Abstract
The contributions of three proteinase genes (rgpA, rgpB, and kgp) to the virulence of Porphyromonas gingivalis W50 were investigated in the murine periodontitis model. Mice were orally inoculated with eight doses (1 × 1010 cells per dose) of rgpA, rgpB, kgp, rgpA rgpB, or rgpA rgpB kgp isogenic mutants, and the level of alveolar bone loss, immune response induced, and number of bacterial cells per half maxilla were compared with those of animals inoculated with wild-type P. gingivalis. The kgp, rgpB, rgpA rgpB, and rgpA rgpB kgp isogenic mutants induced significantly (P < 0.05) less bone loss than the rgpA isogenic mutant and the wild type did, and the virulence of the rgpA isogenic mutant and the wild type were not significantly different. Mice inoculated with the wild type or the rgpA isogenic mutant exhibited significantly (P < 0.01) more P. gingivalis cells per half maxilla than mice inoculated with rgpB, kgp, rgpA rgpB, and rgpA rgpB kgp isogenic mutants or nonchallenged mice did, as determined using real-time PCR. A significant positive correlation was found between the number of P. gingivalis cells detected per half maxilla and the amount of alveolar bone loss induced. Enzyme-linked immunosorbent assay results showed that each isogenic mutant and the wild type induced a predominant P. gingivalis antigen-specific immunoglobulin G3 (IgG3) response. Furthermore, the kgp and rgpA rgpB kgp isogenic mutants induced significantly (P < 0.05) lower IgG3 antibody responses than the responses induced by the wild type or the rgpA, rgpB, and rgpA rgpB isogenic mutants. The results suggest that the order in which the proteinases contribute to the virulence of P. gingivalis in the murine periodontitis model is Kgp ≥ RgpB ≫ RgpA.
Chronic periodontitis is an inflammatory disease of the supporting tissues of teeth involving alveolar bone resorption, which can lead to eventual tooth loss (13, 33). The presence of a consortium of gram-negative bacteria in subgingival plaque has been associated with the development of chronic periodontitis (47). In this consortium, Porphyromonas gingivalis has been identified as a major pathogen (31). A number of virulence factors have been reported to contribute to the pathogenicity of P. gingivalis (reviewed in reference 31). Among these virulence factors, the Arg- and Lys-specific cysteine proteinases and their associated adhesins are considered major virulence determinants in the onset and progression of chronic periodontitis (14, 27). The Arg-specific proteinase activity of P. gingivalis is encoded by two genes designated rgpA and rgpB, and the Lys-specific proteinase activity is encoded by one gene designated kgp (8). The rgpA gene encodes a polyprotein consisting of an N-terminal preprofragment followed by a 45-kDa Arg-specific, calcium-stabilized cysteine proteinase RgpAcat (catalytic domain of RgpA) and four sequence-related adhesin domains RgpAA1, RgpAA2, RgpAA3, and RgpAA4 (37, 45, 50). The rgpB gene encodes an N-terminal preprofragment followed by the Arg-specific proteinase RgpB without the C-terminal adhesin extension of RgpA (43). The kgp gene encodes a polyprotein with an N-terminal preprofragment followed by a 48-kDa Lys-specific cysteine proteinase Kgpcat and five C-terminal adhesin domains KgpA1, KgpA2, KgpA3, KgpA4, and KgpA5 (44, 50). The proteins encoded by rgpA and kgp of P. gingivalis strain W50 have been characterized as cell surface complexes of noncovalently associated proteinases and adhesins designated the RgpA-Kgp complex (27, 36).
The pathogenicity of P. gingivalis and its virulence factors has been examined using a variety of experimental animal models (reviewed in references 12 and 38). Among these experimental animal models, the murine lesion model has been widely used as a model to study the virulence of P. gingivalis (10, 24). Spontaneous P. gingivalis mutants with reduced Arg- and Lys-specific proteinase activity and wild-type P. gingivalis treated with an inhibitor of trypsin-like proteinases have been reported to be avirulent in the murine lesion model (18). Further, P. gingivalis W50 isogenic mutants lacking either rgpA, rgpB, or kgp exhibited significantly reduced pathogenicity compared to the wild-type strain in this model (29). In these experiments, the kgp isogenic mutant was the least virulent, followed by rgpB and rgpA mutants, respectively. Yoneda et al. (51) have also reported that an rgpA rgpB double mutant and kgp mutant induced significantly smaller lesions in mice than wild-type P. gingivalis did, while the rgpA rgpB kgp triple mutant did not induce lesions at all. These studies suggest that the rgpA, rgpB, and kgp gene products are important for the virulence of P. gingivalis.
Despite being widely used, the murine lesion model, where the bacteria are injected subcutaneously, is not an ideal model for the study of periodontitis because it does not assess the abilities of the bacteria to colonize intra-orally and to induce periodontal (alveolar) bone resorption. For these reasons, animal models of periodontitis have been developed. As the rodent molar periodontal anatomy resembles that of humans (12, 22, 35), mice and rats have been used as models for periodontal bone loss induced by pathogenic bacteria introduced into the oral cavity. The pathological processes of periodontal destruction in the rodent model, including alterations in epithelia, destruction of connective tissue of the gingiva and periodontal ligament, and resorption of alveolar bone that occurs on subgingival implantation of a periodontal pathogen, are very similar to those in humans, making the rodent a valuable model to study Porphyromonas gingivalis-induced disease (12, 30, 34). We have developed and modified the rodent periodontitis model (3, 11) to show that oral inoculation with a defined, viable inoculum of P. gingivalis produces reproducible periodontal bone loss in BALB/c mice and Sprague-Dawley rats (30, 40). Furthermore, in both the mouse and rat periodontitis models, the RgpA-Kgp complex when used as a vaccine prevented P. gingivalis-induced bone loss (30, 40). In these models, it was shown that a Th2 antibody response (immunoglobulin G2 [IgG2] and IgG1) directed towards the RgpA-Kgp complex inhibited bone loss and P. gingivalis intra-oral colonization, respectively, as analyzed by DNA probe analysis of subgingival plaque samples (30, 40). This protection was attributed to specific antibodies directed towards adhesin binding motifs of the RgpA-Kgp complex blocking binding of P. gingivalis to subgingival plaque microorganisms and host tissue, hence preventing colonization (30). These results suggest that the Arg- and Lys-specific proteinases of P. gingivalis and their associated adhesins may play a significant role in the establishment of P. gingivalis in subgingival plaque and in the induction of alveolar bone loss.
