Abstract
Candida albicans is a polymorphic opportunistic fungus that can cause life-threatening systemic infections following hematogenous dissemination in patients susceptible to nosocomial infection. Neutrophils form part of the innate immune response, which is the first line of defense against microbes and is particularly important in C. albicans infections. To compare the transcriptional response of leukocytes exposed to C. albicans, we investigated the expression of key cytokine genes in polymorphonuclear and mononuclear leukocytes after incubation with C. albicans for 1 h. Isolated mononuclear cells expressed high levels of genes encoding proinflammatory signaling molecules, whereas neutrophils exhibited much lower levels, similar to those observed in whole blood. The global transcriptional profile of neutrophils was examined by using an immunology-biased human microarray to determine whether different morphological forms or the viability of C. albicans altered the transcriptome. Hyphal cells appeared to have the broadest effect, although the most strongly induced genes were regulated independently of morphology or viability. These genes were involved in proinflammatory cell-cell signaling, cell signal transduction, and cell growth. Generally, genes encoding known components of neutrophil granules showed no upregulation at this time point; however, lactoferrin, a well-known candidacidal peptide, was secreted by neutrophils. Addition to inhibitors of RNA or protein de novo synthesis did not influence the killing activity within 30 min. These results support the general notion that neutrophils do not require gene transcription to mount an immediate and direct attack against microbes. However, neutrophils exposed to C. albicans express genes involved in communication with other immune cells.
Candida albicans is the major human fungal pathogen. It is a commensal of mucosal surfaces in over 50% of individuals, but as an opportunistic pathogen it can cause life-threatening systemic infections in patients with major risk factors for nosocomial infections such as severely immunocompromised people (22). In systemic disease, C. albicans is typically disseminated via the bloodstream. In this environment the invading organisms face an array of cells and molecules that may act against C. albicans and other microbes. The innate immune response is the primary and immediate response against invasion, which has been shown to be of particular importance in defense against C. albicans (1), as part of a type 1 immune response (37). The leukocytes of the innate immune response include polymorphonuclear cells (PMNs; mostly neutrophils, but also eosinophils and basophils) and monocytes. Monocytes secrete a large array of cytokines in response to infection to amplify and coordinate the overall host response, including activation of T cells. The response of monocytes to a C. albicans infection over an 18-h period was recently dissected by using transcript profiling (16).
The leukocytes shown to have the most pronounced and immediate effect on C. albicans are the neutrophils (10). As the first line of defense of the innate immune response, neutrophils capture, phagocytose, and kill invading microbes with a cocktail of potent hydrolytic enzymes, antimicrobial peptides and oxidative species (reviewed recently in references 18 and 35). Many of these molecules are contained within intracellular granules called azurophil (primary) granules, specific (secondary) granules, and gelatinase (tertiary) granules.
In order to dissect the host-fungus interactions of C. albicans bloodstream infections, we have begun to analyze the global transcriptional profiles of both fungal and human cells involved. On the fungal side, C. albicans has been shown to adapt very quickly to the challenges presented by neutrophils and other blood components when exposed to human blood. Within 10 min of incubation in blood, genes involved in protein synthesis are upregulated in the fungal cells. This enables the creation of gene products necessary for the cells to survive under the hostile conditions imposed (11). Furthermore, C. albicans has been shown to upregulate genes involved in the responses to oxidative stress, as well as carbon and nitrogen starvation (10). C. albicans growth was arrested after a 30-min coincubation period with purified PMNs, with only 4% of cells having undergone yeast-hypha morphogenesis (at a host/fungal cell ratio of 1:1.5) (10). After a 1-h incubation, PMNs had killed 62% of the fungal cells. Early time points in experimental C. albicans infections of blood seem to be important since the fungal cells can be “cleared” from the circulating blood within a very short time, as quickly as 5 to 15 min in animal models (22). However, it is not clear whether these cells are in fact removed from the bloodstream or simply not detectable with standard technical procedures such as blood taking.
The ability of C. albicans to switch between a yeast and a hyphal mode of growth is one of the most discussed virulence attributes of this fungus, with both morphological forms playing a role. For example, morphogenesis is vital for the survival of C. albicans in macrophages as, after phagocytosis, fungal cells form germ tubes that can burst host cells, leading to release of the fungus (20), thus allowing persistent infection. Although yeast cells are often considered better suited for dissemination via the blood, whereas hyphal cells are adapted to tissue invasion (3, 4, 38), 40% of C. albicans cells were hyphal after 30 min in whole blood (10), indicating that both forms of C. albicans are likely to be found during blood infection. Clearly, yeast and hyphal cells each express specific sets of genes encoding cell wall proteins and virulence factors, including adhesins and hydrolytic enzymes, suggesting that it is not just the morphological form which is important for distinct functions during infection, but the transcriptional program associated with the transition (9, 17, 30, 43). It has been shown that after 15 h of incubation with neutrophils, C. albicans filaments (and yeast cells, but at a much lower levels) can release a substance, part of which is similar to the known immune modulator adenosine, which can inhibit various activities of PMNs (36), such as the respiratory burst and the release of lactoferrin and β-glucuronidase. However, purified neutrophils clearly inhibit hyphal formation (10, 33). The exact mechanism by which neutrophils block the transition is unknown.
The global transcriptional response to the phagocyte-C. albicans interaction from the fungal perspective has been analyzed (11, 21, 33), showing a high level of transcriptional reprogramming. The host side, too, has been investigated during granulocyte-Candida interactions (25), although using a cell line induced to form granulocytoids instead of primary neutrophils. In that study, Mullick et al. (25) showed that C. albicans can substantially modulate the induction of genes encoding antimicrobial compounds at high ratios of pathogen to host cells.
