Abstract
Systemic pathogens have developed numerous strategies for evading the defenses of the host, permitting dissemination and multiplication in various tissues. One means of survival in the host, particularly in the bloodstream, has been attributed to the ability to avoid phagocytosis via capsular polysaccharide. To further define the virulence capacity of Streptococcus iniae, a zoonotic pathogen with the ability to cause severe systemic disease in both fish and humans, we performed an analysis of the capsule locus. The initial analysis included cloning and sequencing of the capsule synthesis operon, which revealed an approximately 21-kb region that is highly homologous to capsule operons of other streptococci. A genetic comparison of S. iniae virulent strain 9117 and commensal strain 9066 revealed that the commensal strain does not have the central region of the capsule operon composed of several important capsule synthesis genes. Four 9117 insertion or deletion mutants with mutations in the beginning, middle, or end of the capsule locus were analyzed to determine their capsule production and virulence. Virulence profiles were analyzed for each mutant using three separate criteria, which demonstrated the attenuation of each mutant in several tissue environments. These analyses also provided insight into the different responses of the host to each mutant strain compared to a wild-type infection. Our results demonstrate that capsule is not required for all host environments, while excess capsule is also not optimal, suggesting that for an “ideal” systemic infection, capsule production is most likely regulated while the bacterium is in different environments of the host.
Survival of systemic pathogens in vivo depends on their ability to rapidly adapt to constantly changing environments while disseminating from tissue to the bloodstream and then to other sites or organs throughout the body. Even the organisms that remain and replicate at the initial site of infection must possess potent strategies for defense against the artillery of the host innate immune response that is trafficked to the site of infection. Exposure to diverse environments and situations requires a systemic pathogen to have a large repertoire of survival mechanisms and virulence factors, some of which may be required in certain niches but are dispensable in other environments.
In an effort to further define the virulence mechanisms used by systemic pathogens, we recently developed an infectious disease model using a prototypic streptococcal pathogen, Streptococcus iniae, and a natural host, the zebrafish (Danio rerio), in order to analyze the mechanisms of systemic infection in a natural host-pathogen system (20). S. iniae is an important pathogen in both marine animals and humans, causing systemic infections in these hosts. The symptoms of infection are very similar to the symptoms of infections caused by human-specific streptococcal pathogens, such as Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus pneumoniae, with dissemination to multiple organs, including the heart, liver, spleen, and brain (8, 28). A phylogenetic tree of 16S rRNA sequences of streptococcal species revealed that there is a close genetic relationship between S. iniae and S. agalactiae (35). Therefore, not surprisingly, multiple genes recently identified in a large-scale virulence screen of S. iniae in the zebrafish (17) have homologs that encode virulence factors found in S. agalactiae (32). The virulence of S. iniae has also been analyzed in murine models of systemic infection (9, 10, 18), verifying that the clinical pathologies that we observe in our zebrafish system are identical to the clinical pathologies in a mammalian model.
One significant virulence factor found in nearly all systemic pathogens is polysaccharide capsule, although the specific molecular components that make up the capsule can differ from organism to organism. Experimental analyses of multiple pathogens have shown that the principal role of capsule is to protect the organism from phagocytosis, one of the primary safeguards of the innate immune system for clearance and elimination of invading pathogens. The ability of systemic pathogens to survive in the bloodstream has been credited to one of several mechanisms of phagocytosis inhibition. The sialylated polysaccharide capsule of S. agalactiae blocks phagocytosis by inhibiting the binding of the activated complement factor C3b to the surface of the pathogen, preventing the activation of the alternative complement pathway (16), thereby interfering with neutrophil opsonophagocytic killing mechanisms. Furthermore, the amount of capsule expressed on the surface can also affect the activation of this pathway, and decreased capsule synthesis is correlated with increased C3b deposition (16).
The diversity of environments encountered by a systemic pathogen requires both versatility of mechanisms and an ability to adapt rapidly to changing situations. The capacity to regulate capsule production may be very important for the survival of systemic pathogens in different host environments. One of the first steps in colonization by S. pneumoniae occurs in the nasopharynx, and several studies have shown that capsule is not required for this initial step (15, 19). Adamou et al. investigated the adherence of S. pneumoniae to bronchoepithielial cells and found that an unencapsulated mutant bound to a much greater extent than the encapsulated parent strain bound, and they proposed that higher levels of adherence by S. pneumoniae may correlate with enhanced invasive capacity (1). Recently, Hammerschmidt et al. elegantly illustrated that there was a reduction in S. pneumoniae capsule on contact with epithelial cells by scanning electron microscopy and transmission electron microscopy, showing that there was specific modulation of the capsule through interaction with a host environment; however, the mechanism of this regulation was not reported (12).