In the present study, the contributions of RgpA, RgpB, and Kgp to intra-oral colonization, alveolar bone loss, and the immune response induced by P. gingivalis were investigated using rgpA, rgpB, kgp, rgpA rgpB, and rgpA rgpB kgp isogenic mutants in the murine periodontitis model.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lyophilized cultures of the P. gingivalis W50 wild-type strain and isogenic mutants, rgpA (W501), kgp (W50KIA), rgpB (W50D7), and rgpA rgpB (W50AB) mutants, were obtained from the culture collection of the Cooperative Research Centre for Oral Health Science, University of Melbourne, Australia, and have been described before (29, 50). The rgpA rgpB kgp triple mutant (W50ABK) was generated for this study (see below). Bacteria were maintained in an anaerobic chamber (MK3 anaerobic workstation; Don Whitley Scientific Ltd., Shipley, England) at 37°C on horse blood agar plates supplemented with 10% (vol/vol) lysed horse blood. Bacterial colonies were used to inoculate brain heart infusion medium containing 5 μg/ml hemin and 0.5 μg/ml cysteine; for growth of the rgpA (W501), rgpB (W50D7), and kgp (W50KIA) P. gingivalis W50 isogenic mutants, the medium also contained 10 μg/ml erythromycin (29). For the rgpA rgpB (W50AB) isogenic mutant, the medium also contained 1 μg/ml tetracycline and 10 μg/ml chloramphenicol. Batch culture growth was monitored at 650 nm using a spectrophotometer (model 295E; Perkin-Elmer). Culture purity was routinely checked by Gram staining and by colony morphology.
Generation of the rgpA rgpB kgp P. gingivalis isogenic mutant (W50ABK).
The P. gingivalis W50 isogenic mutant (W50ABK) lacking RgpA, RgpB, and Kgp was generated using the rgpA rgpB isogenic mutant (W50AB) and pNS1 (1) containing kgp insertionally disrupted with an ErmF/AM cassette. The kgp::ermA insert of pNS1 was amplified by PCR and electroporated into P. gingivalis W50AB to generate W50ABK. Disruption of kgp in W50ABK was confirmed by Southern blot analysis whereby chromosomal DNA was probed with a 2.1-kb KpnI-BamHI ErmF/AM cassette and with a 3.3-kb BamHI fragment from pNS1 encoding the catalytic domain of Kgp (44). Whole-cell assays of W50ABK using Arg- and Lys-chromogenic substrates showed that the mutant was devoid of RgpA/B and Kgp proteolytic activity (29).
Murine periodontitis model.
The mouse periodontitis experiments were performed as described previously (30) and were approved by the University of Melbourne Ethics Committee for Animal Experimentation. BALB/c mice 6 to 8 weeks old (10 mice per group) were given kanamycin (Sigma-Aldrich, New South Wales, Australia) at 1 mg/ml in deionized water ad libitum for 7 days. Three days after the antibiotic treatment, mice were orally inoculated four times 2 days apart with 1 × 1010 viable P. gingivalis W50 or P. gingivalis rgpA, rgpB, kgp, rgpA rgpB, or rgpA rgpB kgp isogenic mutants (25 μl) in PG buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM CaCl2, and 5 mM cysteine-HCl, pH 8.0) containing 2% (wt/vol) carboxymethyl cellulose (CMC; Sigma-Aldrich, New South Wales, Australia), and a control group was sham infected with PG buffer containing 2% (wt/vol) CMC alone. The inocula were prepared in the anaerobic chamber and then immediately applied to the gingival margin of the maxillary molar teeth. Two weeks later, mice received another four doses (2 days apart) of 1 × 1010 cells of viable P. gingivalis W50 or the isogenic mutants (25 μl) in PG buffer containing 2% (wt/vol) CMC. The number of viable bacteria in each inoculum was verified by enumeration on blood agar. Mice were fed a soft powdered diet (Barastock, Australia) and housed in cages fitted with a raised wire mesh bottom to prevent access to bedding. Four weeks after the last dose, mice were bled from the retrobulbar plexus and killed, and the maxillae were removed and cut in half with one half (right) used for alveolar bone loss measurement and the other half (left) used for real-time PCR.
The right half maxillae were boiled (1 min) in deionized water, mechanically defleshed, and immersed in 2% (wt/vol) potassium hydroxide (16 h, 25°C). The half maxillae were then washed (two times with deionized water) and immersed in 3% (wt/vol) hydrogen peroxide (6 h, 25°C). After the half maxillae were washed (two times with deionized water), they were stained with 0.1% (wt/vol) aqueous methylene blue, and a digital image of the buccal aspect of each half maxilla was captured with an Olympus DP12 digital camera mounted on a dissecting microscope, using OLYSIA BioReport software version 3.2 (Olympus Australia Pty Ltd., New South Wales, Australia) to assess horizontal bone loss. Horizontal bone loss is loss occurring in a horizontal plane, perpendicular to the alveolar bone crest (ABC) that results in a reduction of the crest height. Each half maxilla was aligned so that the molar buccal and lingual cusps of each image were superimposed, and the image was captured with a micrometer in frame, so that measurements could be standardized for each image. The area from the cementoenamel junction to the ABC for each molar tooth was measured using OLYSIA BioReport software version 3.2 imaging software. Bone loss measurements were determined twice by a single examiner using a randomized and blinded protocol.
