Abstract
Nucleotide-binding oligomerization domain (NOD) protein 1 (NOD1) and NOD2 are pathogen recognition receptors that sense breakdown products of peptidoglycan (PGN) (muropeptides). It is shown that a number of these muropeptides can induce tumor necrosis factor alpha (TNF-α) gene expression without significant TNF-α translation. This translation block is lifted when the muropeptides are coincubated with lipopolysaccharide (LPS), thereby accounting for an apparently synergistic effect of the muropeptides with LPS on TNF-α protein production. The compounds that induced synergistic effects were also able to activate NF-κB in a NOD1- or NOD2-dependent manner, implicating these proteins in synergistic TNF-α secretion. It was found that a diaminopimelic acid (DAP)-containing muramyl tetrapeptide could activate NF-κB in a NOD1-dependent manner, demonstrating that an exposed DAP is not essential for NOD1 sensing. The activity was lost when the α-carboxylic acid of iso-glutamic acid was modified as an amide. However, agonists of NOD2, such as muramyl dipeptide and lysine-containing muramyl tripeptides, were not affected by amidation of the α-carboxylic acid of iso-glutamic acid. Many pathogens modify the α-carboxylic acid of iso-glutamic acid of PGN, and thus it appears this is a strategy to avoid recognition by the host innate immune system. This type of immune evasion is in particular relevant for NOD1.
The ability to rapidly recognize microbial components and respond by initiating an acute inflammatory response is a crucial first line of defense against a microbial challenge. Overactivation of this inflammatory response can, however, lead to the clinical symptoms of septic shock and results in 100,000 deaths annually in the United States (33, 34). It has been estimated that 1% of hospitalized patients and 20 to 30% of patients in intensive care units develop sepsis. The advent of new antimicrobial resistance patterns, the increasing use of chemotherapeutic agents, and the emergence of diseases characterized by immunosuppression have caused the incidence of septic shock to increase dramatically.
Lipopolysaccharide (LPS), peptidoglycan (PGN), and lipoteichoic acid are three principal bacterial cell wall components implicated in inducing the clinical manifestations of septic shock (47). These components exert their biological effects by stimulating the host's monocytes and macrophages to produce proinflammatory mediators, such as tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), and IL-6. These mediators in turn elicit a variety of inflammatory responses in the host.
LPS, a vital component of the outer leaflet of the gram-negative bacterial outer membrane, comprises three structural units: an outer polysaccharide component, a core oligosaccharide region, and the innermost portion, lipid A (5, 32). The lipid A region is largely responsible for the proinflammatory activity of LPS and generally consists of a hexa-acylated bis-1,4′-phosphorylated glucosamine disaccharide. The results of recent studies indicate that the lipid A region (11, 15, 32) of LPS initiates inflammatory responses by interacting with the Toll-like receptor 4 (TLR4)/MD2 complex (1, 2, 30, 41) on the surfaces of mononuclear phagocytes.
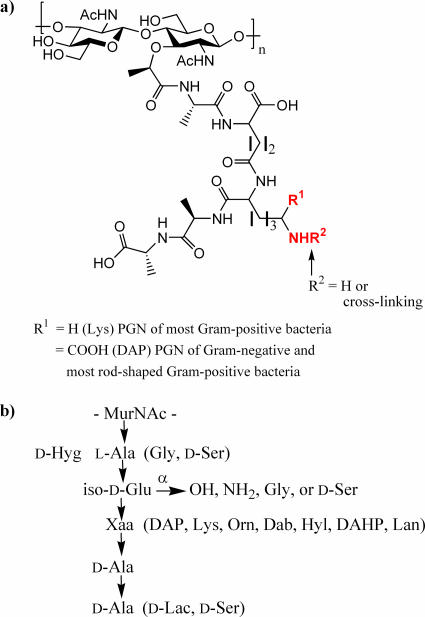
PGN is particularly abundant in gram-positive bacteria, in which it accounts for approximately one-half of the cell wall mass. On the other hand, gram-negative bacteria contain only a relatively thin PGN layer in the periplasmic space (17, 29, 35, 37, 48, 49). PGN is a large polymer composed of alternating β(1-4)-linked N-acetylglucosamine and N-acetylmuramic acid residues, cross-linked by short peptide bridges (Fig. 1). These peptides consist of four or five alternating l and d amino acids that are attached to the carboxylic acid of N-acetylmuramic acid. Among different bacterial species the structure of the sugar chain is rather conserved, while the composition of the peptide subunits varies considerably. For example, lysine is commonly the third amino acid of the peptide moieties of PGN of most gram-positive bacteria, while gram-negative and most rod-shaped gram-positive bacteria have a diaminopimelic acid (DAP) residue at this position. Furthermore, the α-carboxylate of the iso-d-Glu moiety is often amidated or linked to additional amino acids such as Gly or d-Ser (17, 23, 24, 37, 43). Although, it was initially thought that the d-Ala-d-Ala moiety was invariant, it has been discovered that, in antibiotic-resistant gram-positive strains, the terminal d-Ala residue can be replaced by d-Lac or d-Ser (37). There is also considerable heterogeneity in the nature of the interpeptide bridge, which connects the ɛ-amino group of the lysine or DAP moiety to the penultimate d-Ala of another PGN chain, resulting in loss of the terminal d-Ala (37).
FIG. 1.
Structure of PGN. (a) Common primary structure of PGN. (b) Variations in the peptide chain of PGN. Residues in parentheses may replace corresponding amino acids. The α-carboxylic acid of iso-d-Glu may be modified by an amide, Gly, or d-Ser. Dab, 2,4-diaminobutyric acid; DAHP, 2,6-diamino-3-hydroxypimelic acid; Hyg, threo-3-hydroxyglutamic acid; Hyl, hydroxylysine; Lan, lanthionine; Orn, ornithine.