Although capsular polysaccharide has been shown to be an important virulence factor for many streptococcal pathogens (14), little is currently known about the capsular polysaccharide of S. iniae or about the regulation of capsule expression in streptococcal pathogens. Recent reports have confirmed that S. iniae, like other streptococcal pathogens, requires polysaccharide synthesis for a successful infection (5, 17).
Here we describe the sequence and annotation of the polysaccharide capsule synthesis operon of the systemic pathogen S. iniae 9117 (GenBank accession no. AY904444). The virulence profiles of S. iniae 9117 mutants with mutations in the beginning, middle, or end of the capsule locus were analyzed, which revealed multiple phenotypes and diverse susceptibilities to host clearance. The genetic homology of the S. iniae capsule operon to other streptococcal operons demonstrated that capsule synthesis genes and their organization are conserved between species and that certain key elements are necessary for successful systemic disease.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Plasmids were maintained in Escherichia coli Epi300 cells (Epicenter) or E. coli TOP10 cells (Invitrogen). Luria-Bertani medium was used to culture E. coli strains. When necessary, antibiotics were added to Luria-Bertani medium at the following concentrations: erythromycin, 750 μg/ml; and chloramphenicol, 12.5 μg/ml for E. coli strains. Solid media were made by supplementing liquid media with 1.4% agar (BBL). E. coli cultures were incubated at 37°C with shaking unless indicated otherwise. The streptococcal strains used were S. iniae 9117, a human clinical isolate from the blood of a patient with cellulitis, and S. iniae 9066, a commensal fish strain obtained from a swab from a healthy fish (9). S. iniae strains were cultured in Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (BBL) and 2% proteose peptone (BBL) (TP) in airtight conical tubes without agitation at 37°C. S. iniae strains cultured on solid media were incubated in GasPak jars (BBL) with GasPak anaerobic system envelopes (BBL).
Cosmid cloning and sequencing.
Cosmid cloning of large portions of the S. iniae chromosome was performed using a CopyControl fosmid library production kit (Epicenter, Madison, WI) according to the manufacturer's instructions. Genomic DNA from S. iniae capsular mutant strain G1-6 (17) was used for cosmid cloning. This strain has a transposon carrying erythromycin resistance inserted into an open reading frame within the capsule operon, which allowed selection of cosmid clones that contained portions of the capsule operon. Multiple cosmids were sequenced to allow 2× coverage of the entire capsule operon, and the sequence was compiled using the Genetics Computer Group program.
Sequencing of the capsular operon was performed using BigDye Terminator chemistry at the sequencing core at Wayne State University. The analysis of the resultant sequences was performed using the BLAST software (www.ncbi.nlm.nih.gov) (2).
Insertional inactivation of cpsA.
An insertional mutation in the cpsA gene was created using primers 5′cpsA insert-EcoRI#2 (CCG GAA TTC GAA GTA AAC GAA AGA GTT CTT CG) and 3′cpsA insert-SmaI#2 (TCC CCC GGG GTT ATA AAC GCC TGC CTT AGT GAC) to amplify a 685-bp fragment of the cpsA gene. This fragment was then digested with EcoRI and SmaI and inserted into the corresponding EcoRI and SmaI sites of pUC19-ERM (the pUC19 parent vector with the ampicillin resistance gene replaced with an erythromycin resistance gene) to create pUC19-ERM-cpsA. Plasmid pUC19-ERM-cpsA was transformed as described previously (17) into S. iniae wild-type strain 9117 following propagation and recovery from E. coli TOP10 cells (Invitrogen).
Construction of cpsY deletion mutant.
An in-frame deletion in the cpsY gene was constructed with primers CpsY-IFD-5′ (GC ACT TTA TTA TTT AGC AGC ACA ACA TGG) and CpsY-IFD-3′ (GGT TGA AAT CGG ACC ATA AGT ATC AAT ACC), which amplified a 2,950-bp fragment containing the entire cpsY open reading frame with an additional ∼1,000 bp on either side of the cpsY gene. The fragment was inserted into a standard vector, and an 876-bp in-frame deletion was created by performing inverse PCR with primers CpsY-IFD-SmaI-3′-FWD (TCC CCC GGG GGA GAT AAT GTA ATG TAA TTG TTG TAT TCT) and CpsY-IFD-SmaI-5′-REV (TCC CCC GGG GGA CTC TTA AAA GAG GTC AAA TTG ACA CAA AAA AAG). The resulting mutant allele (cpsYΔ11-293) was inserted between the PstI and BamHI sites of the shuttle vector pJRS233 (22) to create pΔcpsY, which was used to replace the wild-type cspY allele in 9117 by a previously described method (26). The chromosomal structure of the resulting mutant, ΔCpsY, was confirmed by PCR.