Determination of subclass antibody by an ELISA.
To determine the subclass antibody responses of mouse sera, enzyme-linked immunosorbent assays (ELISAs) were performed in triplicate using a 5-μg/ml solution of P. gingivalis W50 outer membrane protein (OMP) (see below) in phosphate-buffered saline (PBS) (0.01 M Na2HPO4, 1.5 mM KH2PO4, 0.15 M NaCl), pH 7.0, containing 0.1% (vol/vol) Tween 20 (PBST) to coat wells of flat-bottom polyvinyl microtiter plates (Dynatech Laboratories, McLean, VA). After removal of the coating solution, PBST containing 2% (wt/vol) skim milk powder was added to wells to block the uncoated plastic for 1 h at room temperature. After the wells were washed four times with PBST, serial dilutions of mouse sera in PBST containing 0.5% (wt/vol) skim milk (SK-PBST) were added to each well and incubated for 16 h at room temperature. After the wells were washed six times with PBST, a 1/2,000 dilution of goat IgG to mouse IgM, IgA, IgG1, IgG2a, IgG2b, or IgG3 (Sigma, New South Wales, Australia) was added in SK-PBST and allowed to bind for 2 h at room temperature. Plates were washed six times in PBST, and a 1/5,000 dilution of horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin (Sigma, New South Wales, Australia) in SK-PBST was added to each well and incubated for 1 h at room temperature. After the wells were washed six times with PBST, bound antibody was detected by the addition of 100 μl of ABTS substrate [0.9 mM 2,2′-azino-bis(3-ethylbenz-thiazoline-6) sulfonic acid in 80 mM citric acid containing 0.005% (vol/vol) hydrogen peroxide, pH 4.0] to each well. The optical density at 415 nm was measured using a microplate reader (Bio-Rad microplate reader, model 450).
Western blot of P. gingivalis outer membrane protein preparation probed with P. gingivalis sera.
A P. gingivalis outer membrane protein preparation was probed with sera from mice inoculated with P. gingivalis W50 or with the proteinase isogenic mutants in a Western blot. P. gingivalis W50 OMP was extracted using Triton X-114 as described previously (36). P. gingivalis OMP was precipitated by the addition of trichloroacetic acid to a final concentration of 10% (vol/vol) and incubation for 20 min at 4°C. Precipitated protein was collected by centrifugation (10 min, 16,000 × g) and resuspended in 20 μl of reducing sample buffer (10% [wt/vol] SDS, 0.05% [wt/vol] bromophenol blue, 25% [vol/vol] glycerol, and 0.05% [vol/vol] 2-mercaptoethanol). The pH was adjusted to pH 8.0 with 1.5 M Tris-HCl, and then the solution was heated for 5 min at 100°C. P. gingivalis OMP (10 μg/lane) was loaded onto Novex 12% (wt/vol) Tris-glycine precast mini gels, and electrophoresis was performed using a current of 30 to 50 mA and a potential difference of 125 V using a Novex electrophoresis system (Novex, San Diego, CA). For Western blot analysis, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Problott; Applied Biosystems, New South Wales, Australia) using a transblot cell (Bio-Rad, New South Wales, Australia). The PVDF membrane was wetted in 100% methanol and soaked in transfer buffer [10 mM 3(cyclohexylamino)-1-propanesulfonic acid-10% (vol/vol) methanol, pH 11.5] for 1 minute. Transfer was performed using a potential difference of 60 V for 90 min. The PVDF membrane was blocked with 2% (wt/vol) skim milk powder in Tris-sodium chloride (TN) buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.4) for 2 h at room temperature. The membrane was then incubated overnight at room temperature with P. gingivalis W50 and isogenic mutant sera at a dilution of 1/25 in TN buffer. After incubation, the membrane was washed four times in TN buffer containing 0.05% (vol/vol) Tween 20 and then incubated with a 1/200 dilution of horseradish peroxidase-conjugated rabbit anti-mouse IgG (Sigma, New South Wales, Australia) for 2 h at room temperature. The membrane was then washed four times with TN buffer containing 0.05% (vol/vol) Tween 20, and the bound antibodies were detected by 0.005% (wt/vol) 4-chloro-1-naphthol in TN buffer containing 16.6% (vol/vol) methanol and 0.015% (wt/vol) hydrogen peroxide. A digital image of the Western blot was captured using a FujiFilm LAS 3000 image analyzer (FujiFilm, New South Wales, Australia), and the relative intensity (densitometry) of each lane of the Western blot was measured by FujiFilm multigauge imaging software (FujiFilm, New South Wales, Australia).
Determination of P. gingivalis cell numbers per mouse half maxilla using real-time PCR.
Genomic DNA was isolated from the left half maxillae (including soft and hard tissues) using a DNeasy tissue kit following the animal tissue isolation protocol as described by the manufacturer (QIAGEN Pty Ltd., New South Wales, Australia). The amount and quality of DNA present in the samples was determined using spectrophotometer readings (A260 and A260/280, respectively) and the Quant-iT DNA assay kit (Molecular Probes, Invitrogen, Mt. Waverley, Victoria, Australia) according to the manufacturer's instructions with fluorescence measured on a Wallac 1420 multilabel counter (Perkin-Elmer, Wettesley, MA). Each half maxilla sample was diluted to contain 10 ng/μl DNA and analyzed by real-time PCR. A P. gingivalis standard curve was created using known amounts of P. gingivalis DNA (serial dilutions from 1 × 106 to 1 × 102 cells) added to half maxilla DNA from a noninfected mouse diluted to 10 ng/μl.