It has been proposed that PGN is recognized by TLR2 in combination with TLR1 or TLR6, resulting in the production of cytokines (38, 42, 51). Recent studies using highly purified PGN have, however, provided conflicting results, and hence it is unclear whether TLR1/2 or TLR2/6 can initiate cellular activation after binding PGN (14, 45). In addition, it has been shown that peptidoglycan recognition proteins (PGRPs) (3, 12, 13, 19, 39) can either bind or enzymatically cleave PGN. There are convincing data that nucleotide-binding oligomerization domain (NOD) protein 1 (NOD1) and NOD2 are pattern recognition receptors for PGN (6, 13, 19, 25). Structure-activity relationship studies have indicated that NOD1 recognizes muramyl tripeptide containing DAP (MTP-DAP) (7, 18), whereas NOD2 mediates cellular activation induced by muramyl dipeptide (MDP) and lysine-containing muramyl tripeptide (MTP-Lys) (20, 26).
Recently, we demonstrated that MDP induces TNF-α gene expression without significant TNF-α protein production (50). This block in translation is, however, removed in the presence of LPS and PGN, thereby accounting for an apparently synergistic effect of MDP with LPS or PGN on TNF-α protein production. It is unclear whether any PGN breakdown products other than MDP can induce a synergistic effect with LPS and PGN. In this respect, it has been found that Bordetella pertussis, the causative agent of the respiratory disease pertussis, releases relatively large quantities of the muropeptide tracheal cytotoxin, a subunit of gram-negative bacterial PGN (16). When applied to hamster trachea epithelial cells, muropeptide tracheal cytotoxin and LPS were found to be highly synergistic in the induction of IL-1α and nitric oxide synthase. Others, however, have found that this compound and other PGN part structures do not induce a synergistic effect (44).
As part of a study to determine in detail the immunological properties of PGN part structures, we have examined the ability of a relatively large range of synthetic muropeptides to induce TNF-α mRNA and protein production in a human monocytic cell line. Furthermore, TNF-α secretion induced by LPS alone was compared to that of LPS coincubated with the muropeptides. The resulting data have been correlated with cellular activation studies using HEK293T cells transfected with NOD1 or NOD2 and a reporter gene.
MATERIALS AND METHODS
Reagents.
The synthesis of compounds 1 to 9 has been reported elsewhere (9, 10, 22, 28, 36, 40). Escherichia coli 055:B5 LPS was obtained from List Biological Laboratories, polymyxin B (PMB) from Bedford Laboratories, and recombinant human TNF-α from Endogen. All data presented in this study were generated using the same batch of E. coli 055:B5 LPS. Plasmids expressing human NOD1 and NOD2 (pcDNA3-NOD1-HA and pcDNA3-NOD2-HA, respectively) were generously provided by G. Nunez (University of Michigan, Medical School, Ann Arbor).
Cell culture.
Mono Mac 6 (MM6) cells, provided by H. W. L. Ziegler-Heitbrock (University of Munich, Munich, Germany), were cultured in RPMI 1640 medium with l-glutamine (BioWhittaker) supplemented with penicillin (100 U/ml)-streptomycin (100 μg/ml; Mediatech), OPI supplement (containing oxaloacetate [1 mM], pyruvate [0.45 mM], and bovine insulin [0.2 U/ml; Sigma]), and fetal bovine serum (10%; HyClone). New batches of frozen cell stock were grown up every 2 months, and growth morphology was evaluated. Before each experiment, MM6 cells were incubated with calcitriol (10 ng/ml; Sigma) for 2 days for differentiation into macrophage-like cells. The human embryonic kidney cell line (HEK293T), obtained from ATCC, was grown in Dulbecco's modified Eagle's medium (ATCC) with l-glutamine (4 mM), glucose (4.5 g/liter), and sodium bicarbonate (1.5 g/liter) supplemented with penicillin (100 U/ml)-streptomycin (100 μg/ml; Mediatech) and fetal bovine serum (10%; HyClone). All cells were maintained in a humid 5% CO2 atmosphere at 37°C.
TNF-α ELISA.
Differentiated cells were harvested by centrifugation, gently suspended (106 cells/ml) in prewarmed (37°C) medium, and incubated with different combinations of stimuli for 5 h in the presence or absence of PMB as described below. To study the synergistic effect of the muropeptides, cells were first preincubated with the muropeptides for 30 min. Concentrations of TNF-α in culture supernatants were determined in duplicate by a solid-phase sandwich enzyme-linked immunosorbent assay (ELISA). Briefly, 96-well plates (Nalge Nunc International) were coated with purified mouse anti-human TNF-α monoclonal antibody (MAb; Pharmingen). TNF-α in standards and samples was allowed to bind to the immobilized MAb for 2 h at room temperature, after which biotinylated mouse anti-human TNF-α MAb (Pharmingen) was added. After addition of avidin-horseradish peroxidase conjugate (Pharmingen) and ABTS [2,2′-azino-di(3-ethylbenthiazoline-6-sulfonate)] peroxidase substrate (Kirkegaard & Perry Laboratories), the absorbance was measured at 405 nm using a microplate reader (Molecular Devices). LPS concentration-response data were analyzed using nonlinear least-squares curve fitting in Prism (GraphPad Software, Inc.). These data were fit with the following logistic equation: Y = Emax/[1 + (EC50/X)Hill slope], where Y is the TNF-α response, X is the LPS concentration, Emax is the maximum response, and EC50 is the concentration of LPS producing 50% stimulation. All TNF-α values are normalized for either the Emax or EC50 of LPS (100%) and presented as the means ± standard deviations (SD) of duplicate cultures, with each experiment being repeated three times.
Preparation of RNA and quantification of TNF-α mRNA by real-time PCR analysis.
Cells were harvested by centrifugation, gently suspended (1.25 × 106 cells/ml) in prewarmed (37°C) medium, and incubated with stimuli in the presence or absence of polymyxin B. After 1.5 h the cells were harvested and total RNA isolated using the Absolutely RNA miniprep kit (Stratagene). TNF-α gene expression was quantified in a two-step reverse transcription-PCR (RT-PCR) using random hexamers from TaqMan RT reagents, the Taqman universal PCR master mixture, and TaqMan predeveloped assay reagents for human TNF-α (Applied Biosystems). Measurements were performed using the ABI Prism 7900 HT sequence detection system (Applied Biosystems). As an endogenous control, 18S rRNA gene expression was measured using the TaqMan rRNA control reagents (Applied Biosystems). Results represent means ± SD of quadruplicate measurements. Each experiment was repeated three times.