In analyzing the reactions of C. albicans in blood, we realized that the responses were largely due to the PMNs and presumably to the effects of the constitutively present granular molecules, which are secreted locally or fused with the phagosome of engulfed fungal cells (10). Here, our main aim was to determine whether PMNs elicit a specific transcriptional program in response to C. albicans infection required to directly kill C. albicans and/or to attract additional immune cells. The monocytes are less efficient in killing C. albicans and, rather, seem to be involved in cell-cell signaling, attracting further leukocytes, or inducing differentiation of critical cell types. Therefore, in the present study, we used quantitative reverse transcription-PCR (qRT-PCR) to compare the expression of key cytokine genes by the different blood fractions. We then examined the global transcriptional response of the PMN fraction to C. albicans conditions similar to those used to analyze the transcriptional response of the fungal cells. To determine whether the interaction was an active, two-way process, we analyzed the response of neutrophils to both living and dead cells and to the morphological forms of C. albicans, yeast and hyphae. Furthermore, we investigated whether a de novo synthesis of RNA or protein was essential for killing. By determining the host response, we sought to better understand the reaction of the pathogen and the complex host-pathogen relationship during bloodstream infections.
MATERIALS AND METHODS
Growth of C. albicans.
The well-characterized clinical strain SC5314 (12) was used throughout the present study. C. albicans was grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) overnight at 37°C. Cells were harvested, washed twice in phosphate-buffered saline (PBS), and adjusted to 5 × 107 cells ml−1 in PBS. Cells were either untreated prior to incubation with leukocytes (yeast), exposed to UV radiation for 45 min (UV killed), or induced to undergo yeast-to-hypha morphogenesis by incubation in 10 volumes of donor's plasma for 30 min at 37°C (hyphal). Hyphal induction was confirmed by microscope.
Blood and cellular fractionation.
Human blood was collected from healthy donors as described previously (11). PMNs and mononuclear cells (MNCs) were isolated from the whole blood by gradient density centrifugation in Histopaque 1077 and Histopaque 1119 (Sigma) and then resuspended at their physiological concentrations in the donor's plasma as described previously (10). The purity of the cell types within each fraction has been determined (10).
Incubation of C. albicans with whole blood, plasma, PMNs, or MNCs.
Whole blood, plasma, PMNs, or MNCs were inoculated with C. albicans cells with leukocyte/C. albicans ratios as in whole blood (1:1), in the PMN fraction (1:1.5), and in the MNC fraction (1:3) (to allow for the physiological concentrations of the different fractions). Cells were incubated for 60 min at 37°C with shaking and harvested by centrifugation for 2 min at 4°C. The supernatants were decanted, and the pellets were snap-frozen in liquid nitrogen. Both were stored at −70°C.
RNA preparation and labeling.
The frozen cell pellets were homogenized, the total RNA was isolated, and the mRNA was purified as previously described (10). From mRNA, which was purified from 5 μg of total RNA, Cy5-labeled cRNA was generated by using a linear amplification kit according to the manufacturer's instructions (Agilent Technologies). Cy3 was used to label a common reference RNA (taken from an independent experiment where the PMN fraction was incubated with UV-killed C. albicans yeast cells for 1 h at 37°C) in the same manner.
Microarray hybridization and analysis.
For transcript profiling, custom-made human microarrays were used (AMADID 011412; Agilent Technologies), which were synthesized in duplicate on glass as 60mer oligonucleotides by Agilent's SurePrint technology comprising 8,455 features corresponding to immunology-biased genes. Equal quantities (2 μg) of Cy5- and Cy3-labeled cRNA were hybridized onto the arrays at 60°C overnight according to the supplier's instructions. Slides were washed at 37°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.005% Triton X-102 for 10 min and at room temperature in 6× SSC-0.005% Triton X-102 for 10 min, 0.1× SSC, and 0.005% Triton X-102 for 5 min twice and then dried. Slides were scanned at 5 μm using an Agilent Microarray Scanner BA, and image analysis was carried out with Agilent's feature extraction software version A6.1.1.1 using the default settings. The data analysis was carried out on the Rosetta Inpharmatics platform Resolver Built 3.2.2. Per-spot and per-chip Lowess normalization and further analysis was carried out with GeneSpring 7.1 software (Agilent). The data from duplicate samples were analyzed by one-way analysis of variance with a P value cutoff of 0.05. Of these, genes with a change of regulation of at least twofold compared to the untreated PMNs were further analyzed by using a Venn diagram to isolate groups of genes according to their regulation.
cDNA synthesis and qRT-PCR.
The expression level of selected genes was analyzed by qRT-PCR. The RNA samples analyzed were pooled from three independent experiments, using blood from a single donor. mRNA isolated from total RNA was used to generate cDNA using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Cytokine analysis was carried out on 20 ng of cDNA in a LightCycler as described previously (34). mRNA of key cytokine genes (tumor necrosis factor alpha [TNF-α], granulocyte-macrophage colony-stimulating factor [GM-CSF], gamma interferon [IFN-γ], Toll-like receptor [TLR2], TLR4, interleukin-1A [IL-1A], IL-1B, IL-6, IL-8, and IL-10) was quantified relative to the levels of six housekeeping genes (aldolase, β2-macroglobulin, glucose-6-phosphate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, β-actin, and tyrosine 3-monooxygenase/tryptophane 5-monooxygenase activation protein, zeta polypeptide) and normalized to the expression levels of fractions incubated without C. albicans. Similar data were obtained with pooled cDNAs from six blood donors (data not shown).
Western blot analysis.
For supernatants from leukocytes, C. albicans coincubations were denatured and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane as described previously (10), and then immunoblotted with an antihuman lactoferrin (Dako) or antihuman myeloperoxidase (HyCult Biotechnology) monoclonal antibody and alkaline phosphatase-conjugated anti-rabbit immunoglobulins. Membranes were stripped and reprobed with an anti-human immunoglobulin G antibody (Sigma) as a loading control.
Killing assay.
To investigate the survival of C. albicans exposed to neutrophils, PMNs were incubated for 1 h at 37°C in plasma before the addition of fungal cells. To investigate whether de novo RNA or protein synthesis was necessary for killing, 1 μg of RNA synthesis inhibitor actinomycin D (Sigma) ml−1 or 10 μg of the protein translation inhibitor cycloheximide (Sigma) ml−1 was added to the plasma. After 30 to 120 min of coincubation at 37°C, cells were centrifuged, resuspended in a large volume of water, vigorously vortexed, plated on YPD plates, and incubated overnight at 37°C. The CFU were counted, and the percentage of survival was determined by comparing each time point of incubation to the time zero. As a positive control of C. albicans survival, PMNs were incubated for 30 min in PBS buffer containing 2% formalin and washed three times before the addition of C. albicans cells. Three independent experiments were performed.