Comparative PCR with 9117 and 9066.
PCR were performed with genomic DNA of S. iniae strains 9117 and 9066 using the following cycling parameters: 95°C for 5 min, followed by 20 cycles of 95°C for 30 s, 60°C for 30 s with a 0.5°C/cycle decrease in the annealing temperature, and 72°C for 1 min and then an additional 10 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 1 min. PCR products were loaded on a 0.8% agarose gel and visualized by ethidium bromide staining. Gene-specific primers used in this experiment are listed in Table 1.
TABLE 1.
Primers for S. iniae 9117 capsule genes
| Primer | Sequence (5′-3′) |
|---|---|
| deoD 5′ | CCT GGT GAG ATT GCT GAT AAA ATT CTT CTT CC |
| deoD 3′ | GGT TTC AAG GCC AAC TTG CAT CAT GTC AGT GAA GG |
| hk 5′ | CCT AAA TGA GAG GAT TGA AGA AGA GC |
| hk 3′ | GCT TGT GAC GAG AAA TGT GTG GTA CG |
| cpsY 5′ | TCC CCC CGG GTT ACG CTA GAC AAA TCA TTG AAC AAA CC |
| cpsY 3′ | AAC TGC AGT TAA AAA GTG ACG CAC GGT CAC TAA CG |
| cpsA 5′ | GAG TTA AAA CTT GAA GAA GTT TGC TTC TTA TCC |
| cpsA 3′ | CCT AGC ATA TCA ATT AAG TTC AAA AAG G |
| cpsB 5′ | CGA ACC ATT GTT TCA ACA TCT CAC AGA CG |
| cpsB 3′ | CGT CAC ATG TTG ATA AGC TTC TTG C |
| cpsC 5′ | GCT ACT AGC TGG TTT TTT AGC CTT GGT GG |
| cpsC 3′ | GGA TAA CTG CAA ATA TAC TTA AAA CAG C |
| cpsD 5′ | CCT GGT GAA GGA AAG TCA ACC ACA TCG |
| cpsD 3′ | GCA CAA AAC GAC GTT TAA TAG AAC CTG C |
| cpsE 5′ | CCA TCT TTC AGC CTA TGC TTT TTT AAT GG |
| cpsE 3′ | GCT TAC TCC TGT TTA GCG TCA TTT ATT AAG G |
| cpsF 5′ | GCA TGT ATC CTT ATA TTA AAC GAC TAT TAG C |
| cpsF 3′ | CCT TAT TTT GCT CTG GTC CAC CCT CTA AGA CG |
| cpsG 5′ | GGT GCA AAT TCT TAT ATA GGA ACT TCC |
| cpsG 3′ | CCG ATA TTC GTA TTG ACT CTT CAA ATC C |
| cpsH 5′ | GGA GCA GAC GTT ATT TCT TTG ATG AAC G |
| cpsH 3′ | CCT GTT TAA TCC CTA AAG TAT CGT ATA ACC |
| cpsI 5′ | GCT TAC AAA AAT TTT TTT CAA AAA AGC TCG |
| cpsI 3′ | GCT GGT GAT CCT ATT GCA ATT GAA TTT GCT GG |
| cpsJ 5′ | GGT GCA AAT GGA TAC TTA GGA AAA GGT GTC G |
| cpsJ 3′ | GCT TTT CAA CTT TTC TGA ATT CCC CCA TAC TGC |
| cpsK 5′ | GAT AAC TGT TTG TAT GGC AAC TTT TAA TGG |
| cpsK 3′ | CCT CTT GTT GTT TTT TAC AGT TAG TCC |
| orf276 5′ | CGT GCA GTT TAT ATT AGG CAT ATG ATT CC |
| orf276 3′ | GGA ACA CAT TCT TCA ACT CTA GC |
| orf193 5′ | GGT CAA TAG TAT TAA GTT GTA TAT TAC TAC |
| orf193 3′ | CCT AAA AAA ACA ATC ATC ATC ATA ATG CC |
| orf151 5′ | GGA ATT TTC TCT AAC AAA TTT TCA ATT C |
| orf151 3′ | GGA TTA ATT GAA TCT AAA TAA AAC G |
| cpsL 5′ | CGT TTT ATT TAG ATT CAA TTA ATC C |
| cpsL 3′ | CGA ACA TAT TTG TAA AAT TGT ATT GGT GG |
| orf-1 5′ | GCT ACT CAA AAG AAT TTA AAG AAA CC |
| orf-1 3′ | GCT AAA GTT TGT ATG GTT TGA GCC ATA TCC |
| orf-2 5′ | GGA TAT GGC TCA AAC CAT ACA AAC TTT AGC |
| orf-2 3′ | CCT TTT CAA ATT GAT TAG GTG TAA GAT ACC |
| cpsM 5′ | CGA CTC CAT ATC TAT CAA GGG TTT TAT CG |
| cpsM 3′ | CAG CAA CTG CTA TTT TTT TCA TGG TAT CC |
| cpsN 5′ | CCA TGA AAA AAA TAG CAG TTG CTG GTA CG |
| cpsN 3′ | CCA AGA CAC TCG GAT TAA TCC CGG CCG AAA AGG |
| orf183 5′ | GGT CGA AAG AAC TCC ACT ATC TTA GG |
| orf183 3′ | GCA GTT GTT AAT GAC TGT ATT AGA CTT CC |
