Abstract
The cytolethal distending toxins (CDTs) are secreted virulence proteins produced by several bacterial pathogens, and the subunit CdtB has the ability to create DNA lesions, primarily DNA single-strand breaks (SSBs) in vitro, and cause cell cycle arrest, cellular distension, and cell death in both mammalian and yeast cells. To elucidate the components of the mechanisms underlying the response to CdtB-induced DNA lesions, a CdtB expression plasmid was transformed into a series of diploid yeast strains harboring deletions in 4,708 nonessential genes. A total of 4,706 of these clones were successfully transformed, which we have now designated as a systematic transformation array (STA), and were subsequently screened. We identified 61 sensitive strains from the STA whose deleted genes can be categorized into a number of groups, including DNA metabolism, chromosome segregation, vesicular traffic, RNA catabolism, protein translation, morphogenesis, and nuclear transport, as well as one unknown open reading frame. However, only 28 of these strains were found to be sensitive to HO endonuclease, which is known to create a DNA double-strand break (DSB), suggesting that CdtB-induced DNA lesion is not similar to the direct DSB. Amazingly, CdtB expression elicits severe growth defects in haploid yeast cells, but only marginal defects in diploid yeast cells. The presence and absence of genes known to be involved in DNA repair in these genome-wide data reveal that CdtB-induced DNA damage is specifically repaired well in the diploid by homologous recombination but not by other repair mechanisms. Our present results provide insights into how CdtB pathogenesis is linked to eukaryotic cellular functions.
The cytolethal distending toxins (CDTs) are secreted virulence proteins produced by a number of bacterial pathogens, including Escherichia coli, Haemophilus ducreyi, Campylobacter spp., Salmonella enterica serovar Typhi, Actinobacillus actinomycetemcomitans, Shigella dysenteriae, and Helicobacter spp. (42, 44, 56). CDTs consist of the three subunits CdtA, CdtB, and CdtC and form a ternary complex (40). CdtB shares conserved residues with the active sites of DNase I-like nucleases, and purified CdtB primarily shows single-strand nicking activity on coiled plasmid DNA and subsequently produces linear DNA in vitro (15, 35, 40). Enzymatically active CdtB induces cell cycle arrest at the G2/M phase, inhibits cell proliferation, and causes cellular enlargement in mammalian cells (28, 40). Biochemical analysis has also demonstrated that DNA damage checkpoint machineries and Rho-type GTPase function are involved in CdtB-induced cell cycle arrest and cellular enlargement (13, 15, 16, 30, 60). However, the entire complement of genes required for the repair of CdtB-induced DNA lesions and also those leading to cell cycle arrest, cellular enlargement, and cell death have not been fully identified. Because Hassane et al. showed that CdtB is active in yeast (Saccharomyces cerevisiae) cells similar to mammalian cells (19), we used the yeast genome to further elucidate and characterize the components of the CdtB response pathway and to illustrate the consequences of CdtB activity in host cells.
Genome-wide deletion strains of yeast have now been used in many studies (33, 50, 61), and genome-wide analyses have a number of advantages over their classical genetic counterparts, not only in terms of the greater ease in obtaining global results but also because of their far greater comprehensiveness. Hence, if all of the yeast deletion strains in a particular set are screened for desirable phenomena, the known genes identified in this screening will reveal novel features of the required functions. Moreover, the absence of specific genes can also disclose features of unrelated functions. The value of identifying absent genes in such screening approaches has not been highly emphasized in traditional genetics, even in genome-wide analysis, because these analyses need to be sufficiently systematic to verify that such an absence is not an artifact or due to leakage from the screening filters. In our present study, we have adopted a systematic transformation method that allows us to analyze each deletion strain one by one for the CdtB-induced growth phenotype. This method also allows us to generate a comprehensive series of results for genes required for the CdtB response, which illustrates a genome-wide view of host-pathogen interactions.
In comparison with the numerous previous studies of double-strand breaks (DSBs) (26), little is currently known about the repair mechanisms for single-strand breaks (SSBs). Even if there is no direct evidence for SSB creation induced by CdtB expression in vivo, yeast genome-wide analysis will reveal the feature of CdtB-induced DNA lesions by comparison with the responses to the direct DSB that can be created by the ectopic expression of HO endonuclease in yeast (47) and the other DNA damage. In the present study, we report the results of our genome-wide screen of genes required for CdtB response in yeast. We show here that evolutionally conserved mechanisms involving components of homologous recombination (HR), DNA replication, chromosome maintenance, and mRNA decay are required for the response to CdtB. The genes that we identified in this analysis also indicate that there are specific features of CdtB response that do not fully overlap with the components required for direct DSB and other DNA damage.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth media.
Complete homozygous diploid deletion strains (#95401.H1R3) were obtained from Research Genetics (Huntsville, AL). BY4743 (MATa/MATα leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 met15Δ0/+ +/lys2Δ0) and BY4740 (MATa leu2Δ0 ura3Δ0 lys2Δ0) were used as the parental strains. The diploid W303 strain and isogenic haploid W303-1A strain were also used (57). YPD medium and dropout synthetic medium were prepared using standard procedures (46). The plasmids used were pDCH-CdtB from Campylobacter jejuni (19), YopM (pCFL140) (29) and pRS315 (52). p315GAL-HO was constructed by insertion of the GAL-HO fragment from pGAL-HO (21) into the pRS315 vector.
Yeast transformation.
Systematic yeast transformation experiments were performed using the S. cerevisiae direct transformation kit (Wako Pure Chemicals, Osaka, Japan), which was originally developed in our laboratory. Yeast deletion strains were grown on YPD square plates and picked using 96-pin QReps (X5052; Genetix, Hampshire, United Kingdom). The selected clones were then inoculated in 25 μl of YPD liquid medium in microplates and grown for 24 h without shaking. Twenty-five microliters of direct transformation solution premixed with plasmid DNA was directly added to each well, and the plates were then vortexed and incubated at 42°C for 2 h. Ten-microliter aliquots of the growth mixtures were spotted on square selection plates using a 96-channel pipetting machine (HT station 500; Cosmotec, Tokyo, Japan).
Screening and characterization of CdtB-sensitive strains.