Adhesion and phagocytosis assays.
Yeast or hyphal cells of C. albicans were incubated with the PMN fraction for 10 to 60 min at 37°C. The percentage of C. albicans cells (i) unbound, (ii) bound to, or (iii) phagocytosed by PMNs was determined as described previously (10). In addition, the percentage of PMNs having interacted with at least one fungal cell was calculated. Fungal cells were also incubated with the MNC fraction for 10 to 60 min at 37°C to determine the percentage of both C. albicans cells bound to MNCs, and MNCs in contact with C. albicans cells. The morphology of C. albicans cells in the different cocultures was examined after 60 min of incubation. The data presented are from three independent experiments.
RESULTS
Having previously determined the global transcriptional response of C. albicans to leukocytes (10), we aimed to determine the response of leukocytes to C. albicans under similar conditions. This was done in three ways: (1) quantitative RT-PCR of key cytokine and immune receptor genes, (2) global transcriptional profiling using arrays containing genes with an immunological bias, and (3) Western analysis of effector molecules in the supernatant.
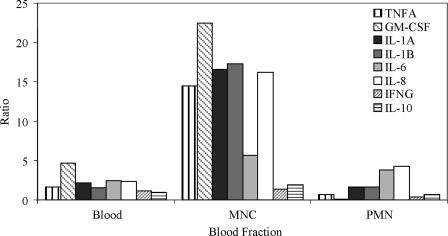
First, to determine the responses of leukocyte fractions from human blood to infections with C. albicans, whole blood, purified MNCs and PMNs were coincubated with C. albicans yeast cells for 1 h. qRT-PCR analysis was carried out on these samples to quantify the levels of eight key cytokine genes and two TLR genes important in fungal infections. In the MNC fraction, the genes encoding TNF-α and IL-1α, IL-1β, and IL-8 were upregulated ∼15-fold in the presence of C. albicans (Fig. 1). Colony-stimulating factor genes for granulocytes and macrophages (GM-CSF) and IL-6 were also upregulated, but at 20- and 5-fold, respectively. In contrast, the PMN fraction showed far lower levels of induction, with all genes upregulated <5-fold. However, certain key cytokine genes, in particular those encoding IL-6 and IL-8, were expressed to higher levels, with the IL-8 gene being upregulated to the highest level at ∼4-fold. Similarly low were the levels of cytokine genes upregulated in whole blood. The highest level observed was an increase of ∼5-fold with the GM-CSF gene.
FIG. 1.
Gene expression levels in blood, MNCs, and PMNs. Determination, by qRT-PCR, of genes encoding proinflammatory cytokines and anti-inflammatory cytokine IL-10 after a 1-h coincubation with C. albicans. The expression levels are normalized to leukocytes incubated without C. albicans. The results are pooled triplicate incubations from a single donor.
The genes encoding both IFN-γ and the anti-inflammatory cytokine IL-10 were not significantly upregulated in MNCs. Neither TLR2 nor TLR4, encoding TLRs for fungal ligands, were significantly altered due to C. albicans in MNCs but were downregulated in both whole blood and PMNs, with the TLR4 gene being more than twofold downregulated (data not shown).
Since PMNs dominate the transcriptional response of C. albicans in blood, we used microarrays representing approximately 8,500 immunobiased genes to further investigate the response of PMNs to C. albicans on a more global level. Therefore, we performed transcript profiling on the PMN fraction incubated with either UV-killed yeast, live yeast, or hyphal C. albicans cells for 1 h. The comparison of live and killed yeast cells was chosen to determine whether there appears to be an active response of the C. albicans cells to which the host cells react rather than a passive interaction with the surface of the cell. Concurrently, yeast and hyphal cell forms were compared to determine whether variations in the cell shape or surface, secreted molecules, or any other attributes associated with these morphological forms have an effect on the response of the PMNs. A 1-h incubation period was chosen to reflect the fact that most of the C. albicans cells were in contact with PMNs (23.2% ± 2.5% attached to and 73.9% ± 2.5% phagocytosed by PMNs), and 65% ± 3.1% of the fungal cells were killed by this time point.
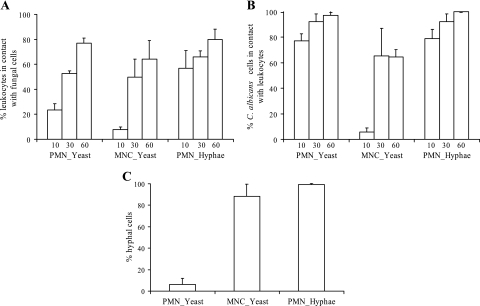
To further investigate the interactions between C. albicans and leukocytes, we calculated (i) the number of leukocytes that were in contact with fungal cells and (ii) the number of fungal cells that interacted with leukocytes. After a 10-min incubation, 56% PMNs were in contact with hyphal cells, whereas only 23% were in contact with yeast cells (Fig. 2A). After 30 min of incubation, the number of PMNs that were in contact with yeast cells had increased, and a similar proportion of PMNs (ca. 75%) were in contact with either yeast or hyphal cells after 60 min of incubation. Surprisingly, almost no differences were observed between the proportions of either yeast or hyphal cells that had contact with PMNs during the 1-h incubation period. In fact, most of the fungal cells were in contact with PMNs by as early as 10 min of incubation time (Fig. 2B). Fewer MNCs were in contact with C. albicans cells. Only 10% of the MNCs were in contact with yeast cells after 10 min, and ca. 65% were in contact with yeast cells at 60 min of incubation. Similarly, fewer C. albicans cells were in contact with MNCs (5 and 65% after 10 and 60 min, respectively) (Fig. 2A and B). It has to be noted that most of the C. albicans cells (>85%) did not change their morphology during the entire incubation time of 60 min (Fig. 2C).