Southern hybridization analysis of S. iniae strains.
Southern hybridization analysis was carried out using KpnI-digested genomic DNA from S. iniae virulent strain 9117 and S. iniae commensal strain 9066 (31). Biotinylated probes were created for each gene tested using a Detector PCR DNA biotinylation kit (KPL). Southern analysis was performed using a Detector AP chemiluminescent blotting kit (KPL).
Competitive assays.
Competitive assays were performed as described previously (17). Briefly, S. iniae strain 9117 (wild type) and mutant strains were cultured separately in TP at 37°C overnight. Cells from these cultures were diluted 1:100 and grown to the mid-log phase (optical density at 600 nm, 0.225). The concentrations of all cultures were then adjusted to 107 CFU/ml. Cells were mixed at a 1:1 ratio, pelleted, and then resuspended to obtain a final concentration of 2 × 107 CFU/ml. Zebrafish were infected intramuscularly with 10 μl of this mixture, resulting in an infectious dose of 2 × 105 CFU. Cultures were serially diluted and plated onto plates with TP and TP supplemented with 2 μg/ml erythromycin to determine the input ratio of mutant to wild type.
After 24 h, hearts and brains were aseptically recovered from euthanized fish and homogenized, and the homogenates were plated onto TP and TP-Erm2 plates to determine the output ratio of mutant to wild type. The competitive index (CI) was calculated by dividing the output ratio (mutant/wild type) by the input ratio (mutant/wild type). Eight inoculated fish were used in all competitive assays. The CI of the wild type was 1. All strains with CI less than 1 were considered attenuated.
Zebrafish survival assays.
S. iniae strain 9117 (wild type) and mutant strains were cultured separately in TP at 37°C overnight. Cultures were diluted 1:100 and grown to the mid-log phase (optical density at 600 nm, 0.225). The final concentrations of all cultures were then adjusted to 1 × 107 CFU/ml. Zebrafish were infected intramuscularly with 10 μl of an individual strain, resulting in an infectious dose of 1 × 105 CFU. A minimum of 12 fish were infected with each strain. The time of survival of zebrafish was recorded over a 4-day period, after which all remaining fish were euthanized.
Gradients for measuring buoyant densities.
Linear hypotonic gradients were prepared using a previously described method, with a few modifications (17). First, a 1.13-g/ml Percoll (Sigma) solution was diluted according to the manufacturer's instructions using double-distilled sterile water to construct gradients that covered a density range from 1.01 to 1.13 g/ml. After incubation overnight, 108 CFU of each bacterial strain was resuspended in 100 μl of phosphate-buffered saline and gently added to the top of a gradient. Density marker beads (Amersham Biosciences) (vials 1 to 9) were combined and added to the gradient. The gradients were centrifuged at 3,220 × g for 4 h at 4°C in a swinging bucket rotor. Bacteria banded at the level of a gradient at which the densities of the gradient and the bacteria were equal.