Transformant arrays were generated in a 96-well format on square selection plates. The transformed yeast deletion clones were picked, transferred to 50-μl volumes of leucine-dropout medium in 96-well microplates, and incubated at 28°C for 24 h without shaking. These cell cultures were then spotted onto synthetic medium containing 2% glucose, 2% galactose, or 2% raffinose and 2% galactose.
For growth assays, deletion strains were grown for 24 h and cell concentrations were adjusted to optical density at 600 nm (OD600) values of 1, 0.1, 0.01 and 0.001. Eight-microliter aliquots of these serial dilutions were then spotted onto galactose or glucose plates. For alternative cell growth and survival assays, the yeast cells were precultured in synthetic dropout liquid medium containing 2% raffinose and then transferred to dropout liquid medium containing 2% raffinose and 2% galactose to give an OD600 of 0.1. After 24 h, OD600 values were measured to compare cell growth, cell suspensions were adjusted to an OD600 value of 0.001, and 100 μl was spread onto YPD plates to compare their survival frequency with that of the CdtB-transformed BY4743 control parental strain. Results were obtained from three independent experiments. For drug sensitivity assays, YPD medium containing 0.25 M hydroxyurea (HU), and 0.2% methyl methanesulfonate (MMS) was used. UV sensitivities were examined by irradiation of spotted serial dilutions of the yeast strains according to a previously described procedure (39).
For flow cytometric analysis, cells were fixed with 70% ethanol, washed with phosphate-buffered saline, and incubated with 1 mg/ml RNase. Cells were then suspended in 20 μg/ml of propidium iodide in phosphate-buffered saline. Flow cytometry was performed using FACS Calibur (Becton Dickinson, Franklin Lakes, NJ).
RESULTS
The impact of ploidy upon CdtB-induced toxicity in yeast.
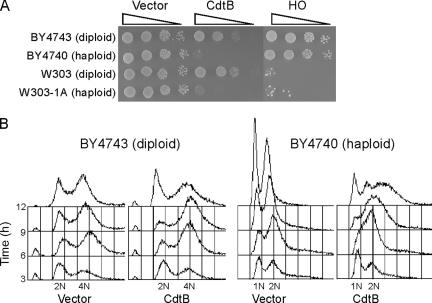
CdtB induces cell cycle arrest during G2/M phase and also causes cellular enlargement in both mammalian and yeast cells (19, 60). We further compared the defects in growth and cell cycle progression in haploid and diploid yeast strains expressing CdtB and found that even in strains of different backgrounds, the CdtB-induced growth defects were severe in haploid cells but not in diploid cells (Fig. 1A). For comparison, HO endonuclease was also expressed under the control of the same GAL promoter in these yeast strains, but the resulting growth defects in this case were found not to be ploidy dependent but strain dependent (Fig. 1A). To next compare cell cycle progression in the haploid and diploid strains expressing CdtB, flow cytometric analyses were performed (Fig. 1B). In haploid yeast, S-phase progression was delayed, and there was a detectable accumulation of cells with a higher DNA content. In diploid yeast, an S-phase delay was not observed but cells were found to accumulate in G2/M phase. These results suggest that CdtB-induced DNA lesions are repaired effectively in diploid cells but not in their haploid counterparts during the transition from S to G2/M. To elucidate the identity of the CdtB response components in diploid yeast, we next screened an array of CdtB-sensitive yeast strains from an established genome-wide set of diploid nonessential deletion mutants (61).
FIG. 1.
Ploidy-dependent phenotypes of CdtB. (A) CdtB-induced growth defects are severe in haploid (BY4740 and W303-1A) but not in diploid (BY4743 and W303) yeast cells. The HO endonuclease (HO)-induced growth defect was also found to be strain dependent but not ploidy dependent. (B) CdtB induces slow S-phase progression, G2/M arrest, and the accumulation of overreplicated cells with a higher DNA content in haploid but not in diploid strains.
Genome-wide screening of CdtB sensitive yeast deletion strains.
Homozygous nonessential deletion strains were inoculated onto YPD plates, but 84 clones could not be recovered from the frozen stocks. The remaining 4,708 strains were successfully recovered and were transformed with a CdtB-expression plasmid (pDCH-CdtB) via our systematic transformation method (see Materials and Methods). A total of 4,602 strains have been successfully transformed by the initial transformation procedure (Fig. 2A). A total of 106 clones were subjected to transformation once more using the same method, and 98 of these strains were successfully transformed. The remaining eight strains were then transformed one by one, and only the ilv1 and leu3 clones that are auxotrophic for leucine could not be transformed but were successfully transformed with a URA3 plasmid (data not shown). This indicates that there are no transformation-incompetent yeast strains in this set of nonessential gene deletion mutants.
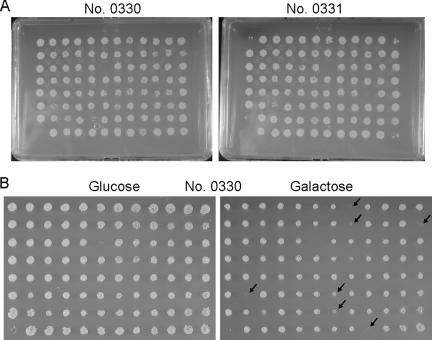
FIG. 2.
The systematic transformation array. (A) Deletion strains were transformed with CdtB plasmid, and transformants were then obtained by growth on selection plates. (B) Transformed deletion strains (plate no. 0330) were screened on glucose (repression) and galactose (induction) plates. Arrows indicate candidate CdtB-sensitive strains.
The transformant array was then transferred to liquid synthetic dropout medium in 96-well microplates and grown for a further 24 h. The cultures were then spotted onto synthetic glucose (repression) or galactose (induction) plates, except for the 0370, 0371, 0372, and 0380 plates, where 2% raffinose-2% galactose plates were used instead because of the presence of many strains that are slow growing on galactose. In our initial screening, 242 sensitive deletion strains were selected (Fig. 2B) and several criteria were used for further qualifying these sensitive clones. Cells from each of these strains, which were not transformed, were spotted onto synthetic glucose or galactose plates, supplemented with the required nutrients, to examine their growth. Seventy-six strains were found to grow slowly on galactose. The remaining 166 strains were transformed again with either pDCH-CdtB or an empty vector control, and serially diluted cultures of each were then spotted onto galactose plates. Among these strains, 76 had a strongly sensitive phenotype, but the remaining 90 were only weakly sensitive or not sensitive. Two of the 76 strains transformed with an empty vector grew slowly on galactose medium. The growth characteristics of the remaining 74 sensitive strains were then examined on synthetic minimal medium supplemented with only leucine, uracil, and histidine (LUH) to determine the auxotrophic mutations of the parental diploid strain. Eleven strains (cog1Δ, fyv10Δ, mto1Δ, npl6Δ, rad54Δ, swi4Δ, swi6Δ, tos1Δ, ydl041wΔ, yil039wΔ, and ypl208wΔ) did not grow on the LUH medium. Moreover, two strains (ybr099cΔ and ynr068cΔ) mated with a mating-tester strain. Hence, these strains may not be diploid or may be affected by unknown problems that arose during the construction process. Finally, 61 strains were determined to be CdtB-sensitive deletion strains (Fig. 3 and Table 1) .