FIG. 2.
Interaction of C. albicans with leukocytes. (A) Percentage of PMNs or MNCs in contact with at least one C. albicans cell after 10, 30, and 60 min of incubation with yeast or hyphal cells. (B) Percentage of C. albicans cells in contact with PMNs or MNCs after yeast or hyphal cells were coincubated in PMN or MNC fractions. (C) Percentage of hyphal cells observed after 1 h of incubation of PMNs or MNCs with yeast or hyphal cells. All values in all three panels correspond to the means of three independent experiments.
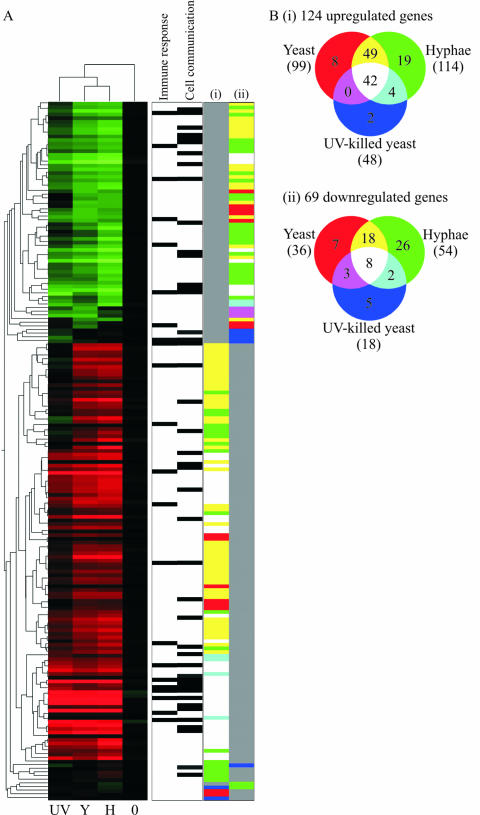
The transcript profiling showed that 191 genes were significantly regulated (Fig. 3). A total of 124 genes were upregulated >2-fold under at least one condition, and 69 genes were downregulated to the same extent. Several genes (the genes shown in Fig. 1) were also quantified by qRT-PCR under these conditions, which showed similar expression patterns and relative levels (data not shown), confirming our microarray data. Cluster analysis by condition showed that transcript profiles due to incubation with the live yeast and hyphae shared more similar expression patterns with each other than with the profile due to dead yeast (Fig. 3), indicating that viability caused a greater difference than cellular morphotype.
FIG. 3.
Gene expression profiles of PMNs in response to C. albicans under different conditions after 60 min of coincubation. (A) Heat map of gene expression depicting the 191 genes regulated at least twofold in PMNs in response to incubation with nil (0), UV-killed yeast (UV), live yeast (Y), or with live hyphal (H) C. albicans cells. The vertical dendrogram indicates similarities between expression patterns of the genes, while the horizontal tree shows the clustering of the different conditions. Genes annotated by gene ontology as being involved in the immune response and cell communication are marked. Columns i and ii are colored according to the appropriate Venn diagram in panel B. (B) Venn diagrams indicating the number of genes upregulated (i) or downregulated (ii) under each condition or combination of conditions.
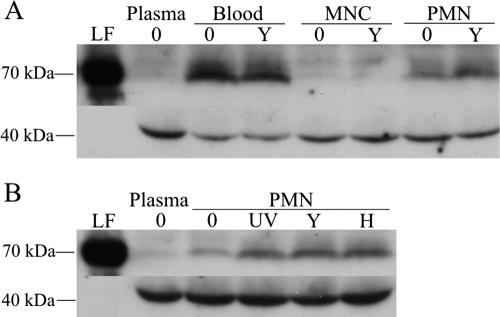
The significantly upregulated genes did not encode proteins present in the granules of neutrophils, with the exception of the gene encoding the urokinase-type plasminogen activator (PLAU; see supplemental data at http://www.galarfungail.org/data.htm). Lactoferrin, which is localized to the specific granules of neutrophils, was observed only at the protein level in the supernatant when neutrophils were present (whole blood and PMN fraction; Fig. 4A), indicating degranulation by neutrophils. A slight increase in lactoferrin levels in response to incubation with C. albicans can be observed in Fig. 4, which compares the response of PMNs to different C. albicans cell types. However, no differences were observed in the presence of the different morphological forms of C. albicans or between living and dead yeast cells. Trace levels of lactoferrin observed in MNCs and plasma and high levels of myeloperoxidase (localized to the azurophil granules) in all samples (data not shown) indicate that the neutrophils were activated, to an extent, by the process of blood fractionation.
FIG. 4.
Western blots showing levels of lactoferrin (LF; ∼80 kDa) in the supernatant after incubation of whole blood or blood fractions with or without C. albicans (A) and PMNs incubated with nil (lanes 0), UV-killed yeast (lanes UV), live yeast (lanes Y) or live hyphal (lane H) C. albicans cells (B). Human immunoglobulin G (∼40 kDa) was detected for a loading control. Replicate samples gave similar results.
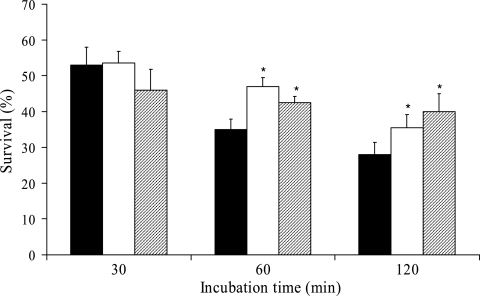
To investigate whether a de novo synthesis of RNA or proteins was necessary for PMNs to kill C. albicans, PMNs were treated with the RNA synthesis inhibitor actinomycin D or the protein synthesis inhibitor cycloheximide before the addition of C. albicans cells. Neither inhibitor affected the survival of C. albicans after 30 min of incubation; however, a moderately, but significantly, reduced killing of C. albicans was observed at 60 and 120 min of incubation (Fig. 5). In contrast, almost all C. albicans cells had formed hyphae at 30 min of incubation when coincubated with formalin-killed PMNs and were still alive after 120 min of incubation (data not shown).