Evaluation of materials for LPS contamination.
To ensure that any increase in TNF-α production was not caused by LPS contamination of the solutions containing the various stimuli, the experiments were performed in the absence and presence of PMB, an antibiotic that avidly binds to the lipid A region of LPS, thereby preventing LPS-induced monokine production (46). TNF-α concentrations in supernatants of cells preincubated with PMB (30 μg/ml) for 30 min before incubation with E. coli O55:B5 LPS (10 ng/ml) for 5 h were reduced from 2,330 ± 111 pg/ml to 10 ± 7 pg/ml. Similarly, preincubation with PMB before incubation with E. coli O55:B5 LPS (100 pg/ml) for 90 min decreased TNF-α mRNA induction from 16.65-fold to 1.03-fold. Preincubation with PMB had no effect on either mRNA expression or TNF-α synthesis by cells incubated with synthetic compounds 1 to 9. Therefore, contamination of the latter preparations was inconsequential.
Transfection and NF-κB activation assay.
The day before transfection, HEK293T cells were plated in 96-well cell culture plates (16,000 cells/well). The next day, cells were transiently transfected with expression plasmids using PolyFect transfection reagent (QIAGEN). Briefly, pcDNA3-NOD1-HA (0.05 ng/well) or pcDNA3-NOD2-HA (0.005 ng/well) was cotransfected with pELAM-Luc (NF-κB-dependent firefly luciferase reporter plasmid; 50 ng/well) (8) and pRL-TK (Renilla luciferase control reporter vector; 1 ng/well; Promega) as an internal control to normalize experimental variations. The empty vector pcDNA3 (Invitrogen) was used as a control and to normalize the DNA concentration for all of the transfection reactions (total DNA, 70 ng/well). Forty-four hours posttransfection, cells were exposed to the stimuli for 4 h, after which cell extracts were prepared. The luciferase activity was measured using the dual-luciferase reporter assay system (Promega), according to the manufacturer's instructions, and the Fluoroskan Accent FL combination luminometer/fluorometer (Thermo Electron Corporation). Expression of the firefly luciferase reporter gene was normalized for transfection efficiency with expression of Renilla luciferase. The data are reported as the means ± SD of triplicate treatments. The transfection experiments were repeated at least twice.
Statistical analysis.
All data were analyzed using the unpaired Student t test. A P value of <0.05 was regarded as statistically significant.
RESULTS
Synthetic derivatives of peptidoglycan.
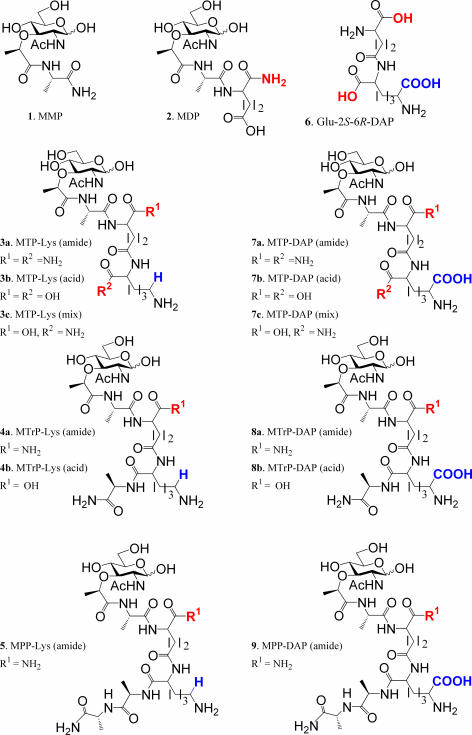
A number of synthetic muropeptides (Fig. 2; compounds 1 to 9) (9, 10, 22, 28, 36, 40) were employed to determine structure-activity relationships for the secretion of TNF-α protein in the absence or presence of LPS, the transcription of TNF-α mRNA, and the activation of HEK293T cells transfected with either NOD1 or NOD2 in combination with a reporter gene. The experiments were designed in such a way to uncover which important PGN part structure can induce synergistic TNF-α production when coincubated with LPS. Furthermore, the data should reveal whether NOD1 and NOD2 are involved in this effect. Also, it was expected that novel structure activity relationships for the activation of NOD1/2 proteins would be obtained, because a number of muropeptides with a unique structure were employed.
FIG. 2.
Structures of synthetic muropeptides.
Muropeptides 1 (MMP) and 2 (MDP) are part structures of PGN of gram-negative and -positive bacteria. Lysine-containing muramyl tripeptides 3a to 3c (MTP-Lys), muramyl tetrapeptide 4a and 4b (MTrP-Lys), and muramyl pentapeptide 5 (MPP-Lys) are derived from PGN of gram-positive bacteria. On the other hand, compounds 7 to 9 are part structures of PGN of gram-negative and rod-shaped gram-positive bacteria and structurally similar to muropeptides 3 to 5, with the exception that the lysine residue is replaced by a DAP moiety. A unique feature of the range of compounds is that the α-carboxylic acid of the iso-d-Glu is either in the acid or amide form. This modification is biologically relevant, because several strains of bacteria modify this functionality as an amide or extend it by amino acids (Fig. 1) (4, 17, 29, 37, 49). Furthermore, the importance of the α-carboxylic acid of the lysine and DAP moieties of the peptides has been investigated by the use of an acid- or amide-modified derivative. Thus, MTP derivatives 3a and 7a have amide-modified α-carboxylic acids at the iso-d-Glu and lysine or DAP moieties, whereas in compounds 3b and 7b these functionalities are in the acid form. MTP derivatives 3c and 7c are mixed structures in which α-carboxylic acid of iso-d-Glu is in the acidic form and the lysine or DAP moiety is modified as an amide. Also, the α-carboxylic acids of the iso-d-Glu's of MTrPs 4 and 8 are either modified as an amide (4a and 8a) or as a carboxylic acid (4b and 8b). In this case, the α-carboxylic acid of the lysine or DAP moiety is extended by a d-Ala residue and thus is in the amide form.