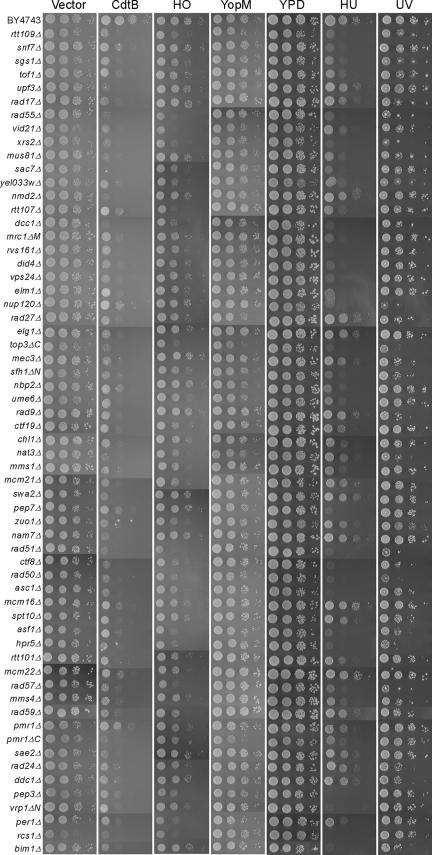
FIG. 3.
CdtB-sensitive strains and their cross-sensitivity properties. Deletion strains were transformed with empty vector and with CdtB, HO endonuclease, and YopM expression constructs, and diluted cultures were then spotted onto galactose plates. CdtB-sensitive strains without plasmid were spotted onto YPD and YPD plates containing 0.25 M HU. The spots on YPD plates were exposed to UV irradiation.
TABLE 1.
CdtB-sensitive genes
| Genea | Function | % of wild typeb:
|
DNA contentc | Cell sized | |
|---|---|---|---|---|---|
| Viability | Growth | ||||
| DNA metabolism | |||||
| DDC1 | DNA damage checkpoint protein | 12 | 67 | = | ++ |
| HPR5 | DNA helicase | 5 | 40 | ≪ | ++ |
| MEC3 | DNA damage checkpoint protein | 7 | 82 | = | + |
| MMS1 | Protection against replication-dependent DNA damage | 14 | 39 | ≪ | ++ |
| MMS4 | Subunit of Mus81-Mms4 endonuclease | 17 | 38 | ≪ | ++ |
| MRC1ΔM/YCL060C | S-phase checkpoint protein | 63 | 33 | G1 | − |
| MUS81* | Subunit of Mus81-Mms4 endonuclease | 10 | 39 | ≪ | ++ |
| RAD9 | DNA damage checkpoint protein | 11 | 81 | = | + |
| RAD17 | DNA damage checkpoint protein | 8 | 72 | = | ++ |
| RAD24* | DNA damage checkpoint protein | 8 | 66 | = | ++ |
| RAD27* | Exonuclease required for Okazaki fragment processing | 23 | 45 | < | ++ |
| RAD50* | Subunit of Mre11-Rad50-Xrs2 protein complex | 7 | 25 | < | ++ |
| RAD51* | Strand exchange protein | 10 | 20 | < | ++ |
| RAD55 | Protein that stimulates strand exchange | 15 | 30 | ≪ | ++ |
| RAD57* | Protein that stimulates strand exchange | 13 | 17 | ≪ | ++ |
| RAD59* | Protein involved in double-strand break repair | 18 | 43 | ≪ | ++ |
| RTT101 | Subunit of a ubiquitin ligase complex | 14 | 42 | < | ++ |
| RTT107 | Regulator of Ty1 transposition | 31 | 82 | < | ++ |
| RTT109 | Regulator of Ty1 transposition | 18 | 51 | < | ++ |
| SAE2 | Protein with a role in double-strand break repair | 16 | 23 | ≪ | +++ |
| SGS1* | Nucleolar DNA helicase | 28 | 45 | < | ++ |
| TOF1 | S-phase checkpoint protein | 83 | 46 | G1 | − |
| TOP3ΔC*/YLR235C | DNA topoisomerase III | 13 | 36 | < | ++ |
| UME6 | Transcriptional regulator of early meiotic genes | 62 | 53 | = | − |
| XRS2 | Subunit of Mre11-Rad50-Xrs2 protein complex | 9 | 24 | ≪ | ++ |
| Chromosome maintenance | |||||
| ASF1* | Nucleosome assembly factor | 16 | 42 | < | − |
| BIM1* | Microtubule-binding protein | 36 | 36 | G1 | + |
| CHL1* | Protein required to establish sister-chromatid pairing | 61 | 38 | G1 | − |
| CTF8 | Component of replication factor C complex | 23 | 32 | = | − |
| CTF19 | Outer kinetochore protein | 55 | 28 | G1 | − |
| DCC1* | Component of replication factor C complex | 31 | 23 | = | − |
| ELG1 | Component of replication factor C complex | 45 | 34 | G1 | ++ |
| MCM16 | Protein involved in minichromosome maintenance | 60 | 34 | G1 | − |
| MCM21 | Protein involved in minichromosome maintenance | 44 | 30 | G1 | − |
| MCM22 | Protein involved in minichromosome maintenance | 85 | 43 | G1 | − |
| NBP2* | Interacts with Nap1 involved in chromatin assembly | 65 | 46 | = | + |
| SFH1ΔN*/VPS65 | Subunit of a chromatin-remodeling complex | 15 | 58 | ≪ | + |
| SPT10 | Putative histone acetylase | 22 | 58 | < | + |
| VID21 | Component of a histone acetyltransferase complex | 37 | 20 | < | ++ |
| Vesicular traffic and ion homeostasis | |||||
| DID4* | Vps protein of the ESCRT-III complex | 15 | 36 | ≪ | ++ |
| PEP3* | Vacuolar peripheral membrane protein | 30 | 30 | ≪ | +++ |
| PEP7* | Protein that facilitates vesicle-mediated protein sorting | 17 | 37 | ≪ | ++ |
| PER1* | Vacuolar membrane protein | 14 | 20 | ≪ | ++ |
| PMR1* | High-affinity Ca2+/Mn2+ P-type ATPase | 54 | 48 | ≪ | ++ |
| PMR1ΔC*/HUR1 | Protein required for hydroxyurea resistance | 14 | 23 | ≪ | +++ |
| RCS1 | Transcription factor involved in iron utilization | 58 | 25 | G1 | ++ |
| RVS161* | Amphiphysin-like lipid raft protein | 11 | 26 | Sub-G1 | ++ |
| SNF7* | Subunit of the ESCRT-III complex | 20 | 47 | ≪ | ++ |
| SWA2* | Auxilin-like protein involved in vesicular transport | 16 | 34 | ≪ | ++ |
| VPS24* | Subunit of the ESCRT-III complex | 19 | 32 | ≪ | ++ |
| RNA catabolism | |||||
| NAM7* | Nonsense-mediated mRNA decay pathway | 13 | 16 | < | +++ |