FIG. 5.
Survival assay of C. albicans exposed to neutrophils treated with RNA and protein synthesis inhibitors. The percentages of survival in the PMN fraction treated with either 1 μg of actinomycin D (□) or 10 μg of cycloheximide (░⃞) ml−1 or in an untreated PMN fraction (▪) were determined as follows: (CFU/CFUtime 0) × 100. The values shown are the means and standard errors for three separate experiments. *, Significantly different compared to untreated PMNs (P < 0.05).
Although the genes encoding the granule proteins were unaffected by the presence of C. albicans, 42 genes, largely involved in signaling, were upregulated in the PMN fraction in response to C. albicans irrespective of cell type [Fig. 3B(i), see supplementary data for all gene lists]. These genes include those involved in processes such as proinflammatory cell-cell signaling (e.g., LIF encoding leukemia inhibitory factor, CCL3 and CXCL2, and macrophage inhibitory proteins 1A and 2A, respectively), cell signal transduction (e.g., NR4A2 and NR4A3, nuclear receptor superfamily members), and cell stimulatory factors (e.g., vascular endothelial growth factor). Only eight genes were downregulated due to the presence of C. albicans, independent of cell type [Fig. 3B(ii) and unpublished data]. These may be involved in the regulation of cell signaling (RGS3 encoding regulator of G-protein signaling and IFN-γ receptor) and growth (inhibin IHBB, cyclin-dependent kinase inhibitor CDKN1B, and zinc finger protein ZFP36L2).
More genes were upregulated in neutrophils after incubation with hyphal cells compared to live yeast (114 and 99, respectively; Fig. 3Bi and see supplemental data at http://www.galarfungail.org/data.htm). Of these genes, 91 (75%) were upregulated in response to both morphological forms. If the genes upregulated due to the UV-killed cells were subtracted, 49 genes were upregulated in response to live cells irrespective of morphological form. These genes were annotated as being involved in stress responses including heat shock protein (HSPCA), receptors (IL7R, IL-7 receptor, and annexin A1), and a kinase (SGK, serum/glucocorticoid regulated kinase). Under the same conditions, the number of downregulated genes was less than half of those upregulated (Fig. 3Bii and unpublished data). These 18 downregulated genes include cell signaling regulators (regulator of G protein signaling RGS19, nuclear factor of kappa light polypeptide gene enhancer, TNF receptor-associated factor TRAF2, and protein phosphatase PPM1F).
Further analysis to identify any genes specifically upregulated after incubation with hyphal cells only (subtraction of genes upregulated in the presence of live or UV-killed yeast cells), yielded a list of 19 genes (Fig. 3Bi and unpublished data) including some genes involved in cell signaling (neuropeptide Y, NPY, and transcription factor PITX1) and in some unknown functions. There were 26 genes downregulated under the same conditions (Fig. 3Bii and unpublished data), many encoding proteins of unknown function but also genes encoding immunological receptors (colony-stimulating factor CSF2RB, leukocyte immunoglobulin-like receptor LILRA2), possible cell signaling regulators (cyclin-dependent kinase CDK5, CDC42 effector protein CDC42EP2, zinc finger protein ZNF238), and IFN regulatory factors (IRF1 and IRF5).
DISCUSSION
Numerous studies in the past have examined aspects of the neutrophil-pathogen response, investigating various microorganisms, including C. albicans (8, 23, 29, 32, 35). Recent studies have also investigated the global transcriptional responses of C. albicans to phagocytes/neutrophils, monocytes, and macrophages (10, 21, 33). However, this is the first study known to us that has examined the large-scale response of freshly isolated human neutrophils to C. albicans. Moreover, we characterized the responses of PMNs to both live and UV-killed yeast cells to determine the unique host response to living fungal cells, and we compared yeast and hyphal forms of C. albicans to determine whether these two morphotypes induced distinct gene expression patterns. Finally, we investigated whether a de novo synthesis of RNA or protein in PMNs is necessary for killing of C. albicans.
We previously showed that the reaction of C. albicans to the different fractions of blood was largely due to the PMNs (10). Therefore, in the present study, we sought to determine, by using immunology-biased human microarrays, the global response of this critical blood cell fraction to C. albicans at early time points, when most fungal cells are in contact with neutrophils and are faced with a hostile environment (as indicated by the fungal transcriptional response). Although PMNs were responsible for most of the transcriptional profile of C. albicans in whole blood, the MNCs had the highest expression levels of genes involved in cell-cell signaling at this time point. This was determined by qRT-PCR of genes encoding the key proinflammatory cytokines TNF-α, GM-CSF, IFNγ, IL-1α, IL-1β, IL-6 and IL-8, anti-inflammatory cytokine IL-10 and the fungal immune receptors TLR2 and TLR4 (Fig. 1). The importance of IFN-γ, IL-1α, and IL-1β in the killing of C. albicans pseudohyphae was demonstrated by the reduced potency of PMNs from mice lacking the gene encoding each of these proteins (42).
The qRT-PCR results suggested that the response profile of whole blood was more similar to that of the PMN fraction compared to the MNC fraction, mirroring the relative proportion of these cells in human blood and the similar effects that whole blood and the PMN fraction have on C. albicans (10). This finding highlights the dominant influence of PMNs in blood but also suggests interactions between the PMN and MNC fractions within the context of whole blood. Genes expressed by PMNs, although at a lower level compared to MNCs, included those involved in cellular communication. Since no GM-CSF gene expression was detected in the PMN fraction, the possibility that low levels of gene expression in the PMN fraction could be due to slight contamination with monocytes can be ruled out. Therefore, our data confirm previous studies which showed the ability of PMNs to express cytokines (5, 15). Genes highly upregulated in the MNCs encoded different proinflammatory cytokines, particularly TNF-α, IL-1α, IL-1β, and IL-8. Consistent with this, neutropenic mice have significantly higher levels of TNF-α and IL-6 produced by non-PMN cells compared to non-neutropenic controls (7). In our study, this response of the MNC fraction is certainly due to monocytes present in this fraction since over this 1-h time period the lymphocytes would not yet be stimulated. Furthermore, in a time course experiment of monocytes incubated with C. albicans, Kim et al. showed similar stimulation (16).