Transcription and translation of TNF-α and activation of NOD1 and NOD2 by lysine-containing peptidoglycan derivatives.
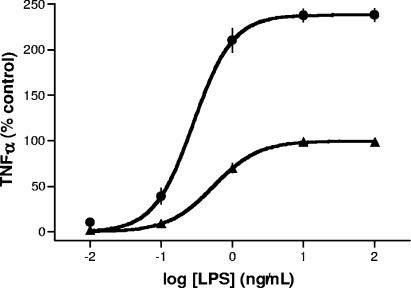
The effect of incubation of MM6 cells with MDP on the TNF-α secretion induced by a wide concentration range of LPS was compared with incubation with LPS alone (Fig. 3). Culturing the cells with muropeptide 2 (MDP; 100 μM) resulted in an LPS dose-response curve having a higher maximal level, whereas the EC50 and Hill slope values did not differ significantly from those obtained in the absence of MDP. Stimulation with MDP alone resulted in virtually no TNF-α protein production.
FIG. 3.
Effect of MDP (muropeptide 2) on dose response of LPS. Mono Mac 6 cells were preincubated with muropeptide 2 (100 μM) (•) or medium (▴) as control for 30 min at 37°C. Increasing concentrations of LPS were added, and after an incubation of 5 h, TNF-α protein was measured by ELISA. TNF-α values were normalized for the Emax of LPS (2,305 pg/ml; 100%). Stimulation with muropeptide 2 alone (100 μM) resulted in a TNF-α concentration of 69 ± 12 pg/ml (3% of control).
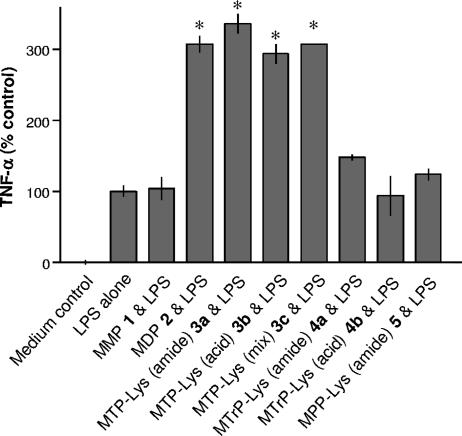
After the dose-response curve for LPS had been established, the MM6 cells were incubated with gram-positive-bacterium-derived muropeptides 1 to 5 (100 μM each) in the presence of LPS at its EC50 value. This concentration was selected because a relatively large synergistic effect would be expected. As can be seen in Fig. 4, muramyl tripeptides 3a to 3c induced a similar synergistic effect as muropeptide 2 (MDP), whereas muropeptides 1 (MMP), 4a and 4b (MTrP-Lys), and 5 (MPP-Lys) were inactive. Compounds 1 to 5 did not produce appreciable levels of TNF-α production when used alone (data not shown).
FIG. 4.
Induction of TNF-α production by MM6 cells treated with muropeptides 1 to 5 and LPS. Mono Mac 6 cells were preincubated with medium as control, peptides 1 and 2, or lysine-containing muropeptides 3 to 5 (100 μM each) for 30 min at 37°C. After 5 h of stimulation with LPS (EC50; 0.5 ng/ml) TNF-α was determined. TNF-α values were normalized for the EC50 of LPS (100%). Asterisks indicate significant differences from cells incubated with LPS alone (P < 0.05).
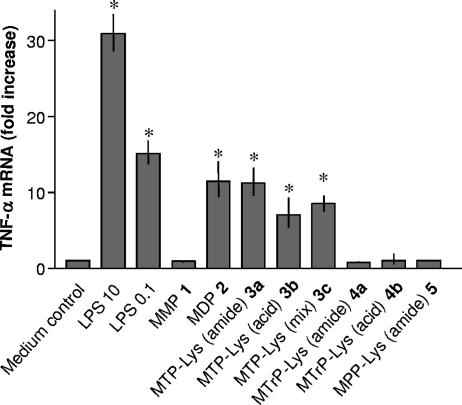
Previously, we had observed that MDP can induce the transcription of TNF-α mRNA without translation to the protein (50). This translational block was, however, lifted when muropeptides were coincubated with another stimulus such as LPS or PGN, resulting in an apparently synergistic effect. As expected, stimulation of MM6 cells for 90 min with LPS and compounds 2 and 3a to 3c resulted in TNF-α mRNA expression, whereas derivatives 1, 4a and 4b, and 5 showed no activity (Fig. 5).
FIG. 5.
Induction of TNF-α mRNA by MM6 cells incubated with muropeptides 1 to 5. Mono Mac 6 cells were stimulated with medium alone (control), medium containing LPS (10 and 0.1 ng/ml), peptides 1 and 2, or lysine-containing muropeptides 3 to 5 (100 μM each) for 1.5 h before RNA was isolated for RT-PCR analysis of TNF-α message (40 PCR cycles). mRNA of the 18S ribosomal gene amplified under the same conditions was used as an internal control. Asterisks indicate significant differences from untreated (control) cells (P < 0.05).