| NMD2* | Nonsense-mediated mRNA decay pathway | 12 | 23 | ≪ | +++ |
| UPF3 | Nonsense-mediated mRNA decay pathway | 9 | 23 | ≪ | +++ |
| Morphogenesis | |||||
| ELM1* | Protein kinase that regulates cellular morphogenesis | 35 | 82 | ≪ | ++ |
| NAT3* | Subunit of the NatB N-terminal acetyltransferase | 18 | 26 | < | − |
| SAC7* | GTPase-activating protein for Rho1p | 11 | 24 | ≪ | ++ |
| VRP1ΔN/YLR338W | Proline-rich, actin-associated protein | 33 | 25 | Sub-G1 | ++ |
| Protein translation | |||||
| ASC1* | Protein involved in translation regulation | 23 | 29 | ≪ | ++ |
| ZUO1* | Cytosolic ribosome-associated chaperone | 22 | 31 | < | +++ |
| Nuclear transport | |||||
| NUP120 | Subunit of the nuclear pore complex | 24 | 70 | = | ++ |
| Unknown | |||||
| YEL033W | Hypothetical protein | 29 | 22 | G1 | +++ |
Genes with human homologs are indicated with asterisks. Short ORF deletions overlapping with longer ORFs on either the same or the opposite strand were classified as the longer ORF with an indication of the deleted regions. ΔN, ΔC, and ΔM indicate N-terminal, C-terminal, and middle part deletions, respectively.
Viability and cell growth were measured at 24 h after CdtB induction and are expressed as a percentage of values divided by the value of BY4743 wild-type cells with a CdtB plasmid. The data shown are the means of three independent experiments.
DNA content is expressed as follows: =, wild-type levels; ≪ and <, higher and slightly higher DNA content, respectively; G1, increased G1 cell population; Sub-G1, appearance of sub-G1 fraction.
+++, ++, and +, very large, large, and slightly large, respectively; −, no enlargement compared with vector-transformed strains. The BY4743 wild-type cell size was designated as +.
To examine the possibility that enhanced gene expression was caused by these gene deletions, the 61 selected CdtB-sensitive strains were transformed with a plasmid harboring galactose-inducible YopM, a Yersinia virulence factor (29). None of these mutants was sensitive to YopM (Fig. 3), however, indicating that enhanced gene expression was not the underlying cause of the CdtB-sensitive phenotype. Some of the mutations in these strains are partial deletions of longer open reading frames (ORFs) in the opposite strands and are classified as the longer ORF in each case. The protein function is listed on the basis of information in the Saccharomyces Genome Database (12), which shows that 32 of the deleted genes among these clones have human homologs (Table 1).
The genes corresponding to our 61 selected CdtB-sensitive deletion strains could be classified into seven groups and one unknown ORF. The major group comprises genes involved in DNA metabolism, including HR repair genes such as RAD50, RAD51, RAD55, RAD57, RAD59, and XRS2 (27). The absence of RAD52, RAD54, and MRE11 in this group, which are known as HR repair genes, was due to incorrect deletions. We therefore constructed diploid strains with homozygous deletions in these three genes and confirmed that all of them display CdtB sensitivity (data not shown). Our screen also identified all three known complexes for nonessential replication factor C (RFC), which associates with proliferating cell nuclear antigen (PCNA) to form the replication fork structure (34). These complexes comprise Rad24-RFC; the PCNA-like clamp consisting of Rad17, Ddc1, and Mec3; Ctf8/Dcc1-RFC; and Elg1-RFC. We also identified the S-phase checkpoint proteins Tof1 and Mrc1 (32); DNA damage mediator Rad9 (32), the Sae2 regulator (3); the Hpr5 (Srs2) helicase required for recovery from a stalled replication fork (59); another helicase, Sgs1 (23); and the Top3 DNA topoisomerase and Mus81-Mms4 endonuclease required for replication fork stability (4). In addition, proteins such as Bim1, Chl1, Ctf19, and Mcm21, which are required for efficient sister chromatid cohesion (36), were also identified. The additional genes that we identified could be assigned to vesicular traffic and ion homeostasis, RNA catabolism, protein translation, morphogenesis, and nuclear transport groups. Only one unknown ORF, YEL033W, which was previously identified in an ionizing radiation screen (6), was also identified in our current CdtB screen.
Cross-sensitivity of CdtB-sensitive strains to other genotoxic agents.
