [DOI] [PubMed]
- 16.Dessus-Babus, S., S. T. Knight, and P. B. Wyrick. 2000. Chlamydial infection of polarized HeLa cells induces PMN chemotaxis but the cytokine profile varies between disseminating and non-disseminating strains. Cell. Microbiol. 2:317-327. [DOI] [PubMed] [Google Scholar]
- 17.Dessus-Babus, S. C., T. L. Darville, F. P. Cuozzo, K. Ferguson, and P. B. Wyrick. 2002. Differences in innate immune responses (in vitro) to HeLa cells infected with non-disseminating serovar E and disseminating serovar L2 of Chlamydia trachomatis. Infect. Immun. 70:3234-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Discher, D. E., P. Janmey, and Y. L. Wang. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science 310:1139-1143. [DOI] [PubMed] [Google Scholar]
- 19.Gabel, B. R., C. Elwell, S. C. van Ijzendoorn, and J. N. Engel. 2004. Lipid raft-mediated entry is not required for Chlamydia trachomatis infection of cultured epithelial cells. Infect. Immun. 72:7367-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guseva, N. V., S. T. Knight, J. D. Whittimore, and P. B. Wyrick. 2003. Primary cultures of female swine genital epithelial cells in vitro: a new approach for the study of hormonal modulation of Chlamydia infection. Infect. Immun. 71:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guseva, N. V., S. C. Dessus-Babus, J. D. Whittimore, C. G. Moore, and P. B. Wyrick. 2005. Characterization of estrogen-responsive epithelial cell lines and their infectivity by genital Chlamydia trachomatis. Microbes Infect. 7:1469-1481. [DOI] [PubMed] [Google Scholar]
- 22.Hopper, S., J. S. Wilbur, B. L. Vasquez, J. Larson, S. Clary, I. J. Mehr, H. S. Seifert, and M. So. 2000. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igietseme, J. U., P. B. Wyrick, D. Goyeau, and R. G. Rank. 1994. An in vitro model for immune control of chlamydial growth in polarized epithelial cells. Infect. Immun. 62:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jutras, I., L. Abrami, and A. Dautry-Varsat. 2003. Entry of the Lymphogranuloma venereum strain of Chlamydia trachomatis into host cells involves cholesterol-rich membrane domains. Infect. Immun. 71:260-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kane, C. D., and G. I. Byrne. 1998. Differential effects of gamma interferon on Chlamydia trachomatis growth in polarized and nonpolarized human epithelial cells in culture. Infect. Immun. 66:2349-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane, C. D., R. M. Vena, S. P. Ouellette, and G. I. Byrne. 1999. Intracellular tryptophan pool sizes may account for differences in gamma interferon-mediated inhibition and persistence of chlamydial growth in polarized and nonpolarized cells. Infect. Immun. 67:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazmierczak, B. I., K. Mostov, and J. N. Engel. 2001. Interaction of bacterial pathogens with polarized epithelium. Annu. Rev. Microbiol. 55:407-435. [DOI] [PubMed] [Google Scholar]
- 28.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 29.Linzmeier, R. M., and T. Ganz. 2005. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics 86:423-430. [DOI] [PubMed] [Google Scholar]
- 30.Merz, A. J., D. B. Rifenbery, C. G. Arvidson, and M. So. 1996. Traversal of a polarized epithelium by pathogenic neisseriae: facilitation by type IV pili and maintenance of epithelial barrier function. Mol. Med. 2:745-754. [PMC free article] [PubMed] [Google Scholar]
- 31.Michelini, M., A. Rosellini, V. Mandys, T. Simoncini, and R. P. Revoltella. 2005. Cytoarchitecture modifications of the human uterine endocervical mucosa in long-term three-dimensional organotypic culture. Pathol. Res. Pract. 201:679-689. [DOI] [PubMed] [Google Scholar]
- 32.Miyairi, I., O. S. Mahdi, S. P. Ouellette, R. J. Belland, and G. I. Byrne. 2006. Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J. Infect. Dis. 194:350-357. [DOI] [PubMed] [Google Scholar]
- 33.Moorman, D. R., J. W. Sixbey, and P. B. Wyrick. 1986. Interaction of Chlamydia trachomatis with human epithelium in culture. J. Gen. Microbiol. 132:1055-1067. [DOI] [PubMed] [Google Scholar]
- 34.Nickerson, C. A., and C. M. Ott. 2004. A new dimension in modeling infectious disease. ASM News 70:171-175. [Google Scholar]
- 35.Norkin, L. C., S. A. Wolfrom, and E. S. Stuart. 2001. Association of caveolin with Chlamydia trachomatis inclusions at early and late stages of infection. Exp. Cell Res. 266:229-238. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien, L. E., M. M. Zegers, and K. E. Mostov. 2002. Building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell. Biol. 3:531-537. [DOI] [PubMed] [Google Scholar]
- 37.Paul, T. R., S. T. Knight, J. E. Raulston, and P. B. Wyrick. 1997. Delivery of azithromycin to Chlamydia trachomatis-infected polarized human endometrial cells by polymorphonuclear leucocytes. J. Antimicrob. Chemother. 39:623-630. [DOI] [PubMed] [Google Scholar]
- 38.Pharmacia Fine Chemicals. 1981. Microcarrier cell culture: principles and methods. Almqvist and Wiksell Tryckeri AB, Uppsala, Sweden.