Another cytokine gene expressed at high levels by monocytes in our study was IFN-γ. This cytokine is known to enhance the killing potential of PMNs and macrophages (2, 27). It has been shown that C. albicans stimulates the production of IFN-γ in blood in a IL-18-, IL-12-, and IL-1β-dependent manner (28).
Neither IL-10 nor TLR2 and TLR4 genes were found to be upregulated in our study. The very low levels of IL-10 gene expression at this early time point of host fungus interaction are not surprising since expression of this cytokine leads to suppression of the innate immune response, which is required to fight C. albicans infection as the first line of defense (24). High IL-10 protein levels from PMNs have been observed after a 48-h incubation with either heat-killed yeast or hyphae (40). It is possible that the IL-10 gene is transcribed after the 1-h time point investigated here or that the protein could be presynthesized. Although involved in the host response against C. albicans (31, 44), as cell surface receptors, TLR2 and TLR4 are most likely to be present on the cell surface prior to activation (and/or regulated at a posttranslational level) and so would not be upregulated at the mRNA level at early time points.
There was a long-held belief that PMNs are terminally differentiated and incapable of gene expression in response to environmental changes. However, recent studies have disproved this notion (5, 15). Consistent with this, we observed the regulation of numerous genes in PMNs in response to C. albicans. However, the regulation of gene expression in PMNs does not directly correspond with the gene expression response in C. albicans (10). The response of C. albicans (notably the upregulation of genes involved in nitrogen and carbon starvation and those involved in oxidative stress) appears to be due to the environmental changes caused by the oxidative burst and/or the proteins generated during myelopoiesis and stored in the granules found within neutrophils (6). Our data suggest that these components are already present in when cells come into contact with C. albicans and therefore do not need to be newly synthesized, at least not within the first 30 min of contact. This view is supported by the fact that RNA and protein synthesis inhibitors have no effect on the killing of C. albicans by PMNs within the first 30 min of coincubation. However, since blockage of de novo synthesis of either RNA or protein does cause a minor but significant reduced capacity of neutrophils to kill C. albicans after 30 min (60 to 120 min), it can be concluded that certain RNAs and/or proteins need to be produced for full killing efficiency at later time points. Components potentially involved in killing include lactoferrin, which is secreted in the PMN fractions. Several previous studies have demonstrated the candidacidal properties of lactoferrin (29, 39, 41). The only gene representing the neutrophil granule proteins, which was significantly upregulated, was the protease urokinase-type plasminogen activator (i.e., PLAU) (13). However, PLAU has been detected not only in the specific granules but also in the cell plasma membrane (14) and may have functions additional to the killing of microbes, which may explain the expression observed. The expression of the PLAU gene was also detected in granulocytoids after incubation with C. albicans (25). In a study by Mullick et al. (25), the levels of genes encoding myeloperoxidase, defensins, and neutrophil elastase, all known antimicrobials, were reduced in response to C. albicans; however, this effect was only detected in PMNs at a high ratio of C. albicans to PMNs (5:1), higher than that used in the present study. As we were investigating the host side of the interaction previously observed (10), we did not explore the PMN/C. albicans response at higher ratios.
Although genes involved in antimicrobial activities were not upregulated in our study, we identified a number of genes that were up- or downregulated in PMNs when exposed to C. albicans. The genes upregulated in the PMN fraction in response to C. albicans (independent of cell viability and morphology) include those encoding cell stimulatory factors, vascular endothelial growth factor and early growth factor 1, proinflammatory cell-cell signaling leukemia inhibitory factor (LIF), and the chemokines macrophage inhibitory proteins 1A and 2A (CCL3 and CXCL2, respectively). Our results mirror the transcript profiles observed previously in granulocytoids and neutrophils (15, 25).
In addition, genes upregulated in a C. albicans viability-dependent manner encoded a significant proportion of stress response proteins; this was not the case for the response to UV-killed cells, indicating a direct effect of live cells upon the PMNs. These genes included heat shock proteins (HSPA8, HSPCA, HSPCB, and HSPH1), which participate in the regulation of CXC-type chemokines (26). This connection links the host cells' stress in mounting an antimicrobial response to the amplification of the overall immune response by attracting further reactive cells.
After exposure to the PMN fraction for 30 min, C. albicans cells were 96% restricted to the yeast form compared to 60% in whole blood and only 15 to 20% in plasma and with MNCs (10). Note that the majority of genes upregulated due to the yeast or hyphal form were shared in both conditions. Hyphal cells specifically induced only 19 genes in PMNs and yeast cells induced eight genes. Hence, under the conditions investigated, the responses of PMNs to the different morphological forms did not vary significantly despite the fact that fewer PMNs were contacting yeast cells than hyphae at early time points. The few differences observed may be due to the specific transcriptomes of yeast and hyphal cells resulting in specific cell wall proteins, as well as secreted factors (9, 19, 31). In contrast, heat-killed yeast and hyphae induced different responses in MNCs on the protein level after a 24-h incubation, which included specific TLR and cytokine differences between cell types (40). It is possible that this occurs for PMNs, too; perhaps a longer incubation time would increase the divergence in expression profiles.