To compare the CdtB mode of action with the activity of other genotoxic agents, we examined the sensitivity of our selected CdtB-sensitive strains to HO endonuclease expression, UV irradiation, and hydroxyurea treatment (Fig. 3 and Table 2). As shown in Fig. 3, not all of the CdtB-sensitive strains are HO endonuclease sensitive, indicating that these particular clones respond differently to HO endonuclease-induced DSB. Strains that harbor deletions in genes involved in HR repair were found to be sensitive to all forms of induced DNA damage. In contrast, deletion strains for the RFC complexes (rad24Δ, rad17Δ, ddc1Δ, mec3Δ, ctf8Δ, dcc1Δ, and elg1Δ) are susceptible to different forms of DNA damage. Similarly, the rad9Δ strain is specifically sensitive to UV irradiation, whereas the mrc1ΔΜ strain is sensitive to HU. Table 2 also indicates strains that are sensitive to camptothecin, which is known to produce SSB in the presence of DNA topoisomerase I (22). Nearly all genes (22/25) categorized in DNA metabolism in Table 1 were listed as camptothecin sensitive (43), but in the other categories there are not many overlapped genes (8/36).
TABLE 2.
Cross-sensitivity of CdtB-sensitive strains to other DNA-damaging agents
| Sensitivitya | Gene/ORFb |
|---|---|
| UVs HUs HOs | ASC1, ASF1, CTF8, DCC1, HPR5, RAD50, RAD51, RAD55, RAD57, RTT107, RTT109, TOP3ΔC, VID21, XRS2, YEL033W |
| UVs HUs HOr | BIM1, MMS4, MUS81, NUP120, SGS1, TOF1, SFH1ΔN |
| UVs HUr HOs | NAT3 |
| UVs HUr HOr | DDC1, MEC3, RAD17, RAD24, RAD27, RAD59, RAD9, SPT10 |
| UVr HUs HOs | CTF19, MMS1, PER1, PMR1ΔC, RCS1, SAC7, RTT101, PEP3, UME6 |
| UVr HUs HOr | CHL1, DID4, ELM1, MRC1ΔM, NBP2, PEP7, PMR1, RVS161, SNF7, VPS24, VRP1ΔN |
| UVr HUr HOs | MCM21, MCM22, ZUO1 |
| UVr HUr HOr | ELG1, MCM16, NAM7, NMD2, SAE2, SWA2, UPF3 |
s, sensitivity; r, resistance; HO, HO endonuclease.
Entries in boldface indicate that the strain was camptothecin sensitive (43).
Phenotypic analyses of CdtB-sensitive strains.
To examine the phenotypic consequences of CdtB activity, we investigated the growth and survival frequency and the DNA content and cell size distributions (Table 1 and see Fig. S1 in the supplemental material) at 24 h after CdtB induction in our selected CdtB-sensitive yeast strains. In all of our CdtB-sensitive deletion strains, the cell growth and survival frequency were reduced (Table 1). In the DNA damage checkpoint deletion strains (ddc1Δ, mec3Δ, rad9Δ, rad17Δ, and rad24Δ), however, the cell growth was close to wild-type levels but the overall viability was significantly reduced. Consistent with this, these DNA damage checkpoint deletion strains show similar DNA content distributions to CdtB-expressing wild-type cells (Table 1 and see Fig. S1A in the supplemental material). In contrast, deletion strains for S-phase checkpoint (mrc1ΔM and tof1Δ), Elg1-RFC (elg1Δ), and chromosome maintenance proteins (bim1Δ, ctf19Δ, mcm16Δ, mcm21Δ, and mcm22Δ) have higher survival frequencies that are accompanied by the accumulation of cells in G1. Deletion strains for Nam7, Upf3, and Nmd2, which are three tightly interacting proteins required for nonsense-mediated RNA decay (20), accumulated a considerable number of enlarged cells. In addition, deletion strains for vesicular traffic, ion homeostasis, morphogenesis (except for NAT3), protein translation, and nuclear transport genes accumulate large cells (Table 1 and see Fig. S1B in the supplemental material).
The absence of known DNA repair gene deletions in the selected list of CdtB-sensitive strains.
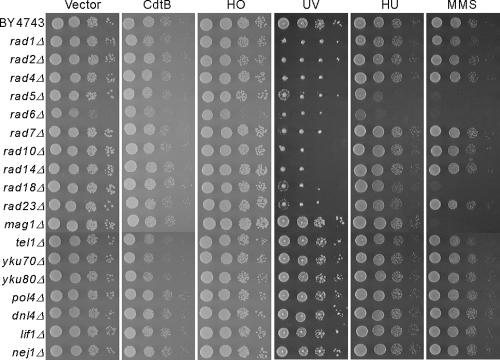
We speculated that if our genome-wide screen had identified all of the components required for the CdtB response, the lack of known DNA repair genes would implicate a role for unused repair mechanisms in this pathway. We confirmed again that 18 strains harboring deletions in genes responsible for nucleotide excision repair (NER) (RAD1, -2, -4, -7, -10, -14, and -23) (45), postreplication repair (PRR) (RAD5, 06, and -18) (9), base excision repair (BER) (MAG1) (37), and nonhomologous end joining (NHEJ) (TEL1, YKU70, YKU80, POL4, DNL4, LIF1, and NEJ1) (41) were not sensitive to CdtB except for the tel1Δ, yku70Δ, and yku80Δ deletion strains, which showed very weak growth defects (Fig. 4). All of the rad deletion strains examined are sensitive to UV, the rad5 strain is sensitive to HU, and the rad5, rad6, and rad18 strains are sensitive to MMS, as is mag1. This indicates that although these genes are necessary for the cellular response to selective DNA damage pathways, they do not function in the response to CdtB-induced DNA lesions.
FIG. 4.
Unused DNA repair mechanisms operate during the response to CdtB-induced DNA damage. Deletion strains that had not been identified by the CdtB screen but have been associated with DNA repair mechanisms were examined for their sensitivity to CdtB, HO endonuclease, UV, HU, and MMS. The DNA repair mechanisms in these deletion strains were nucleotide excision (rad1Δ, rad2Δ, rad4Δ, rad7Δ, rad10Δ, rad14Δ, and rad23Δ), postreplication (rad5Δ, rad6Δ, and rad18Δ), base excision (mag1Δ), and nonhomologous end joining (tel1Δ, yku70Δ, yku80Δ, pol4Δ, dnl4Δ, lif1Δ, and nej1Δ).
DISCUSSION
STA.