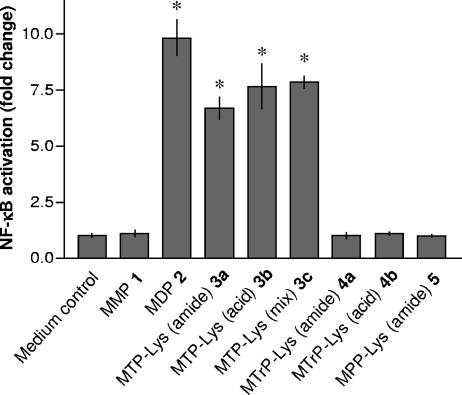
Several studies have indicated that the NOD proteins are involved in synergistic TNF-α production, when particular muropeptides are cocultured with LPS. In this respect, MM6 cells express NOD1 and NOD2 mRNA as determined by RT-PCR and agarose gel electrophoresis (data not shown). Furthermore, recent studies with HEK293T cells transiently transfected with NOD1 or NOD2 in combination with an NF-κB-luciferase reporter construct have demonstrated that these proteins can induce NF-κB activation when stimulated with particular PGN fragments (7, 18, 20, 26). Thus, it was expected that incubation of HEK293T cells transfected with NOD2 with compounds 1 to 5 would yield similar structure-activity relationships as observed for TNF-α mRNA transcription and a synergistic effect for TNF-α secretion. Indeed, compounds 2 and 3a to 3c induced approximately a 10-fold increase in NF-κB activation in cells transfected with NOD2, whereas compounds 1, 4, and 5 were inactive (Fig. 6). None of these derivatives showed activity in cells transfected with NOD1 (data not shown).
FIG. 6.
Response of HEK293T cells expressing NOD2 to muropeptides 1 to 5. Induction of NF-κB activation was determined in triplicate cultures of HEK293T cells transiently transfected with the pcDNA-NOD2 expression vector in the presence of pELAM-Luc, pRL-TK, and pcDNA3 plasmids. Forty-four hours posttransfection, cells were treated with synthetic compounds 1 to 5 (5 μM each) or were left untreated (control). Forty-eight hours posttransfection, NF-κB activation, as firefly luciferase activity relative to Renilla luciferase activity, was determined. In the transfection experiment shown, human TNF-α (10 ng/ml) induced 14.0-fold ± 1.7-fold activation of NF-κB. Asterisks indicate significant differences from untreated (control) cells (P < 0.05).
Transcription and translation of TNF-α and activation of NOD1 and NOD2 by DAP-containing peptidoglycan derivatives.
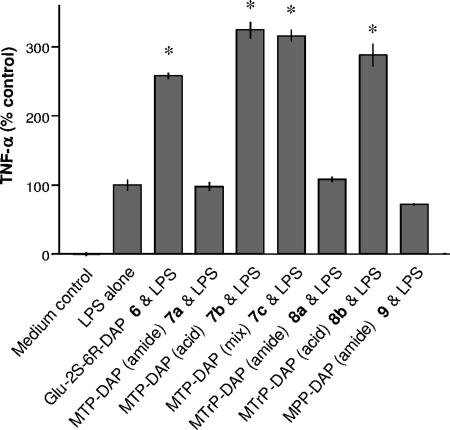
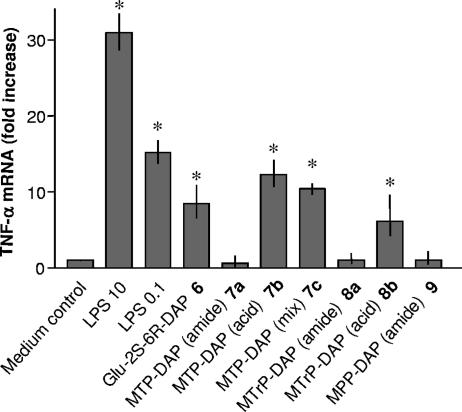
Next, similar cellular activation studies were performed with compounds 6 to 9, which contain the unusual amino acid DAP, unique to PGN from gram-negative bacteria. Although each compound displayed similar activities for synergistic effects (Fig. 7), TNF-α mRNA transcription (Fig. 8), and activation of NOD1 (Fig. 9), the structure-activity relationship was unusual. In this case, dipeptide 6, MTP-DAPs 7b and 7c, and MTrP-DAP 8b displayed activity in each assay. Interestingly, MTP-DAP 7a, which has amide functions at the α-carboxylic acids of both the iso-glutamic acid and DAP moieties, was inactive. Thus, it appears that for NOD1 ligands the chemical nature of the α-carboxylic acid of the iso-glutamic acid residue is critical for biological activity, whereas this is not the case for lysine-containing derivatives. As expected, none of the DAP-containing derivatives showed activity in cells transfected with NOD2 (data not shown).
FIG. 7.
Induction of TNF-α production by MM6 cells treated with muropeptides 6 to 9 and LPS. Mono Mac 6 cells were preincubated with medium as control or DAP-containing muropeptides 6 to 9 (100 μM each) for 30 min at 37°C. After 5 h of stimulation with LPS (EC50; 0.5 ng/ml) TNF-α was determined. TNF-α values were normalized for the EC50 of LPS (100%). Asterisks indicate significant differences from cells incubated with LPS alone (P < 0.05).
FIG. 8.
Induction of TNF-α mRNA by MM6 cells incubated with muropeptides 6 to 9. Mono Mac 6 cells were stimulated with medium alone (control), medium containing LPS (10 and 0.1 ng/ml), or DAP-containing muropeptides 6 to 9 (100 μM each) for 1.5 h before RNA was isolated for RT-PCR analysis of TNF-α message (40 PCR cycles). mRNA of the 18S ribosomal gene amplified under the same conditions was used as an internal control. Asterisks indicate significant differences from untreated (control) cells (P < 0.05).
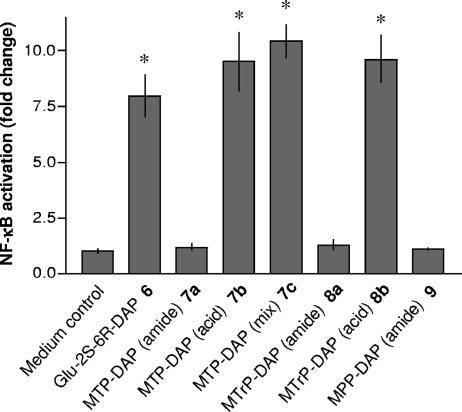
FIG. 9.