Due to the complexity and variability of host-microbe interactions it is difficult to define a reliable in vivo infection model (discussed in relation to genomics (19). Rather, we can only dissect specific interactions and investigate these in detail in an attempt to understand the complex host-pathogen relationship in vivo. Our data indicate that freshly isolated human PMNs do not depend on gene transcription to respond to and kill C. albicans cells under the conditions examined here since (i) the responsible proteins are already present in the cells' granules and (ii) de novo synthesis inhibitors do not increase the potential of C. albicans to survive PMNs. However, PMNs do upregulate numerous genes in response to C. albicans, after 1 h of contact with the fungus, and these are largely involved in recruitment or activation of additional immune cells and increase the overall effect of the immune response rather than representing a unique pathogen-specific response. This includes genes encoding cytokines and cytokine receptors. Therefore, PMNs are prepared for immediate killing of C. albicans but contribute to the immune response by the expression of genes involved in cellular communication. The consequence of this response may be the recruitment of other immune cells or additional PMNs. In the present study, a higher number of PMNs contacted C. albicans yeast cells at 30 min compared to 10 min of incubation, whereas almost the same proportion of C. albicans cells were in contact with PMNs at both incubation times. Being highly motile cells, neutrophils can quickly congregate after recruitment at the sites of PMN-C. albicans interactions, as is observed in vivo during infection.
Although screening the global transcription can give fascinating insights into cellular activities, it cannot provide a complete picture of PMN-C. albicans interactions. Comprehensive proteomic studies are necessary to determine the actual secretion of factors that will ultimately cause the effects. MNCs also express genes of key cytokines in response to C. albicans but at much higher levels. Many of the highly upregulated genes in PMNs are expressed irrespective of fungal morphology or viability. However, there are also unique PMN responses to live C. albicans and a few increases that are morphology dependent, which indicate that active processes by live cells vary between the yeast and hyphal growth forms. Overall, the cross talk observed is highly complex; each component of the infection process has the ability to modulate other components—different host cell types and different pathogen cell types contribute to the spectrum of inputs, resulting in a large variety of responses. Elucidation of these dissected responses will hopefully aid our understanding of the overall response and thus promote development of novel therapies to supplement or substitute the host response in susceptible patients.
Acknowledgments
C.F. and A.L.M. were supported by the EU projects Galar Fungail (QLK2-2000-00795) and Galar Fungail 2 (MCRTN-2003-504148). M.S. and G.W. were supported by the Deutsche Forschungsgemeinschaft (Sch 897/1 3 and Sch 897/3 1).
We thank Mihai Netea for critical reading of the manuscript, all blood donors, and Antje Erler and Wiebke Thoma for taking blood.
Editor: A. Casadevall
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Ashman, R., C. Farah, S. Wanasaengsakul, Y. Hu, G. Pang, and R. Clancy. 2004. Innate versus adaptive immunity in Candida albicans infection. Immunol. Cell Biol. 82:196-204. [DOI] [PubMed] [Google Scholar]
- 2.Ashman, R. B., and J. M. Papadimitriou. 1995. Production and function of cytokines in natural and acquired immunity to Candida albicans infection. Microbiol. Rev. 59:646-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, J., and P. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 4.Brown, A., and N. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 5.Cassatella, M. A. 1995. The production of cytokines by polymorphonuclear neutrophils. Immunol. Today 16:21-26. [DOI] [PubMed] [Google Scholar]
- 6.Christin, L., D. R. Wysong, T. Meshulam, S. Wang, and R. D. Diamond. 1997. Mechanisms and target sites of damage in killing of Candida albicans hyphae by human polymorphonuclear neutrophils. J. Infect. Dis. 176:1567-1578. [DOI] [PubMed] [Google Scholar]
- 7.Daley, J. M., J. S. Reichner, E. J. Mahoney, L. Manfield, W. L. Henry, Jr., B. Mastrofrancesco, and J. E. Albina. 2005. Modulation of macrophage phenotype by soluble product(s) released from neutrophils. J. Immunol. 174:2265-2272. [DOI] [PubMed] [Google Scholar]
- 8.Diamond, R. D., R. A. Clark, and C. C. Haudenschild. 1980. Damage to Candida albicans hyphae and pseudohyphae by the myeloperoxidase system and oxidative products of neutrophil metabolism in vitro. J. Clin. Investig. 66:908-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebanks, R. O., K. Chisholm, S. McKinnon, M. Whiteway, and D. M. Pinto. 2006. Proteomic analysis of Candida albicans yeast and hyphal cell wall and associated proteins. Proteomics 6:2147-2156. [DOI] [PubMed] [Google Scholar]
- 10.Fradin, C., P. De Groot, D. MacCallum, M. Schaller, F. Klis, F. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397-415. [DOI] [PubMed] [Google Scholar]
- 11.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillardin, C. d'Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47:1523-1543. [DOI] [PubMed] [Google Scholar]
- 12.Gillum, A., E. Tsay, and D. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 13.Gyetko, M. R., D. Aizenberg, and L. Mayo-Bond. 2004. Urokinase-deficient and urokinase receptor-deficient mice have impaired neutrophil antimicrobial activation in vitro. J. Leukoc. Biol. 76:648-656. [DOI] [PubMed] [Google Scholar]
- 14.Heiple, J. M., and L. Ossowski. 1986. Human neutrophil plasminogen activator is localized in specific granules and is translocated to the cell surface by exocytosis. J. Exp. Med. 164:826-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasama, T., Y. Miwa, T. Isozaki, T. Odai, M. Adachi, and S. L. Kunkel. 2005. Neutrophil-derived cytokines: potential therapeutic targets in inflammation. Curr. Drug Targets Inflamm. Allergy 4:273-279. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H., E. Choi, J. Khan, E. Roilides, A. Francesconi, M. Kasai, T. Sein, R. Schaufele, K. Sakurai, C. Son, B. Greer, S. Chanock, C. Lyman, and T. Walsh. 2005. Expression of genes encoding innate host defense molecules in normal human monocytes in response to Candida albicans. Infect. Immun. 73:3714-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumamoto, C. A., and M. D. Vinces. 2005. Contributions of hyphae and hypha-coregulated genes to Candida albicans virulence. Cell Microbiol. 7:1546-1554. [DOI] [PubMed] [Google Scholar]