After centrifugation the distances of the density marker bead and bacterial strain bands from the top of the gradient were determined. Buoyant densities for bacteria strains were calculated from a standard curve for the density marker beads.
Systemic dissemination of bacteria.
Bacterial strains were grown as described above for the zebrafish survival assays to a final concentration of 1 × 107 CFU/ml. Zebrafish were infected intramuscularly with 10 μl of a strain, resulting in an infectious dose of 1 × 105 CFU. At specific times postinfection, spleens, hearts, and brains were aseptically recovered from euthanized fish and homogenized. Homogenates were diluted and plated on TP plates to determine the number of CFU per organ. For each strain, a minimum of eight organs per time were analyzed.
Statistical analysis.
The statistical significance of differences between paired samples was determined by a two-tailed paired t test using the StatView statistical analysis software (SAS Institute Inc., Cary, NC).
RESULTS
Sequencing and genomic organization of the capsular operon of S. iniae.
The entire polysaccharide synthesis operon of S. iniae 9117 was sequenced and compiled (GenBank accession no. AY904444). Similar to capsule operons in other pathogenic bacteria that have been analyzed, the polysaccharide synthesis operon of S. iniae is quite large, with more than 20,000 nucleotides in 21 open reading frames (Fig. 1A). The open reading frames have various degrees of homology to capsule synthesis genes of several other gram-positive bacteria, including Streptococcus suis, S. agalactiae, S. pyogenes, Streptococcus thermophilus, Lactococcus lactis, and Bacillus halodurans (Table 2). We assigned putative names to the open reading frames based on homology to genes found in other gram-positive polysaccharide synthesis operons.
FIG. 1.
Streptococcal capsule operon maps. (A) Organization and identification of genes in the polysaccharide synthesis operon of the 9117 strain of S. iniae (accession no. AY904444). Bent arrows indicate a region containing promoter sequences, and hairpin symbols indicate putative terminator sequences. Genes shown in black are also found in the capsule operon of S. iniae commensal strain 9066, while genes shown in gray are missing. The vertical arrows indicate the sites of insertion of mutations referred to in the text. (B) Organization and identification of genes in the capsule operon of S. iniae commensal strain 9066. The cross-hatched box at the beginning of cpsM indicates the missing 154 bp of the beginning of this gene compared to the 9117 gene. The bar above the sequence indicates the region that was subjected to sequencing. (C) Genes in the capsule operon of S. agalactiae NEM316, a serovar III strain (accession no. AL732656).
TABLE 2.
Homology of capsule genes
| Gene product | Homologa | % Identity (% similarity)/spanb | Predicted function |
|---|---|---|---|
| CpsY | CpsY, S. agalactiae | 78 (89)/305 | Regulator |
| Putative transcriptional regulator, S. pyogenes | 84 (93)/298 | Regulator | |
| CpsA | CpsA, S. agalactiae | 57 (75)/456 | Chain length determination |
| EpsA, S. thermophilus | 56 (76)/458 | ||
| CpsB | CpsB, S. agalactiae | 67 (83)/243 | Chain length determination |
| EpsB, S. thermophilus | 70 (83)/243 | ||
| CpsC | EpsC, S. thermophilus | 56 (79)/229 | Chain length determination |
| CpsC, S. pneumoniae | 54 (72)/227 | ||
| CpsD | CpsD, S. agalactiae | 63 (82)/212 | Chain length determination and export |
| EpsD, S. thermophilus | 63 (80)/209 | ||
| CpsE | Cps9E, S. suis | 61 (73)/600 | Glycosyl transferase |
| CpsF | Cps9F, S. suis | 69 (80)/196 | Glycosyl transferase |
| CpsG | UDP glucose-4-epimerase, B. halodurans | 48 (65)/281 | UDP glucose epimerase |
| CpsH | Eps11J, S. thermophilus | 32 (54)/337 | Unknown |
| CpsI | O-Acetyl-transferase, Aeromonas hydrophila | 52 (70)/173 | Acetyl transferase |
| CpsJ | Eps11I, S. thermophilus | 59 (73)/271 | UDP glucose epimerase |
| CpsK | Eps11K, S. thermophilus | 50 (69)/208 | Glycosyl/rhamnosyl transferase |
| ORF 276 | Hypothetical protein, Bacillus thetaiotamicron | 24 (44)/234 | Unknown |
| ORF 193 | Unknown, no domain homology | ||
| ORF 151 | Unknown, no domain homology | ||
| CpsL | Glycosyl transferase, Clostridium acetylbutylicum | 22 (42)/278 | Glycosyl transferase |
| ORF-1 (IS981) | Transposase of IS981, L. lactis | 100 (100)/86 | |
| ORF-2 (IS981) | Transposase of IS981, L. lactis | 92 (92)/279 | |
| Hypothetical protein, S. thermophilus | 91 (92)/284 | Unknown | |
| CpsM | EpsU, S. thermophilus | 33 (56)/435 | Polysaccharide synthesis and export |
| CpsN | Eps11H, S. thermophilus | 74 (86)/409 | Putative UDP glucose dehydrogenase |
| ORF 183 | Conserved hypothetical protein, S. pyogenes | 79 (89)/174 | Unknown |
Based on BLASTX results. Only the matches with the highest levels of homology are listed.