In our present study, we transformed a CdtB expression plasmid into 4,706 nonessential diploid deletion strains and the resulting array was used as a comprehensive screen for genes required for the CdtB response (Fig. 2). The transformation method is simple and reliable: i.e., yeast cell cultures grown in a microplate can be directly mixed with the direct transformation solution (Wako Pure Chemicals) containing plasmid DNA (0.5 to 1 μg/μl) that has been prepared by an alkaline-sodium dodecyl sulfate method without removing E. coli RNA (7). The mixtures were then incubated at 42°C for 2 h (1) and spotted on selection plates. Transformation-incompetent deletion strains were not found among the 4,708 strains that we utilized in our screen, indicating that none of the nonessential genes are necessary for plasmid transformation. Uptake and maintenance of plasmids may thus be essential processes in yeast. The use of the systematic transformation array (STA) thus enabled us to perform a genome-wide functional screen and analyze the mechanisms underlying the CdtB response in 4,706 yeast deletion strains. As shown here, the STA apparently can be used for genome-wide functional analyses of other virulence factors which elicit phenotypes when expressed in yeast (2, 29, 33, 49, 54).
Genes required for the CdtB response.
Our genome-wide analysis of the response to CdtB identified genes involved in HR repair, the DNA damage checkpoint, S-phase checkpoint, Mus81-Mms4 endonuclease, DNA topoisomerase, and DNA helicases (Table 1), but not for the other DNA repair mechanisms such as NER, PRR, and BER (Fig. 4). The contribution of the NHEJ repair pathway to the CdtB-induced DNA lesion is difficult to interpret because the NHEJ repair pathway is less important than HR in yeast (26). Although there may be a possible partial role of NHEJ for the repair of CdtB-induced DNA lesions (Fig. 4), CdtB-induced DNA lesions appear to be predominantly repaired by the HR repair mechanism, with the involvement of DNA checkpoint and DNA metabolism proteins in yeast. These genome-wide findings thus indicate that CdtB-induced DNA lesions are different from other lesions of this type, such as those induced by IR, MMS, or UV, which require RAD genes for NER and PRR (6, 8, 11, 17, 18). In addition, not all CdtB-sensitive strains are sensitive to HO endonuclease (Fig. 3 and Table 2), indicating that the DNA damage events caused by CdtB are not direct DSBs.
It was thought previously that HR repair utilizes sister chromatids in haploid cells but can utilize homologous chromosomes in addition to sister chromatids in diploid cells, and thus this response may be optimal in strains with greater ploidy. However, there are only a few reports that describe different sensitivities to DNA-damaging agents between haploid and diploid yeast cells (38, 48). HO endonuclease-induced DSB is preferably repaired by HR, but there is no phenotypic difference between haploid and diploid cells in this response (Fig. 1). HR repair of DSBs is known to prefer sister chromatids over homologous chromosomes in yeast (24). Therefore, it has been unclear whether the presence of homologous chromosomes in diploid organisms provides a significant advantage in terms of the DNA repair process. In this study, we show that CdtB-induced growth defects and an S-phase delay were severe in haploid but not in diploid yeast cells (Fig. 1). Identification of chromosome maintenance genes in this study also suggests that SSB repair requires proteins for chromosome behavior. If homologous chromosomes but not sister chromatids are used for repair substrate during S phase, sister chromatid cohesion should be released during repair with homologous chromosomes. Chromosome maintenance genes may be necessary for SSB repair in diploid cells due to the use of homologous chromosomes during S phase as is the case for the postreplicative DSB repair (53, 55, 62). Due to the strong CdtB effects in haploid yeast, we could not compare the CdtB sensitivities of haploid deletion strains even though CdtB expression can be reduced by the addition of small amounts of glucose together with galactose. However, evolutionally conserved genes in this study and the genes specifically required for CdtB response will provide clues toward our further understanding of DNA repair mechanisms for SSB and also the importance of homologous chromosomes in diploid cells.
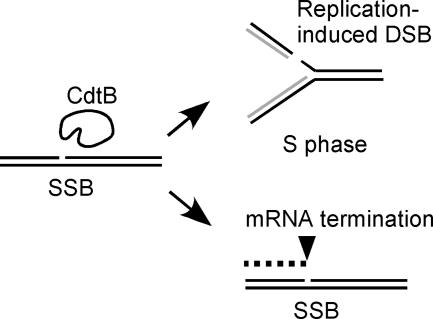
Our selected list of CdtB-sensitive deletion strains includes many components that function during S phase. In particular, three nonessential RFC complexes (34) were all found to be sensitive to CdtB, in which only Ctf8/Dcc1-RFC was sensitive to HO endonuclease. In addition, the individual deletion strain of Elg1-RFC did not show significant sensitivity to UV, HU, or HO endonuclease, as shown in our study, nor did it show sensitivity to MMS (5, 25). This suggests that Elg1 is required at least for SSB and replication-induced DSB, but not for other types of DNA damage. Identification of many components that function during S phase suggest that SSB is repaired during S phase, possibly due to the creation of DSBs by the replication of SSBs (Fig. 5). Moreover, this is not similar to DNA damage caused by the replication fork stall, because HU sensitivity is not fully overlapped among the CdtB-sensitive strains (Table 2).
FIG. 5.
CdtB-induced DNA lesion. CdtB primarily shows SSB activity which produces a DSB when the nicked strand is replicated during the S phase of the cell cycle. Homologous recombination mechanism is exclusively required for the repair. The SSB also produces an aberrantly terminated mRNA molecule. The mRNA decay pathway is required for the response and associated with the cellular distension.
The DNA-damage checkpoint deletion strains (ddc1Δ, mec3Δ, rad9Δ, rad17Δ, and rad24Δ) showed growth close to wild-type levels after the CdtB induction, but the overall viability was significantly reduced (Table 1). Consistent with this, these DNA-damage checkpoint deletion strains show DNA content distributions similar to those of CdtB-expressing wild-type cells (Table 1 and see Fig. S1A in the supplemental material). Growth and cell cycle progression in the presence of CdtB-induced DNA lesions may cause lethal damage in the DNA damage checkpoint deletion strains. In contrast, the strains with deletions for S-phase checkpoint (mrc1ΔM and tof1Δ), Elg1-RFC (elg1Δ), and chromosome maintenance proteins (bim1Δ, ctf19Δ, mcm16Δ, mcm21Δ, and mcm22Δ) have higher viability that is accompanied by the accumulation of cells in G1. We could not explain the accumulation of G1 cells in these strains, but the G1 phase may not be susceptible to the CdtB-induced DNA lesion.