The cell numbers of P. gingivalis per half maxilla were quantified using the P. gingivalis-specific 16S RNA forward and reverse primers as described by Kuboniwa et al. (19) and the platinum SYBR green supermix UDG kit (Invitrogen, New South Wales, Australia). Using these primers, only one PCR product was produced as confirmed by melting curve analysis and agarose gel electrophoresis.
Real-time PCR was carried out in triplicate using a Rotor-Gene 3000 system (Corbett Research, Australia). A reaction mixture volume of 25 μl contained half maxilla DNA (80 ng), 12.5 μl of SYBR green supermix, 400 nM concentration of each primer, and 5.0 mM MgCl2 and was dispensed using a CAS 1200 liquid-handling robot system (Corbett Research, New South Wales, Australia). The following PCR cycling conditions were used: an initial hold at 50°C for 2 min, followed by a denaturation step at 95°C for 2 min. The following cycling conditions were repeated for 50 cycles: denaturation at 95°C for 15 s, annealing at 63°C for 30 s, and then extension at 72°C for 30 s. Fluorescence data were collected immediately following the extension step of each cycle. The specificity of the primer pairs was confirmed by melting curve analysis by heating from 72°C to 95°C in 0.2°C increments. Melting peaks were compared with the bands obtained following agarose gel electrophoresis. The detection limit for the assay using the standard curve was 5 × 102 P. gingivalis cells.
Statistical analysis.
The bone loss data were statistically analyzed using a one-way analysis of variance (ANOVA) and Dunnett's T3 test and Cohen's effect size (SPSS for Windows, release 6.0; SPSS). Effect sizes, represented as Cohen's d (6) were calculated using the effect size calculator provided online by the United Kingdom Evidence-Based Education website (http://cem.dur.ac.uk/ebeuk/research/effectsize). According to Cohen (6), a small effect size is 0.2 ≤ d < 0.5, a moderate effect size is 0.5 ≤ d < 0.8, and a large effect size is d ≥ 0.8. The number of P. gingivalis cells per half maxilla as determined by real-time PCR was analyzed using a one-way ANOVA. The IgA, IgM, and IgG subclass antibody titers were statistically analyzed using Student's t test using SPSS software (SPSS for Windows, version 12).
RESULTS
Induction of alveolar bone loss in the murine periodontitis model by the P. gingivalis W50 wild-type strain and proteinase isogenic mutants.
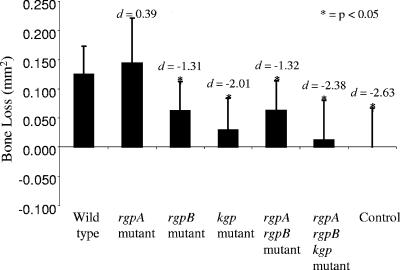
To evaluate the virulence of P. gingivalis W50 and the proteinase isogenic mutants, BALB/c mice were orally inoculated with eight doses (1 × 1010 viable cells per dose) of bacteria, and the resulting periodontal bone loss was measured (Fig. 1). Mice inoculated with P. gingivalis rgpB, rgpA rgpB, kgp, or rgpA rgpB kgp isogenic mutants exhibited significantly (P < 0.05) less alveolar bone loss than mice inoculated with the P. gingivalis W50 wild-type strain or the rgpA isogenic mutant did. No significant difference in bone loss was observed between mice inoculated with the wild type and the rgpA isogenic mutant. The smaller effect size values for the rgpB isogenic mutant (d = −1.31; 95% confidence interval [95% CI], −2.19 to −0.07) and the rgpA rgpB isogenic mutant (d = −1.32; 95% CI, −2.17 to −0.27) suggest that these mutants may be more virulent than the kgp (d = −2.01; 95% CI, −2.83 to −0.76) and rgpA rgpB kgp (d = −2.38; 95% CI, −2.89 to −0.80) isogenic mutants (Fig. 1).
FIG. 1.
Alveolar bone loss induced by P. gingivalis W50 and proteinase isogenic mutants in the murine periodontitis model. Mice were orally inoculated with eight doses (1 × 1010 viable cells per dose) of bacteria, and the resulting alveolar bone loss was measured. Measurement of bone loss is the mean area measured in mm2 from the cementoenamel junction to the ABC of the buccal aspect of each molar of the right half maxilla. Data are presented as means plus standard deviations (error bars) (n = 10) and were analyzed by one-way ANOVA with Dunnett's T3 test and Cohen's effect size (d). Values that were significantly different (P < 0.05) from the value for the group inoculated with the P. gingivalis W50 wild-type strain are indicated by an asterisk.
Determination of P. gingivalis cell numbers per mouse half maxilla using real-time PCR.
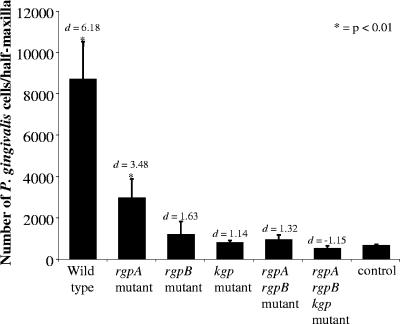
The number of P. gingivalis W50 and proteinase isogenic mutant cells per half maxilla, determined using real-time PCR, is presented in Fig. 2. Mice inoculated with P. gingivalis W50 and the rgpA isogenic mutant had significantly (P < 0.01) more P. gingivalis cells per half maxilla than did mice inoculated with the nonchallenged control. Further, there were significantly (P < 0.01) less P. gingivalis cells per half maxilla in mice inoculated with the rgpA isogenic mutant than in mice inoculated with P. gingivalis W50 (wild type) (Fig. 2). The number of P. gingivalis cells detected per half maxilla in mice challenged with kgp, rgpB, rgpA rgpB, or rgpA rgpB kgp isogenic mutants were not significantly higher than those detected in the nonchallenged control. However, the results of effect size analysis suggested that there were more P. gingivalis cells per half maxilla detected in mice challenged with rgpB (d = 1.63; 95% CI, 0.56-2.56) or rgpA rgpB (d = 1.32; 95% CI, 0.31-2.23) isogenic mutants than in mice challenged with the kgp or rgpA rgpB kgp isogenic mutants or the nonchallenged control. Regression analysis showed a significant positive linear relationship (P < 0.02; r2 = 0.77) between the number of P. gingivalis cells detected per half maxilla and the amount of alveolar bone loss induced by P. gingivalis W50 and the proteinase isogenic mutants.