Amino acid identity and similarity for the span indicated.
The transcriptional regulatory region of the group B streptococcus (GBS) capsule operon is reported to be between the divergently transcribed cpsY gene (also called mtaR [30]) and the cpsA gene (13) (Fig. 1C). The start site of transcription in this intergenic region has been mapped using primer extension analysis (6). Analysis of the intergenic region between the cpsY and cpsA genes in S. iniae revealed potential −10 and −35 sites for transcription of both cpsY and cpsA that overlap and are similar to the −10 and −35 sites that have been proposed for the intergenic region of the GBS capsule operon (data not shown) (13). The genes encoding CpsY and CpsA are also highly homologous to the genes found in S. agalactiae, and the levels of identity to MtaR (CpsY) and CpsA are 78% and 65%, respectively, at the amino acid level.
Comparison of the capsule operons of a virulent strain and a commensal strain.
The initial analysis of S. iniae strains demonstrated that isolates capable of causing disease in both fish and humans were virtually identical, as shown by pulsed-field gel electrophoresis, while strains isolated from nondiseased fish, a condition suggesting a commensal relationship, were genetically diverse (34). A previous study designed to identify virulence genes of S. iniae virulent strain 9117 using zebrafish as an infectious disease model showed that 10 of the attenuated mutants had insertions in putative capsule synthesis genes (17). When S. iniae virulent strain 9117 and S. iniae commensal strain 9066 were compared, phenotypic differences were observed during growth in broth culture (17). While virulent strain 9117 is buoyant in broth culture and forms chains approximately 6 to 10 cocci long, commensal strain 9066 aggregates out of broth culture and forms chains that are approximately fourfold longer than 9117 chains. Previous reports have shown that capsule production in streptococci is inversely related to buoyant density (29). S. iniae 9117 mutants with insertions in capsule synthesis genes, which proved to be attenuated for virulence, mimicked the phenotype displayed by commensal strain 9066 when they were grown in broth culture and applied to a density gradient (17) (Fig. 2). We therefore hypothesized that the capsule operon of strain 9066 may have different genetic components than the capsule operon of strain 9117, which may play some role in the ability to cause disease. To test this hypothesis, we performed PCR with genomic DNA from 9117 and 9066 using primers specific for each gene identified in the capsule operon of 9117, followed by Southern hybridization (data not shown).
FIG. 2.
Buoyant density profiles of S. iniae wild-type strains 9117 and 9066 and four 9117 capsular mutants in a hypotonic Percoll gradient with endpoints of 1.01 to 1.13 g/ml. Individual strains were added to the top of a gradient and centrifuged, facilitating sedimentation to a specific density which correlated with the amount of capsule expressed. Strains with greater encapsulation have low buoyant densities, while strains with little or no encapsulation have high buoyant densities. The data are the results of one of three Percoll gradient experiments performed. WT, wild-type strain 9117.
Based on the PCR results, S. iniae commensal strain 9066 appears to be missing genes cpsF through cpsL, as well as orf276, orf193, and orf151. Genes on either side of the missing genomic segments are still present in the 9066 capsule operon as primers specific for 9117 capsule genes were able to produce products during PCR amplification (data not shown). However, since PCR amplification results cannot be used as conclusive results for determining missing genetic regions due to subtle differences in sequence that may affect primer annealing and subsequent amplification, we performed Southern hybridization using biotinylated probes created from strain 9117 chromosomal DNA for the genes missing as determined by the 9066 PCR to confirm the absence of these genes from the 9066 chromosome. In addition, PCR amplification revealed the presence of the IS981 locus in 9066 DNA, but it did not confirm that the IS981 genes are found in the capsule operon of 9066 as insertion sequences can be found in multiple regions throughout the genome (23, 24). Southern hybridization analysis confirmed the PCR results by demonstrating the absence of the cpsF, cpsG, cpsH, cpsI, cpsJ, cpsK, and cpsL genes, as well as orf276 and orf151, in strain 9066 (Fig. 1B).