Response of HEK293T cells expressing NOD1 to muropeptides 6 to 9. Induction of NF-κB activation was determined in triplicate cultures of HEK293T cells transiently transfected with the pcDNA-NOD1 expression vector in the presence of pELAM-Luc, pRL-TK, and pcDNA3 plasmids. Forty-four hours posttransfection, cells were treated with the synthetic compounds 6 to 9 (5 μM each) or were left untreated (control). Forty-eight hours posttransfection, NF-κB activation, as firefly luciferase activity relative to Renilla luciferase activity, was determined. In the transfection experiment shown, human TNF-α (10 ng/ml) induced 10.8-fold ± 0.9-fold activation of NF-κB. Asterisks indicate significant differences from untreated (control) cells (P < 0.05).
DISCUSSION
The results presented here demonstrate that lysine- and DAP-containing muropeptides 2, 3a to 3c, 6, 7b and 7c, and 8b induce TNF-α gene expression without significant TNF-α translation. The block in translation is, however, removed in the presence of LPS, thereby accounting for an apparent synergistic effect of these compounds on TNF-α protein production. Importantly, MTP-DAP 7a did not induce TNF-α gene expression nor did it exhibit a synergistic effect with LPS. The structure of muropeptide 7a is similar to that of the muramyl tripeptide 7b, except that the α-carboxylic acids of the iso-glutamic acid and DAP moiety are modified as amides. The observation that mixed compound 7c is active indicates that the iso-glutamic acid moiety needs a free α-carboxylic acid for TNF-α mRNA transcription. The importance of the acidic nature of this functionality was also observed for MTrP derivatives 8a and 8b. Interestingly, amidation of the α-carboxylic acids of the iso-glutamic acid moiety of lysine-containing MTP (muropeptides 3a to 3c) did not affect the biological properties. However, for this series of compounds no activity was observed for the lysine-containing MTrPs (MTrP-Lys 4a and 4b).
The observation that the DAP- and lysine-containing muramyl tripeptides display different structure-function relationships indicates that they initiate cellular activation through different receptors. In this respect, TLR2, PGRPs, and the NOD proteins have been implicated in PGN-mediated innate immune responses (13). Initially, it was thought that PGN is recognized by TLR2 in combination with TLR1 or TLR6, resulting in the production of cytokines (38, 42, 51). However, recent studies have cast doubt on the involvement of this receptor, because extensive purification of PGN resulted in a loss of activity when PGN was exposed to cells transfected with CD14 and TLR2 (45). However, others have not observed loss of activity upon extensive purification of PGN (14), and hence the involvement of TLR2 in PGN recognition is still controversial. It has been shown that PGRPs (3, 12, 13, 19, 39) can either bind or enzymatically cleave PGN. Little is, however, known about the mode of cellular activation mediated by mammalian PGRPs. Crystal structure and binding studies are beginning to shed light on the ligand requirements of these PGRPs (22, 28), which may provide important clues for determining their biological functions. There are convincing data to support the hypothesis that NOD receptors (NOD1 and NOD2) are pattern recognition receptors that sense PGN (6, 13, 19, 25). For example, previous studies have shown that NOD1 recognizes DAP-containing MTP (7, 18), and NOD2 has been implicated in cellular activation by MDP and lysine-containing MTP (20, 26). Furthermore, it has been shown that mice pretreated with MDP are sensitized to endotoxic shock induced by LPS, whereas NOD2-deficient mice were resistant to such a challenge (27). In addition, it has been observed that a synergistic effect of lipopeptide Pam3CS(K)4 and MDP in wild-type mice is absent in mouse macrophages deficient in NOD2. Also, down regulation of NOD2 in vascular endothelial cells by interfering RNA reduced synergistic effects (31). Thus, these studies indicate that the NOD proteins are involved in the apparently synergistic effect of muropeptides with LPS.
In this study, we have observed similar structure-activity relationships of compounds 1 to 9 for TNF-α mRNA production, synergistic TNF-α secretion on costimulation with LPS, and NF-κB activation mediated by NOD1 or NOD2, further implicating the NOD proteins in synergistic TNF-α secretion. Thus, it appears that activation of the NOD proteins leads to TNF-α mRNA transcription without translation to the protein. This translation block is lifted when the muropeptides are coincubated with a TLR agonist.
The structure-activity relationship that we have established is unusual. Previously, it was proposed that the NOD1 sensing system requires an exposed DAP moiety (7, 18). However, we have found that MTrP-DAP 8b can activate NF-κB in a NOD1-dependent manner, demonstrating that an exposed DAP moiety is not essential for sensing by NOD1. The activity was, however, lost when the α-carboxylic acid of the iso-glutamic acid moiety was modified as an amide. On the other hand, amidation of this function does not affect the activity of MDP (muropeptide 2) and MTP-Lys (muropeptides 3a to 3c), which are compounds sensed by NOD2.
Many bacteria modify the α-carboxylic acid of iso-glutamic acid of PGN by amidation (4, 17, 29, 37, 49) or by addition of Gly or d-Ser. For example, Staphylococcus aureus, Staphylococcus epidermidis, Arthrobacter crystallopoietes, and various strains of lactobacilli, micrococci, and streptococci are only a few of many gram-positive bacterial strains that have the α-carboxyl group of glutamic acid present as an amide (37, 43). Also, many strains of gram-negative bacteria modify the carboxylic acids of glutamic acid or DAP as amides, and based on this modification three subtypes of PGN have been identified (37). Thus, E. coli and Bacillus megaterium have none of the carboxyl groups amidated. In Bacillus licheniformis, the α-carboxyl group of glutamic acid is modified as an amide, whereas most of the peptide subunits of B. subtilis and B. licheniformis have an amide substituent on the carboxyl group of the d-asymmetric carbon of DAP. Cell walls of Lactobacillus plantarum and Corynebacterium diphtheriae have both amidated carboxyl groups.