FIG. 2.
Enumeration of P. gingivalis cells per left half maxilla by real-time PCR. Data are presented as means plus standard deviations (error bars) (n = 10) and were statistically analyzed by ANOVA and Cohen's effect size (d). Values that were significantly different (P < 0.05) from the value for the control (nonchallenged) group inoculated with the P. gingivalis W50 wild-type strain are indicated by an asterisk.
Antibody subclass and Western blot analysis of sera from mice orally inoculated with P. gingivalis W50 (wild type) and proteinase isogenic mutants in the mouse periodontitis model.
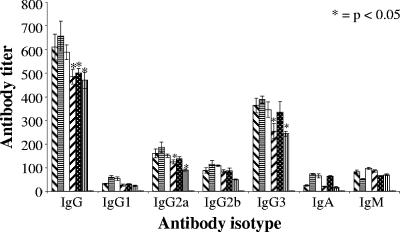
Mice inoculated with P. gingivalis W50 and proteinase isogenic mutant strains were bled, and serum IgG, IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM titers were determined by an ELISA using a P. gingivalis W50 outer membrane protein preparation as the adsorbed antigen (Fig. 3). Mice orally inoculated with P. gingivalis W50 and proteinase isogenic mutants induced a significant serum IgG response (Fig. 3). P. gingivalis kgp, rgpA rgpB, and rgpA rgpB kgp isogenic mutants induced a significantly (P < 0.05) lower IgG response than that induced by the P. gingivalis W50 wild-type strain. However, the serum IgG response induced by the rgpA or rgpB isogenic mutants was not significantly different from that induced by the wild type. Regression analysis showed a significant positive linear relationship (P < 0.007; r2 = 0.863) between the serum IgG response and the level of alveolar bone loss induced by P. gingivalis W50 and the proteinase isogenic mutants. A significant exponential relationship (P < 0.05; r2 = 0.790) was also observed between the serum IgG levels and the number of P. gingivalis W50 and proteinase isogenic mutant cells detected per half maxilla. The predominant serum IgG subclass induced by each P. gingivalis strain was IgG3, followed typically by IgG2a > IgG2b > IgM > IgA = IgG1 responses (Fig. 3). P. gingivalis kgp and rgpA rgpB kgp isogenic mutants induced a significantly (P < 0.05) lower IgG2a and IgG3 responses than that induced by the P. gingivalis W50 wild-type strain. However, the IgG2a and IgG3 responses induced by the rgpA, rgpB, and rgpA rgpB isogenic mutants were not significantly different from those induced by the wild type. No antibody response to P. gingivalis OMP was detected in sera from nonchallenged (control) mice. Regression analysis showed a significant positive linear relationship (P < 0.01; r2 = 0.843) between the serum IgG3 response and the level of alveolar bone loss induced by P. gingivalis W50 (wild type) and the proteinase isogenic mutants. Furthermore, a significant exponential relationship (P < 0.05; r2 = 0.78) was observed between the serum IgG3 levels and the number of P. gingivalis W50 wild-type and proteinase isogenic mutant cells detected per half maxilla.
FIG. 3.
Serum antibody subclass responses of mice challenged with either wild-type P. gingivalis W50 or proteinase isogenic mutants. Antibody subclasses in anti-P. gingivalis W50 (wild type) sera ( ), anti-rgpA mutant sera (
), anti-rgpA mutant sera ( ), anti-rgpB mutant sera (□), anti-kgp mutant sera (
), anti-rgpB mutant sera (□), anti-kgp mutant sera ( ), anti-rgpA rgpB mutant sera (
), anti-rgpA rgpB mutant sera ( ), and anti-rgpA rgpB kgp mutant sera (
), and anti-rgpA rgpB kgp mutant sera ( ) and in sera from nonchallenged mice (
) and in sera from nonchallenged mice ( ) were analyzed by an ELISA. Antibody titers are expressed as the dilution factor which produced an absorbance fivefold greater than background in the ELISA. Each titer represents the mean plus standard deviation (error bar) of four values. Antibody titers that were significantly different (P < 0.05) from the antibody titer for the group inoculated with the P. gingivalis W50 wild-type strain are indicated by an asterisk.
) were analyzed by an ELISA. Antibody titers are expressed as the dilution factor which produced an absorbance fivefold greater than background in the ELISA. Each titer represents the mean plus standard deviation (error bar) of four values. Antibody titers that were significantly different (P < 0.05) from the antibody titer for the group inoculated with the P. gingivalis W50 wild-type strain are indicated by an asterisk.