All of the genes shown to be missing in the 9066 capsule operon except orf193 are arranged contiguously and have a putative function in polymerization of the capsular polysaccharide chain (Fig. 1). Notably, 8 of the 10 attenuated capsule mutants previously isolated from the S. iniae 9117 strain in a large-scale signature-tagged mutagenesis screen (17) are found in this region of the capsule operon, giving credence to the hypothesis that this region may play some role in the commensal and virulent phenotypes exhibited by S. iniae strains 9066 and 9117, respectively.
Thus, the results revealed that several genes that are present in the 9117 capsule operon are not present in the 9066 genome; however, they did not reveal if additional genes are present in the 9066 capsule operon. The data also did not confirm if the IS981 locus, which was present as determined by PCR, is actually in the capsule operon of 9066, as a probe for IS981 hybridized to multiple segments of 9066 DNA, as determined by Southern analysis (data not shown). To further define the elements that make up the 9066 capsule operon region, a segment of DNA was amplified using primers that hybridized to the 3′ region of the 9117 cpsE gene going toward the end of the operon and to the 5′ region of the 9117 cpsN gene going in reverse toward the beginning of the operon. These genes were shown to be present in 9066 by the PCR analysis. An approximately 2-kb fragment was amplified, cloned into a vector, and subjected to sequencing. Sequence analysis revealed that the sequence of the 3′ end of the cpsE gene and the 5′ end of the cpsN gene were identical to the sequences found in 9117, as well as the majority of the cpsM gene. However, the first 154 nucleotides encoding CpsM were absent in 9066, leaving in question whether CpsM is actually expressed or functional. Between the end of the cpsE gene and the truncated cpsM gene is 672 bp with approximately 99% homology to a partial region of orf193 of 9117, although multiple changes in sequence result in no putative open reading frames in this region of the 9066 genome (Fig. 1B). As suspected, the IS981 locus was not present in this segment of the 9066 capsule operon, suggesting that the presence of multiple capsule genes in 9117 that are missing in the 9066 capsule locus may have been caused by an event mediated by the insertion element.
Virulence profiles of strains with mutations in the capsule locus.
To determine if the loss of a portion of the capsule locus in 9066 may play some role in the commensal phenotype observed in nature and aquaculture, we used our zebrafish infectious disease model to compare infections with S. iniae virulent strain 9117 and infections with commensal strain 9066. While only 8% of the zebrafish inoculated with 9117 were alive 4 days postinjection, 27% of the fish inoculated with the 9066 strain survived this long. Although the 9066 and 9117 strains of S. iniae have the same 16S rRNA sequence, previous work in which protein profiles of the two strains were examined showed that there are multiple differences between the strains (J. D. Miller and M. N. Neely, unpublished data). Furthermore, data from a previous study demonstrated that the 9066 commensal strain is genetically diverse from 9117 as determined by pulsed-field gel electrophoresis, suggesting that there may be multiple differences between the strains outside the capsule operon (34). Therefore, in an effort to analyze virulence with respect to capsule alone with all other factors being the same, we used isogenic mutants of the 9117 strain with differences only in the capsule locus. Mutations were constructed in the genes at the beginning of the S. iniae capsule operon, cpsY and cpsA, which are the two genes on either side of the putative capsule promoter region (Fig. 1A). Both the cell morphology and the virulence profiles of these mutants were examined and compared to the cell morphology and the virulence profiles of the wild-type strain and two previously isolated S. iniae capsule mutants, F12-1 and E6-9 (Fig. 1A). The long-chain phenotype and aggregation of bacterial cells in broth culture which were observed previously for S. iniae capsule mutants (17) were also observed for the cpsA insertion mutant and F12-1, while the cpsY deletion mutant and E6-9 grew like the wild-type strain (data not shown).