Our results suggest that modification of the structure of PGN can be a strategy to avoid recognition by the host innate immune system. Previously, it was found that PGN fragments from B. subtilis also do not activate NOD1 (7, 18). Interestingly, PGN of this gram-positive bacterial strain contains DAP as the third amino acid, which is 95% amidated (20, 37). Thus, modification of the δ-carboxylic acid of DAP may be another way to avoid detection by NOD1.
It is well established that gram-negative bacteria avoid detection by the host immune system by modification of the lipid A component of LPS (11, 15, 32). For example, pathogens such as Helicobacter pylori and Chlamydia trachomatis, which are associated with chronic infections, contain LPSs of low biological activity. Reduction in toxicity of LPS is due to removal or modification of acyl chains or changes in phosphorylation state. It appears that bacteria employ a similar strategy to avoid host detection of PGN. In particular, there is evidence supporting modification of carboxylic acids of iso-glutamic acid and DAP as an effective strategy to avoid immune detection by NOD1. Interestingly, NOD2 is not affected by modification of the iso-glutamic acid moiety. It may well be possible that NOD2, which is a general sensor for PGN, has been under greater evolutionary pressure to sense a wide range of PGN modifications. However, it is important to realize that almost every amino acid of PGN can be modified (4, 17, 29, 37, 49). Furthermore, a number of pathogens change their PGNs' carbohydrate backbones by acetylation or 1,6-anhydropyranose formation. Thus, a larger range of PGN derivatives need to be prepared to determine their effects on NOD1 and NOD2 sensing. It is also important to realize that small structural differences of muropeptides may be the origin of conflicting reports (16, 44).
Finally, the stem peptides of peptidoglycan contain usually four or five amino acids, which can be cross-linked with peptide fragments of various lengths (Fig. 1), and thus a key issue is how biologically active muropeptides, such as the various muramyl tripeptides discussed in this paper, are formed in the host. Serum enzymes of the host, including amidases and lysosyme, are involved in PGN processing; however, to the best of our knowledge no mammalian enzyme that can produce muramyl tripeptides has been identified. On the other hand, bacteria express a relatively large variety of enzymes that can hydrolyze peptidoglycan, including those that can produce immunologically relevant muramyl tripeptides (17, 23, 24). In this respect, d,l-carboxypeptidases can cleave the peptide bond between alanine and isoglutamine of muramyl tetrapeptides, leading to the formation of muramyl tripeptide-containing fragments. Furthermore, the carbohydrate backbone can be hydrolyzed by N-acetylmuramidases and N-acetylglucosaminidase, and finally, endopeptidases, such as lysostaphin, can cleave the peptide bond between lysine- and glycine-rich bridging peptides. Peptidoglycan biosynthesis is highly dynamic, and as much as 50% is turned over per generation. It has been estimated that during this process as much as 6 to 8% of peptidoglycan is lost due to the release of short fragments (21), which can be released in the host, taken up by macrophages and related cells, and then sensed by NOD proteins.
Acknowledgments
This research was supported by the Institute of General Medicine of the National Institutes of Health (GM065248).
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Beutler, B. 2004. Innate immunity: an overview. Mol. Immunol. 40:845-859. [DOI] [PubMed] [Google Scholar]
- 3.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 4.Bugg, T. D. H. 1999. Bacterial peptidoglycan. Biosynthesis and its inhibition, p. 241. In B. M. Pinto (ed.), Comprehensive natural products chemistry. Carbohydrates and their derivatives including tannins, cellulose, and related lignins, vol. 3. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 5.Caroff, M., D. Karibian, J. M. Cavaillon, and N. Haeffner-Cavaillon. 2002. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 4:915-926. [DOI] [PubMed] [Google Scholar]
- 6.Chamaillard, M., S. E. Girardin, J. Viala, and D. J. Philpott. 2003. NODs, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell. Microbiol. 5:581-592. [DOI] [PubMed] [Google Scholar]
- 7.Chamaillard, M., M. Hashimoto, Y. Horie, J. Masumoto, S. Qiu, L. Saab, Y. Ogura, A. Kawasaki, K. Fukase, S. Kusumoto, M. A. Valvano, S. J. Foster, T. W. Mak, G. Nunez, and N. Inohara. 2003. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4:702-707. [DOI] [PubMed] [Google Scholar]
- 8.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury, A. R., and G. J. Boons. 2005. The synthesis of diaminopimelic acid containing peptidoglycan fragments using metathesis cross coupling. Tetrahedron Lett. 46:1675-1678. [Google Scholar]
- 10.Chowdhury, A. R., A. Siriwardena, and G. J. Boons. 2002. A highly convergent approach for the synthesis of disaccharide repeating units of peptidoglycan. Tetrahedron Lett. 43:7805-7807. [Google Scholar]
- 11.Darveau, R. P. 1998. Lipid A diversity and the innate host response to bacterial infection. Curr. Opin. Microbiol. 1:36-42. [DOI] [PubMed] [Google Scholar]
- 12.Dziarski, R. 2004. Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 40:877-886. [DOI] [PubMed] [Google Scholar]
- 13.Dziarski, R. 2003. Recognition of bacterial peptidoglycan by the innate immune system. Cell. Mol. Life Sci. 60:1793-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziarski, R., and D. Gupta. 2005. Staphylococcus aureus peptidoglycan is a toll-like receptor 2 activator: a reevaluation. Infect. Immun. 73:5212-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erridge, C., E. Bennett-Guerrero, and I. R. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4:837-851. [DOI] [PubMed] [Google Scholar]
- 16.Flak, T. A., L. N. Heiss, J. T. Engle, and W. E. Goldman. 2000. Synergistic epithelial responses to endotoxin and a naturally occurring muramyl peptide. Infect. Immun. 68:1235-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghuysen, J. M. 1968. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 32:425-464. [PMC free article] [PubMed] [Google Scholar]