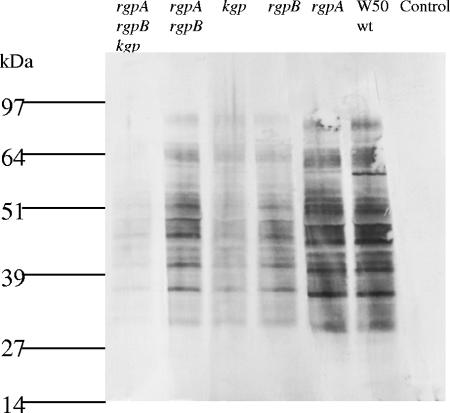
Sera from mice orally inoculated with the P. gingivalis W50 wild-type strain and proteinase isogenic mutants were used to probe a P. gingivalis W50 OMP preparation in a Western blot (Fig. 4). Western blot densitometric analysis (relative intensity) of the individual lanes was used to compare the antibody response induced by P. gingivalis W50 (wild type) to that of each of the proteinase isogenic mutants. The highest intensity was observed for wild-type P. gingivalis W50, and this intensity was set at 100% (Fig. 4). The relative intensity observed for the rgpA isogenic mutant was 95% of that observed for P. gingivalis W50. A decrease in the relative intensity compared to that of wild-type P. gingivalis W50 was found for the rgpB isogenic mutant (64%), and a similar decrease was found for the rgpA rgpB isogenic mutant (70%). The kgp and rgpA rgpB kgp isogenic mutants induced the weakest antibody response with relative intensities of 37% and 31%, respectively, compared with that of the P. gingivalis W50 wild-type strain. Statistical analysis indicated a significant positive linear relationship (P < 0.01; r2 = 0.934) between the relative intensity of the antibody response as determined by Western blot densitometry and the level of alveolar bone loss induced by the P. gingivalis W50 wild-type strain and the proteinase isogenic mutants. Furthermore, a significant exponential relationship (P < 0.01; r2 = 0.89) was observed between the relative intensity of the antibody response as determined by Western blot densitometry and the number of P. gingivalis W50 wild-type and isogenic mutant cells detected per half maxilla by real-time PCR.
FIG. 4.
Western blot analysis of P. gingivalis outer membrane preparation using anti-P. gingivalis sera. P. gingivalis outer membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a PVDF membrane, and probed with anti-rgpA rgpB kgp mutant sera, anti-rgpA rgpB mutant sera, anti-kgp mutant sera, anti-rgpB mutant sera, anti-rgpA mutant sera, P. gingivalis W50 (wild type [wt]) sera, and nonchallenged control sera. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel.
DISCUSSION
In this study, the ability of the P. gingivalis W50 wild-type strain and kgp, rgpA, rgpB, rgpA rgpB, and rgpA rgpB kgp isogenic mutants to induce alveolar bone loss, colonize the maxilla, and induce an immune response was studied using the murine periodontitis model. The kgp and rgpB isogenic mutants induced significantly (P < 0.05) less alveolar bone loss than the wild type did. The results of effect size analysis suggested that the kgp isogenic mutant was less virulent than the rgpB isogenic mutant was. In contrast, inactivation of the rgpA gene did not affect the ability of P. gingivalis to induce alveolar bone loss. The results of effect size analysis suggested that the rgpA rgpB kgp isogenic mutant was the least effective at inducing alveolar bone loss, suggesting a possible additive effect between Kgp and RgpB in the virulence of P. gingivalis. However, no additive effect between RgpB and RgpA in inducing alveolar bone loss was observed, as mice inoculated with the rgpA rgpB isogenic mutant were found to have levels of alveolar bone loss similar to those of the mice inoculated with the rgpB isogenic mutant. These data suggest that the order in which the proteinases contribute to the induction of alveolar bone loss in the murine periodontitis model is Kgp ≥ RgpB ≫ RgpA, which is consistent with the contribution of these proteinases to the virulence of P. gingivalis in the murine lesion model (29).
The number of P. gingivalis cells per half maxilla of each mouse was determined using real-time PCR. Previous studies have used real-time PCR to quantify periodontopathic bacteria from human subgingival plaque samples, and the technique is a more reliable, sensitive and rapid method of enumerating bacteria than culturing is (2, 25, 26, 42, 46). In this study, the number of P. gingivalis cells per half maxilla in mice inoculated with the kgp isogenic mutant or rgpB isogenic mutant were found not to be significantly different from that of nonchallenged mice, thus suggesting that Kgp and RgpB are essential for intra-oral colonization. Furthermore, the smaller number of P. gingivalis cells per half maxilla of mice inoculated with the kgp isogenic mutant compared to the number of mice inoculated with the rgpA isogenic mutant indicates that Kgp has a greater role in facilitating P. gingivalis colonization than RgpA does. A role for RgpA in colonization is also possible, as the number of rgpA isogenic mutant cells detected per half maxilla was significantly (P < 0.05) lower than that for wild-type cells (Fig. 2). RgpA and Kgp have been implicated in the adherence of P. gingivalis to oral epithelial cells (4, 5) and have also been reported to play a significant role in the coaggregation of P. gingivalis with other plaque bacteria, such as Prevotella intermedia and streptococcal species (15-17). This may help explain the role suggested in this study for these proteinase-adhesin complexes in the intra-oral colonization of P. gingivalis.
The greater role for RgpB over RgpA may be due in part to the role attributed to RgpB as a processing enzyme, involved in the processing of a number of P. gingivalis outer membrane virulence factors, for example, fimbriae, which may also aid intra-oral colonization (39). The significance of Kgp in colonization and pathogenicity may also be partly explained by its role in hemoglobin binding, degradation, and heme accumulation, which is essential for growth and virulence of P. gingivalis (9, 21, 23, 32). Thus, a reduction in the ability of the P. gingivalis kgp isogenic mutant to accumulate heme would result in reduced growth and virulence and consequently therefore may help explain the lower intra-oral recovery of this mutant.
In this study, we found that there was a highly significant correlation between the number of P. gingivalis cells per half maxilla and the severity of alveolar bone loss. These findings support previous human studies where the level of P. gingivalis in subgingival plaque has been associated with the severity of periodontitis as measured by periodontal pocket depth and loss of clinical attachment (20, 26, 28, 47).