Features of CdtB-induced DNA lesions.
As shown in the lists in Tables 1 and 2, CdtB analysis in yeast demonstrated specific features for DNA repair machineries. If CdtB creates an SSB in vivo, it produces a DSB during the S phase (Fig. 5). As expected, many S-phase repair machineries including all nonessential RFC components were identified in this study. CdtB-sensitive genes categorized in DNA metabolism are well overlapped with the genes identified in the camptothecin-sensitive screen (43). However, there are many nonoverlapping genes in the categories of vesicular traffic, RNA catabolism, and protein translation. Camptothecin is thought to produce SSB mainly during the S phase in the presence of DNA topoisomerase I; thus, the nonoverlapping genes are thought to be required for the problems that occurred during the other cell cycle phases. Figure 5 illustrates another possible problem caused by CdtB (i.e., aberrantly terminated mRNA). The transcription of genomic regions harboring SSBs will produce incomplete short mRNAs due to termination at the nick points. This seems problematic during all phases of the cell cycle. Accordingly, we identified NAM7, NMD2, and UPF3 in our screen as CdtB-specific genes which encode closely interacting factors required for the nonsense-mediated mRNA decay pathway (20). Interestingly, the accumulation of small RNAs in response to CdtB has been observed in yeast (19). Therefore, our data suggest that the mRNA decay pathway plays a role in the degradation of accumulated aberrant RNAs caused by CdtB. It is also noteworthy that the cell size of the three deletion strains for the mRNA decay pathway became significantly larger in response to CdtB (Table 1 and see Fig. S1B in the supplemental material). Cellular distension, the phenotype from which the name of CdtB is derived, is possibly caused by the accumulation of incomplete mRNAs, which will produce truncated proteins that have to be degraded. Deletion strains for vesicular traffic and protein translation also showed accumulation of larger cells in response to CdtB, which may be required for the control of aberrant proteins.
There is a known mammalian SSB repair system that play a role in neurodegenerative diseases (10, 14). This SSB repair machinery involves poly(ADP) ribose, DNA polymerase β, and DNA ligase III, but these have not been found in microorganisms. In neuronal cells, which are nonproliferative and do not enter S phase, a long persistent G1 phase is the only phase of the cell cycle that is evident. Therefore, we speculate that neuronal cells, or cells that remain in G1, require special DNA repair machinery for SSBs, which may be necessary for reducing aberrant mRNAs and the resulting abnormal proteins.
CdtB pathogenesis.
Although many pathogenic bacteria appear to produce CDT (42, 44, 56), there is no clear association between the action of CDT and disease symptoms. Several genes associated with CdtB virulence have already been identified in mammalian cells (13, 16, 44, 56, 60); we have identified a lot of genes required for the CdtB response by our yeast genome-wide analysis. If CdtB-induced DNA lesions are repaired by HR repair machineries that act predominantly during S phase (31, 32, 34), CdtB seems to be a time bomb for proliferating cells such as T cells, becoming harmful when cells replicate their DNA. Since Actinobacillus CdtB has been known as an immunosuppressive factor capable of impairing human lymphocyte function (51), the sensitivity of lymphocytes to CdtB may be due to their capability for proliferation or HR repair. Many identified genes in this study are conserved from yeast to humans and thus must be involved in the pathogenesis in mammalian cells that occurs in response to CdtB.
In conclusion, the yeast genome can be effectively used for the analysis of bacterial virulence factors (58). Furthermore, STA provides geneticists and pathologists with a new tool for the analysis of all yeast and nonyeast genes, and the resulting genome-wide data have the potential to elucidate many of the salient features of gene function and of the associated pathways. In addition, CdtB, which creates a specific DNA lesion, probably an SSB, will become a new tool for the analysis of a novel DNA repair mechanism in diploidy, a common feature of eukaryotic organisms.
Supplementary Material
Acknowledgments
We thank Carol L. Pickett, Samuel I. Miller, and Ira Herskowitz for generously donating plasmids. The authors would also like to acknowledge the technical expertise of the DNA Core Facility of the Center for Gene Research, Yamaguchi University, for their valuable assistance during this study.
Editor: D. L. Burns
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akada, R., M. Kawahata, and Y. Nishizawa. 2000. Elevated temperature greatly improves transformation of fresh and frozen competent cells in yeast. BioTechniques 28:854-856. [PubMed] [Google Scholar]
- 2.Alto, N. M., F. Shao, C. S. Lazar, R. L. Brost, G. Chua, S. Mattoo, S. A. McMahon, P. Ghosh, T. R. Hughes, C. Boone, and J. E. Dixon. 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124:133-145. [DOI] [PubMed] [Google Scholar]
- 3.Baroni, E., V. Viscardi, H. Cartagena-Lirola, G. Lucchini, and M. P. Longhese. 2004. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24:4151-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone, and G. W. Brown. 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22:4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broomfield, S., T. Hryciw, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486:167-184. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott, K. W. 2004. DNA single-strand breaks and neurodegeneration. DNA Repair 3:875-882. [DOI] [PubMed] [Google Scholar]
- 11.Chang, M., M. Bellaoui, C. Boone, and G. W. Brown. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99:16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, K. R., S. Weng, R. Balakrishnan, M. C. Costanzo, K. Dolinski, S. S. Dwight, S. R. Engel, B. Feierbach, D. G. Fisk, J. E. Hirschman, E. L. Hong, L. Issel-Tarver, R. Nash, A. Sethuraman, B. Starr, C. L. Theesfeld, R. Andrada, G. Binkley, Q. Dong, C. Lane, M. Schroeder, D. Botstein, and J. M. Cherry. 2004. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32:D311-D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comayras, C., C. Tasca, S. Y. Pérès, B. Ducommun, E. Oswald, and J. De Rycke. 1997. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing cdc2 protein kinase dephosphorylation and activation. Infect. Immun. 65:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Khamisy, S. F., G. M. Saifi, M. Weinfeld, F. Johansson, T. Helleday, J. R. Lupski, and K. W. Caldecott. 2005. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434:108-113. [DOI] [PubMed] [Google Scholar]