- 18.Lee, W., R. Harrison, and S. Grinstein. 2003. Phagocytosis by neutrophils. Microbes Infect. 5:1299-1306. [DOI] [PubMed] [Google Scholar]
- 19.Liu, M., S. J. Popper, K. H. Rubins, and D. A. Relman. 2006. Early days: genomics and human responses to infection. Curr. Opin. Microbiol. 9:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, H., J. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. Fink. 1997. Nonfilamentous Candida albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz, M., J. Bender, and G. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavor, A., S. Thewes, and B. Hube. 2005. Systemic fungal infections caused by Candida species: epidemiology, infection process, and virulence attributes. Curr. Drug Targets 6:863-874. [DOI] [PubMed] [Google Scholar]
- 23.Mayer-Scholl, A., P. Averhoff, and A. Zychlinsky. 2004. How do neutrophils and pathogens interact? Curr. Opin. Microbiol. 7:62. [DOI] [PubMed] [Google Scholar]
- 24.Mencacci, A., E. Cenci, G. D. Sero, C. Fe d'Ostiani, P. Mosci, G. Trinchieri, L. Adorini, and L. Romani. 1998. IL-10 is required for development of protective Th1 responses in IL-12-deficient mice upon Candida albicans infection. J. Immunol. 161:6228-6237. [PubMed] [Google Scholar]
- 25.Mullick, A., M. Elias, P. Harakidas, A. Marcil, M. Whiteway, B. Ge, T. J. Hudson, A. Caron, L. Bourget, S. Picard, O. Jovcevski, B. Massie, and D. Thomas. 2004. Gene expression in HL60 granulocytoids and human polymorphonuclear leukocytes exposed to Candida albicans. Infect. Immun. 72:414-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagarsekar, A., J. D. Hasday, and I. S. Singh. 2005. CXC chemokines: a new family of heat-shock proteins? Immunol. Investig. 34:381-398. [DOI] [PubMed] [Google Scholar]
- 27.Nathan, C. F., H. W. Murray, M. E. Wiebe, and B. Y. Rubin. 1983. Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158:670-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Netea, M. G., R. J. Stuyt, S. H. Kim, J. W. Van der Meer, B. J. Kullberg, and C. A. Dinarello. 2002. The role of endogenous interleukin (IL)-18, IL-12, IL-1β, and tumor necrosis factor-α in the production of interferon-γ induced by Candida albicans in human whole-blood cultures. J. Infect. Dis. 185:963-970. [DOI] [PubMed] [Google Scholar]
- 29.Okutomi, T., S. Abe, S. Tansho, H. Wakabayashi, K. Kawase, and H. Yamaguchi. 1997. Augmented inhibition of growth of Candida albicans by neutrophils in the presence of lactoferrin. FEMS Immunol. Med. Microbiol. 18:105-112. [DOI] [PubMed] [Google Scholar]
- 30.Pitarch, A., M. Sanchez, C. Nombela, and C. Gil. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell Proteomics 1:967-982. [DOI] [PubMed] [Google Scholar]
- 31.Pivarcsi, A., L. Bodai, B. Rethi, A. Kenderessy-Szabo, A. Koreck, M. Szell, Z. Beer, Z. Bata-Csorgoo, M. Magocsi, E. Rajnavolgyi, A. Dobozy, and L. Kemeny. 2003. Expression and function of Toll-like receptors 2 and 4 in human keratinocytes. Int. Immunol. 15:721-730. [DOI] [PubMed] [Google Scholar]
- 32.Richardson, M. D., and H. Smith. 1981. Resistance of virulent and attenuated strains of Candida albicans to intracellular killing by human and mouse phagocytes. J. Infect. Dis. 144:557-564. [DOI] [PubMed] [Google Scholar]
- 33.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and G. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaller, M., U. Boeld, S. Oberbauer, G. Hamm, B. Hube, and H. C. Korting. 2004. Polymorphonuclear leukocytes (PMNs) induce protective Th1-type cytokine epithelial responses in an in vitro model of oral candidosis. Microbiology 150:2807-2813. [DOI] [PubMed] [Google Scholar]
- 35.Segal, A. W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smail, E. H., B. N. Cronstein, T. Meshulam, A. L. Esposito, R. W. Ruggeri, and R. D. Diamond. 1992. In vitro, Candida albicans releases the immune modulator adenosine and a second, high-molecular weight agent that blocks neutrophil killing. J. Immunol. 148:3588-3595. [PubMed] [Google Scholar]
- 37.Spellberg, B., and J. E. Edwards, Jr. 2001. Type 1/Type 2 immunity in infectious diseases. Clin. Infect. Dis. 32:76-102. [DOI] [PubMed] [Google Scholar]
- 38.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 39.Tanida, T., F. Rao, T. Hamada, E. Ueta, and T. Osaki. 2001. Lactoferrin peptide increases the survival of Candida albicans-inoculated mice by upregulating neutrophil and macrophage functions, especially in combination with amphotericin B and granulocyte-macrophage colony-stimulating factor. Infect. Immun. 69:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Graaf, C. A. A., M. G. Netea, I. Verschueren, J. W. M. van der Meer, and B. J. Kullberg. 2005. Differential cytokine production and Toll-like receptor signaling pathways by Candida albicans blastoconidia and hyphae. Infect. Immun. 73:7458-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viejo-Diaz, M., M. T. Andres, and J. F. Fierro. 2005. Different anti-Candida activities of two human lactoferrin-derived peptides, Lfpep and kaliocin-1. Antimicrob. Agents Chemother. 49:2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vonk, A. G., C. W. Wieland, M. Versteegen, I. C. Verschueren, M. G. Netea, L. A. Joostent, P. E. Verweij, and B. J. Kullberg. 2005. Influence of endogenous pro-inflammatory cytokines on neutrophil-mediated damage of Candida albicans pseudohyphae, quantified in a modified tetrazolium dye assay. Med. Mycol. 43:551-557. [DOI] [PubMed] [Google Scholar]
- 43.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350-357. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, S., J. Li, X. Jia, and Y. Wu. 2004. The expression of Toll-like receptor 2 and 4 mRNA in local tissues of model of oropharyngeal candidiasis in mice. J. Huazhong Univ. Sci. Technol. Med. Sci. 24:639-641. [DOI] [PubMed] [Google Scholar]