- 18.Girardin, S. E., I. G. Boneca, L. A. Carneiro, A. Antignac, M. Jehanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zathringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti, and D. J. Philpott. 2003. NOD1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- 19.Girardin, S. E., and D. J. Philpott. 2004. Mini-review: the role of peptidoglycan recognition in innate immunity. Eur. J. Immunol. 34:1777-1782. [DOI] [PubMed] [Google Scholar]
- 20.Girardin, S. E., L. H. Travassos, M. Herve, D. Blanot, I. G. Boneca, D. J. Philpott, P. J. Sansonetti, and D. Mengin-Lecreulx. 2003. Peptidoglycan molecular requirements allowing detection by NOD1 and NOD2. J. Biol. Chem. 278:41702-41708. [DOI] [PubMed] [Google Scholar]
- 21.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162:391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan, R., A. Roychowdhury, B. Ember, S. Kumar, G. J. Boons, and R. A. Mariuzza. 2004. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 101:17168-17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtje, J. V., and U. Schwarz. 1989. The metabolism of murein in bacteria: a source of bioactive compounds in the host organism, p. 101-112. In M. E. Bushell and U. Grafe (ed.), Bioactive metabolites from microorganisms. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 25.Inohara, N., and G. Nunez. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 26.Inohara, N., Y. Ogura, A. Fontalba, O. Gutierrez, F. Pons, J. Crespo, K. Fukase, S. Inamura, S. Kusumoto, M. Hashimoto, S. J. Foster, A. P. Moran, J. L. Fernandez-Luna, and G. Nunez. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278:5509-5512. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, K. S., M. Chamaillard, Y. Ogura, O. Henegariu, N. Inohara, G. Nunez, and R. A. Flavell. 2005. NOD2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307:731-734. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., A. Roychowdhury, B. Ember, Q. Wang, R. Guan, R. A. Mariuzza, and G. J. Boons. 2005. Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins Iα and S. J. Biol. Chem. 280:37005-37012. [DOI] [PubMed] [Google Scholar]
- 29.Labischinski, H., and N. Maidhof. 1994. Bacterial peptidoglycan: overview and evolving concepts, p. 27. In J.-M. Ghuysen and R. Hakenbeck (ed.), New comprehensive biochemistry, vol. 27. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 30.Miyake, K. 2004. Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD2. Trends Microbiol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 31.Oh, H. M., H. J. Lee, G. S. Seo, E. Y. Choi, S. H. Kweon, C. H. Chun, W. C. Han, K. M. Lee, M. S. Lee, S. C. Choi, and C. D. Jun. 2005. Induction and localization of NOD2 protein in human endothelial cells. Cell. Immunol. 237:37-44. [DOI] [PubMed] [Google Scholar]
- 32.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangel-Frausto, M. S. 2005. Sepsis: still going strong. Arch. Med. Res. 36:672-681. [DOI] [PubMed] [Google Scholar]
- 34.Rice, T. W., and G. R. Bernard. 2005. Therapeutic intervention and targets for sepsis. Annu. Rev. Med. 56:225-248. [DOI] [PubMed] [Google Scholar]
- 35.Rietschel, E. T., J. Schletter, B. Weideman, V. El-Samalouti, T. Mattern, U. Zahringer, U. Seydel, H. Brade, H.-D. Flad, S. Kusumoto, D. Gupta, R. Dziarski, and A. J. Ulmer. 1998. Lipopolysaccaride and peptidoglycan; CD14-dependent bacterial inducers of inflammation. Microb. Drug Resist. 4:37-44. [DOI] [PubMed] [Google Scholar]
- 36.Roychowdhury, A., M. A. Wolfert, and G. J. Boons. 2005. Synthesis and proinflammatory properties of muramyl tripeptides containing lysine and diaminopimelic acid moieties. Chembiochem 6:2088-2097. [DOI] [PubMed] [Google Scholar]
- 37.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 39.Steiner, H. 2004. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 198:83-96. [DOI] [PubMed] [Google Scholar]
- 40.Swaminathan, C. P., P. H. Brown, A. Roychowdhury, Q. Wang, R. Guan, N. Silverman, W. E. Goldman, G. J. Boons, and R. A. Mariuzza. 2006. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs). Proc. Natl. Acad. Sci. USA 103:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda, K., and S. Akira. 2004. TLR signaling pathways. Semin. Immunol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 43.Tipper, D. J., W. Katz, J. L. Strominger, and J. M. Ghuysen. 1967. Substituents on the alpha-carboxyl group of D-glutamic acid in the peptidoglycan of several bacterial cell walls. Biochemistry 6:921-929. [DOI] [PubMed] [Google Scholar]
- 44.Traub, S., N. Kubasch, S. Morath, M. Kresse, T. Hartung, R. R. Schmidt, and C. Hermann. 2004. Structural requirements of synthetic muropeptides to synergize with lipopolysaccharide in cytokine induction. J. Biol. Chem. 279:8694-8700. [DOI] [PubMed] [Google Scholar]
- 45.Travassos, L. H., S. E. Girardin, D. J. Philpott, D. Blanot, M. A. Nahori, C. Werts, and I. G. Boneca. 2004. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 5:1000-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsubery, H., I. Ofek, S. Cohen, and M. Fridkin. 2000. The functional association of polymyxin B with bacterial lipopolysaccharide is stereospecific: studies on polymyxin B nonapeptide. Biochemistry 39:11837-11844. [DOI] [PubMed] [Google Scholar]
- 47.van Amersfoort, E. S., T. J. C. van Berkel, and J. Kuiper. 2003. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin. Microbiol. Rev. 16:379-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 49.Wittmann, V. 2001. Glycoproteins. Occurrence and significance. Peptidoglycan, p. 2278-2280. In B. O. Fraser-Reid, K. Tatsuta, and J. Thiem (ed.), Glycoscience: chemistry and chemical biology, vol. III. Springer, Berlin, Germany. [Google Scholar]
- 50.Wolfert, M. A., T. F. Murray, G. J. Boons, and J. N. Moore. 2002. The origin of the synergistic effect of muramyl dipeptide with endotoxin and peptidoglycan. J. Biol. Chem. 277:39179-39186. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163:1-5. [PubMed] [Google Scholar]