Western blot densitometry of a P. gingivalis W50 OMP preparation, probed with sera from mice orally inoculated with the P. gingivalis W50 wild-type strain and proteinase isogenic mutants, showed that mice inoculated with the wild type and the rgpA isogenic mutant induced the strongest P. gingivalis-specific serum antibody responses. In contrast, compared to mice inoculated with the wild type, mice inoculated with the rgpB and rgpA rgpB isogenic mutants were found to have a weaker P. gingivalis-specific serum antibody response, and the serum antibody response was weaker still in mice inoculated with the kgp and rgpA rgpB kgp isogenic mutants. Furthermore, significant positive correlations were observed between the densitometric values and the level of alveolar bone loss and number of P. gingivalis cells per half maxilla. The induction of a lower P. gingivalis-specific antibody response and alveolar bone loss in mice inoculated with the rgpB, rgpA rgpB, kgp, and rgpA rgpB kgp isogenic mutants compared to those in mice inoculated with the wild type or the rgpA isogenic mutant reflects the poor intra-oral colonization by these isogenic mutants in this study. The results of ELISA analysis of mouse sera showed that challenge with P. gingivalis induced high-titer serum IgG and IgG3 responses, and this was positively correlated with the level of alveolar bone loss and the number of P. gingivalis cells detected per half maxilla. A similar positive correlation between P. gingivalis-specific serum IgG and IgG2 levels and disease severity as measured by probing depth and loss of clinical attachment has been reported in humans with chronic periodontitis (7, 28, 41). Similar to human IgG2, mouse IgG3 is a Th1 cytokine-induced antibody, and a predominantly Th1 response is suggested to be destructive in human periodontal disease (28, 48, 49). Furthermore, it has been shown in the murine periodontitis model that alveolar bone resorption is associated with a predominantly Th1 response (30). Thus, in these respects, the microbiology and immunopathology of the murine periodontitis model are similar to those of human disease.
In summary, the data presented here indicate that P. gingivalis kgp and rgpB isogenic mutants colonize the maxilla poorly, elicit a weak P. gingivalis-specific antibody response, and induce significantly less alveolar bone loss than the wild type in the murine periodontitis model. The order in which the proteinases contributed to the virulence of P. gingivalis in this model was Kgp ≥ RgpB ≫ RgpA.
Acknowledgments
This project is supported by Australian National Health and Medical Research Council (project no. 251708) and the National Institute of Health (grant no. 1R01DE1419801). Neil O'Brien-Simpson is a C.R. Roper Fellow.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Aduse-Opoku, J., N. N. Davies, A. Gallagher, A. Hashim, H. E. Evans, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2000. Generation of Lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146:1933-1940. [DOI] [PubMed] [Google Scholar]
- 2.Asai, Y., T. Jinno, H. Igarashi, Y. Ohyama, and T. Ogawa. 2002. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J. Clin. Microbiol. 40:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 4.Bosshardt, D. D., and N. P. Lang. 2005. The junctional epithelium: from health to disease. J. Dent. Res. 84:9-20. [DOI] [PubMed] [Google Scholar]
- 5.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. 1969. Statistical power analysis for the behavioral sciences. Academic Press, New York, NY.
- 7.Craig, R. G., R. Boylan, J. Yip, D. Mijares, M. Imam, S. S. Socransky, M. A. Taubman, and A. D. Haffajee. 2002. Serum IgG antibody response to periodontal pathogens in minority populations: relationship to periodontal disease status and progression. J. Periodontal Res. 37:132-146. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 9.Dashper, S. G., K. J. Cross, N. Slakeski, P. Lissel, P. Aulakh, C. Moore, and E. C. Reynolds. 2004. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol. Immunol. 19:50-56. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole, J. L., L. Kesavalu, S. L. Schneider, R. L. Machen, and S. C. Holt. 1995. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1:115-128. [DOI] [PubMed] [Google Scholar]
- 11.Evans, R. T., B. Klausen, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, and R. J. Genco. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813-819. [DOI] [PubMed] [Google Scholar]
- 12.Genco, C. A., T. Van Dyke, and S. Amar. 1998. Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 6:444-449. [DOI] [PubMed] [Google Scholar]
- 13.Genco, R. J. 1992. Host responses in periodontal diseases: current concepts. J. Periodontol. 63:338-355. [DOI] [PubMed] [Google Scholar]
- 14.Kadowaki, T., K. Nakayama, K. Okamoto, N. Abe, A. Baba, Y. Shi, D. B. Ratnayake, and K. Yamamoto. 2000. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem. (Tokyo) 128:153-159. [DOI] [PubMed] [Google Scholar]
- 15.Kamaguchi, A., H. Baba, M. Hoshi, and K. Inomata. 1994. Coaggregation between Porphyromonas gingivalis and mutans streptococci. Microbiol. Immunol. 38:457-460. [DOI] [PubMed] [Google Scholar]
- 16.Kamaguchi, A., K. Nakayama, T. Ohyama, T. Watanabe, M. Okamoto, and H. Baba. 2001. Coaggregation of Porphyromonas gingivalis and Prevotella intermedia. Microbiol. Immunol. 45:649-656. [DOI] [PubMed] [Google Scholar]
- 17.Kamaguchi, A., T. Ohyama, E. Sakai, R. Nakamura, T. Watanabe, H. Baba, and K. Nakayama. 2003. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology 149:1257-1264. [DOI] [PubMed] [Google Scholar]