- 39.Raulston, J. E. 1994. Pharmacokinetics of azithromycin and erythromycin in human endometrial epithelial cells and in cells infected with Chlamydia trachomatis. J. Antimicrob. Chemother. 34:765-776. [DOI] [PubMed] [Google Scholar]
- 40.Sansonetti, P. J. 2001. Rupture, invasion, and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol. Rev. 25:3-14. [DOI] [PubMed] [Google Scholar]
- 41.Schachter, J. 1990. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 42.Schachter, J., and P. B. Wyrick. 1994. Culture and isolation of Chlamydia trachomatis, p. 377-390. In V. L. Clark and P. M. Bavoil (ed.), Methods in enzymology: bacterial pathogenesis, vol. 236, part B. Interaction of pathogenic bacteria with host cells. Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 43.Shannon, J. E. 1981. Chlamydiae need the real McCoy. ATCC Quarterly Newsl. 1:2-3. [Google Scholar]
- 44.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, M. M., and J. H. George. 2005. Exploring and engineering the cell surface interface. Science 310:1135-1138. [DOI] [PubMed] [Google Scholar]
- 46.Stuart, E. S., W. C. Webley, and L. C. Norkin. 2003. Lipid rafts, caveolae, caveolin-1, and entry by chlamydiae into host cells. Exp. Cell Res. 287:67-78. [DOI] [PubMed] [Google Scholar]
- 47.Tam, J. E., S. T. Knight, C. H. Davis, and P. B. Wyrick. 1992. Eukaryotic cells grown on microcarrier beads offer a cost-efficient way to propagate Chlamydia trachomatis. BioTechniques 13:374-378. [PubMed] [Google Scholar]
- 48.Unsworth, B. R., and P. I. Lelkes. 1998. Growing tissues in microgravity. Nat. Med. 4:901-907. [DOI] [PubMed] [Google Scholar]
- 49.Webley, W. C., L. C. Norkin, and E. S. Stuart. 2004. Caveolin-2 associates with intracellular chlamydial inclusions independently of caveolin-1. BMC Infect. Dis. 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyrick, P. B., J. Choong., C. H. Davis, S. T. Knight, M. O. Royal, A. S. Maslow, and C. R. Bagnell. 1989. Entry of genital Chlamydia trachomatis into polarized human epithelial cells. Infect. Immun. 57:2378-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyrick, P. B., C. H. Davis, J. E. Raulston, S. T. Knight, and J. Choong. 1994. Effect of clinically relevant culture conditions on antimicrobial susceptibility of Chlamydia trachomatis. Clin. Infect. Dis. 19:931-936. [DOI] [PubMed] [Google Scholar]
- 52.Wyrick, P. B., D. G. Gerbig, S. T. Knight, and J. E. Raulston. 1996. Accelerated development of genital Chlamydia trachomatis serovar E in McCoy cells grown on microcarrier beads. Microb. Pathog. 20:31-40. [DOI] [PubMed] [Google Scholar]
- 53.Wyrick, P. B., S. T. Knight, T. R. Paul, R. G. Rank, and C. S. Barbier. 1999. Persistent chlamydial antigens in antibiotic-exposed infected cells trigger neutrophil chemotaxis. J. Infect. Dis. 179:954-966. [DOI] [PubMed] [Google Scholar]
- 54.Wyrick, P. B. 2000. Intracellular survival by Chlamydia. Cell Microbiol. 2:275-282. [DOI] [PubMed] [Google Scholar]
- 55.Yeaman, C., K. K. Grindstaff, and W. J. Nelson. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73-98. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, L., A. L. Douglas, and T. P. Hatch. 1998. Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect. Immun. 66:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]