- 15.Elwell, C., K. Chao, K. Patel, and L. Dreyfus. 2001. Escherichia coli CdtB mediates cytolethal distending toxin cell cycle arrest. Infect. Immun. 69:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisan, T., X. Cortes-Bratti, E. Chaves-Olarte, B. Stenerlow, and M. Thelestam. 2003. The Haemophilus ducreyi cytolethal distending toxin induces DNA double-strand breaks and promotes ATM-dependent activation of RhoA. Cell. Microbiol. 5:695-707. [DOI] [PubMed] [Google Scholar]
- 17.Game, J. C., G. W. Birrell, J. A. Brown, T. Shibata, C. Baccari, A. M. Chu, M. S. Williamson, and J. M. Brown. 2003. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 160:14-24. [DOI] [PubMed] [Google Scholar]
- 18.Hanway, D., J. K. Chin, G. Xia, G. Oshiro, E. A. Winzeler, and F. E. Romesberg. 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA 99:10605-10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassane, D. C., R. B. Lee, M. D. Mendenhall, and C. L. Pickett. 2001. Cytolethal distending toxin demonstrates genotoxic activity in a yeast model. Infect. Immun. 69:5752-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, F., A. H. Brown, and A. Jacobson. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herskowitz, I., and R. E. Jensen. 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194:132-146. [DOI] [PubMed] [Google Scholar]
- 22.Hsiang, Y., R. Hertzberg, S. Hecht, and L. F. Liu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 27:14873-14878. [PubMed] [Google Scholar]
- 23.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadyk, L. C., and L. H. Hartwell. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132:387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanellis, P., R. Agyei, and D. Durocher. 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13:1583-1595. [DOI] [PubMed] [Google Scholar]
- 26.Krejci, L., L. Chen, S. Van Komen, P. Sung, and A. Tomkinson. 2003. Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 74:159-201. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 28.Lara-Tejero, M., and J. E. Galan. 2002. Cytolethal distending toxin: limited damage as a strategy to modulate cellular functions. Trends Microbiol. 10:147-152. [DOI] [PubMed] [Google Scholar]
- 29.Lesser, C. F., and S. I. Miller. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20:1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, L., A. Sharipo, E. Chaves-Olarte, M. G. Masucci, V. Levitsky, M. Thelestam, and T. Frisan. 2002. The Haemophilus ducreyi cytolethal distending toxin activates sensors of DNA damage and repair complexes in proliferating and non-proliferating cells. Cell. Microbiol. 4:87-99. [DOI] [PubMed] [Google Scholar]
- 31.Lisby, M., A. Antunez De Mayolo, U. H. Mortensen, and R. Rothstein. 2003. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2:479-483. [PubMed] [Google Scholar]
- 32.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532:41-58. [DOI] [PubMed] [Google Scholar]
- 33.Mager, W. H., and J. Winderickx. 2005. Yeast as a model for medical and medicinal research. Trends Pharmacol. Sci. 26:265-273. [DOI] [PubMed] [Google Scholar]
- 34.Majka, J., and P. M. J. Burgers. 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 78:227-260. [DOI] [PubMed] [Google Scholar]
- 35.Mao, X., and J. M. Dirienzo. 2002. Functional studies of the recombinant subunits of a cytolethal distending holotoxin. Cell. Microbiol. 4:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas, T. Kwok, R. Newitt, R. Aebersold, C. Boone, G. W. Brown, and P. Hieter. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Memisoglu, A., and L. Samson. 2000. Base excision repair in yeast and mammals. Mutat. Res. 451:39-51. [DOI] [PubMed] [Google Scholar]
- 38.Mortimer, R. K. 1958. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 9:312-326. [PubMed] [Google Scholar]
- 39.Myung, K., S. Smith, and R. D. Kolodner. 2004. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:15980-15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nes̆ić, D., Y. Hsu, and C. E. Stebbins. 2004. Assembly and function of a bacterial genotoxin. Nature 429:429-433. [DOI] [PubMed] [Google Scholar]
- 41.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294:2552-2556. [DOI] [PubMed] [Google Scholar]
- 42.Oswald, E., J. P. Nougayrede, F. Taieb, and M. Sugai. 2005. Bacterial toxins that modulate host cell-cycle progression. Curr. Opin. Microbiol. 8:83-91. [DOI] [PubMed] [Google Scholar]
- 43.Parsons, A. B., R. L. Brost, H. Ding, Z. Li, C. Zhang, B. Sheikh, G. W. Brown, P. M. Kane, T. R. Hughes, and C. Boone. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22:62-69. [DOI] [PubMed] [Google Scholar]
- 44.Pickett, C. L., and C. A. Whitehouse. 1999. The cytolethal distending toxin family. Trends Microbiol. 7:292-297. [DOI] [PubMed] [Google Scholar]
- 45.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 46.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Rudin, N., and J. E. Haber. 1988. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 8:3918-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saeki, T., I. Machida, and S. Nakai. 1980. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 73:251-265. [DOI] [PubMed] [Google Scholar]
- 49.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scherens, B., and A. Goffeau. 2004. The uses of genome-wide yeast mutant collections. Genome Biol. 5:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shenker, B. J., T. McKay, S. Datar, M. Miller, R. Chowhan, and D. Demuth. 1999. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J. Immunol. 162:4773-4780. [PubMed] [Google Scholar]
- 52.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjögren, C., and K. Nasmyth. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991-995. [DOI] [PubMed] [Google Scholar]
- 54.Skrzypek, E., T. Myers-Morales, S. W. Whiteheart, and S. C. Straley. 2003. Application of a Saccharomyces cerevisiae model to study requirements for trafficking of Yersinia pestis YopM in eucaryotic cells. Infect. Immun. 71:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ström, L., H. B. Lindroos, K. Shirahige, and C. Sjögren. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16:1003-1015. [DOI] [PubMed] [Google Scholar]
- 56.Thelestam, M., and T. Frisan. 2004. Cytolethal distending toxins. Rev. Physiol. Biochem. Pharmacol. 152:111-133. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 58.Valdivia, R. H. 2004. Modeling the function of bacterial virulence factors in Saccharomyces cerevisiae. Eukaryot. Cell 3:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaze, M. B., A. Pellicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 60.Whitehouse, C. A., P. B. Balbo, E. C. Pesci, D. L. Cottle, P. M. Mirabito, and C. L. Pickett. 1998. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect. Immun. 66:1934-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El-Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts. P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 62.Xu, H., C. Boone, and H. L. Klein. 2004. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol. 24:7082-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.