- 18.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 1996. Trypsin-like protease activity of Porphyromonas gingivalis as a potential virulence factor in a murine lesion model. Microb. Pathog. 20:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Kuboniwa, M., A. Amano, K. R. Kimura, S. Sekine, S. Kato, Y. Yamamoto, N. Okahashi, T. Iida, and S. Shizukuishi. 2004. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 19:168-176. [DOI] [PubMed] [Google Scholar]
- 20.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Listgarten, M. A. 1975. Similarity of epithelial relationships in the gingiva of rat and man. J. Periodontol. 46:677-680. [DOI] [PubMed] [Google Scholar]
- 23.Marsh, P. D., A. S. McDermid, A. S. McKee, and A. Baskerville. 1994. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology 140:861-865. [DOI] [PubMed] [Google Scholar]
- 24.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 25.Nonnenmacher, C., A. Dalpke, R. Mutters, and K. Heeg. 2004. Quantitative detection of periodontopathogens by real-time PCR. J. Microbiol. Methods 59:117-125. [DOI] [PubMed] [Google Scholar]
- 26.Nonnenmacher, C., A. Dalpke, J. Rochon, L. Flores-de-Jacoby, R. Mutters, and K. Heeg. 2005. Real-time polymerase chain reaction for detection and quantification of bacteria in periodontal patients. J. Periodontol. 76:1542-1549. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien-Simpson, N., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409-426. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien-Simpson, N. M., C. L. Black, P. S. Bhogal, S. M. Cleal, N. Slakeski, T. J. Higgins, and E. C. Reynolds. 2000. Serum immunoglobulin G (IgG) and IgG subclass responses to the RgpA-Kgp proteinase-adhesin complex of Porphyromonas gingivalis in adult periodontitis. Infect. Immun. 68:2704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien-Simpson, N. M., R. D. Pathirana, R. A. Paolini, Y. Y. Chen, P. D. Veith, V. Tam, N. Ally, R. N. Pike, and E. C. Reynolds. 2005. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 175:3980-3989. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien-Simpson, N. M., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2004. Antigens of bacteria associated with periodontitis. Periodontol. 2000 35:101-134. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 33.Oliver, R. C., and L. J. Brown. 1993. Periodontal diseases and tooth loss. Periodontol. 2000 2:117-127. [DOI] [PubMed] [Google Scholar]
- 34.Page, R. C. 1988. Are there convincing animal models for periodontal diseases? p. 112-122. In B. Guggenheim (ed.), Periodontology today. Karger, Basel, Switzerland.
- 35.Page, R. C., and H. D. Schroeder. 1982. Periodontitis in man and other animals. A comparative review. Karger, Basel, Switzerland.
- 36.Pathirana, R. D., N. O' Brien-Simpson, P. D. Veith, P. Riley, and E. C. Reynolds. 2006. Characterization of proteinase-adhesin complexes of Porphyromonas gingivalis. Microbiology 152:2381-2394. [DOI] [PubMed] [Google Scholar]
- 37.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J. Biol. Chem. 270:1007-1010. [DOI] [PubMed] [Google Scholar]
- 38.Persson, G. R. 2005. Immune responses and vaccination against periodontal infections. J. Clin. Periodontol. 32:39-53. [DOI] [PubMed] [Google Scholar]
- 39.Potempa, J., A. Sroka, T. Imamura, and J. Travis. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 40.Rajapakse, P. S., N. M. O'Brien-Simpson, N. Slakeski, B. Hoffmann, and E. C. Reynolds. 2002. Immunization with the RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis protects against periodontal bone loss in the rat periodontitis model. Infect. Immun. 70:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai, Y., H. Shimauchi, H. O. Ito, M. Kitamura, and H. Okada. 2001. Porphyromonas gingivalis-specific IgG subclass antibody levels as immunological risk indicators of periodontal bone loss. J. Clin. Periodontol. 28:853-859. [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2001. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45:39-44. [DOI] [PubMed] [Google Scholar]
- 43.Slakeski, N., P. S. Bhogal, N. M. O'Brien-Simpson, and E. C. Reynolds. 1998. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology 144:1583-1592. [DOI] [PubMed] [Google Scholar]
- 44.Slakeski, N., S. M. Cleal, P. S. Bhogal, and E. C. Reynolds. 1999. Characterization of a Porphyromonas gingivalis gene prtK that encodes a lysine-specific cysteine proteinase and three sequence-related adhesins. Oral Microbiol. Immunol. 14:92-97. [DOI] [PubMed] [Google Scholar]
- 45.Slakeski, N., S. M. Cleal, and E. C. Reynolds. 1996. Characterization of a Porphyromonas gingivalis gene prtR that encodes an arginine-specific thiol proteinase and multiple adhesins. Biochem. Biophys. Res. Commun. 224:605-610. [DOI] [PubMed] [Google Scholar]
- 46.Smythies, L. E., J. A. Chen, J. R. Lindsey, P. Ghiara, P. D. Smith, and K. B. Waites. 2000. Quantitative analysis of Helicobacter pylori infection in a mouse model. J. Immunol. Methods 242:67-78. [DOI] [PubMed] [Google Scholar]
- 47.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 48.Takeichi, O., J. Haber, T. Kawai, D. J. Smith, I. Moro, and M. A. Taubman. 2000. Cytokine profiles of T-lymphocytes from gingival tissues with pathological pocketing. J. Dent. Res. 79:1548-1555. [DOI] [PubMed] [Google Scholar]
- 49.Taubman, M. A., and T. Kawai. 2001. Involvement of T-lymphocytes in periodontal disease and in direct and indirect induction of bone resorption. Crit. Rev. Oral Biol. Med. 12:125-135. [DOI] [PubMed] [Google Scholar]
- 50.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoneda, M., T. Hirofuji, H. Anan, A. Matsumoto, T. Hamachi, K. Nakayama, and K. Maeda. 2001. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J. Periodontal. Res. 36:237-243. [DOI] [PubMed] [Google Scholar]