Previous work has demonstrated that the amount of capsule present is inversely related to the buoyant density and that the loss of capsule can be demonstrated using buoyant density centrifugation (17, 29). When organisms are added to the top of a linear hypotonic Percoll gradient, the strains with a low level of encapsulation sediment to a high-density region, while the strains with more capsule are found at lower-density regions. To confirm that the cpsA mutant produces less capsule than the wild type produces, a Percoll density gradient analysis was performed with the cpsY, cpsA, F12-1, and E6-9 mutants and the wild-type strain (Fig. 2). As shown previously (17), F12-1 has a high buoyant density, indicating a small amount of capsule, and E6-9 has a low buoyant density and slightly more capsule than the wild-type strain. As expected, the cpsA mutant was similar to the F12-1 mutant, with very little capsule compared to wild-type strain 9117. The cpsY mutant appeared to have only slightly less capsule than the wild-type strain. The 9066 commensal strain has less capsule than wild-type strain 9117 but more capsule than the F12-1 or cpsA mutant strains. However, it should be noted that the capsule production by each of the strains was measured after in vitro growth, and therefore, the results may not completely correlate with the amount of capsule in vivo.
Three different assays were used to examine the virulence profiles of the mutants with mutations in the capsule operon using a zebrafish infection model. Competitive assays, in which the mutant strain was inoculated at a 1:1 ratio along with the wild-type strain into zebrafish, were used to measure the ability of the strain to compete with the wild-type strain, as well as the ability to survive in the presence of the inflammation elicited by the wild-type strain. The results of these assays demonstrated that the two strains having the smallest amount of capsule as determined by the density gradient analysis, the cpsA and F12-1 mutants, exhibited the greatest attenuation, with >3-log attenuation in both the brain and the heart (Fig. 3). These results are in contrast to the results obtained for the cpsY and E6-9 mutants. The cpsY mutant exhibited almost no attenuation in the heart, suggesting that it competed equally with the wild-type strain in this environment, but it was 1.5 log less virulent than the wild-type strain in the brain (Fig. 3). The E6-9 mutant, which has an insertion in the very end of the capsule operon, exhibited approximately 0.5-log, but still significant, attenuation in both organs.
FIG. 3.
CI assays of capsule mutants. CIs were calculated by comparison with S. iniae wild-type strain 9117, as described in Materials and Methods. The CI is defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type). S. iniae wild-type strain 9117 has a CI of 1, and a CI less than 1 indicates a quantitative level of attenuation of virulence for a mutant compared to the wild type. CIs were determined individually for each mutant recovered from both hearts (A) and brains (B). Each triangle represents one fish, while the bar indicates the average for all fish for a mutant. The mean CI for each mutant in each organ is indicated, as is the P value for a comparison to the wild-type value (n = 8).
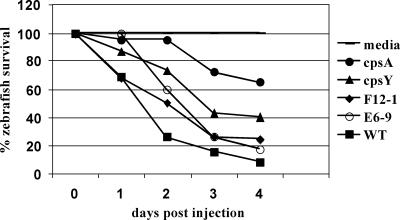
The second assay used to examine virulence was a comparison of the survival of zebrafish over time when the four mutant strains were inoculated individually (in the absence of the wild-type strain) over a 4-day period with the survival of zebrafish either inoculated with the wild-type strain or subjected to a mock infection (Fig. 4). In this assay, the cpsA mutant was still the most attenuated strain; 65% of the zebrafish were alive at day 4 after inoculation of this strain, while only 8% of the zebrafish survived for the same time after inoculation of the wild-type strain. Surprisingly, the F12-1 mutant, which showed high attenuation in the competitive assays in the presence of the wild-type strain and showed the same amount of capsule deficiency as the cpsA mutant, exhibited only about twofold attenuation at 2 days and very little attenuation at 4 days compared to the wild-type strain. These results, compared to the results of the competitive assay, suggest that the F12-1 mutant is much more susceptible to clearance due to host inflammation caused by the presence of the wild-type strain. However, a different scenario occurred with the cpsY deletion strain, which exhibited more attenuation than it exhibited in the competitive assays, with 40% of the fish surviving at day 4 (compare Fig. 3A and B to Fig. 4). The results of the two assays with the cpsY mutant suggest that at least in the heart, the wild-type strain can complement the cpsY mutant, since the attenuation was not significant when the wild-type strain was present in the competitive assays. Finally, the E6-9 mutant exhibited a virulence profile similar to the profile observed in the competitive assays, with only a small amount of attenuation.
FIG. 4.
Zebrafish survival assay. Zebrafish were infected with S. iniae wild-type strain 9117 or one of four capsule mutants separately as described in Materials and Methods and monitored for 4 days. The time of survival was graphed as a percentage of the total number of fish infected with a strain. A minimum of 12 zebrafish were infected with each strain. Zebrafish inoculated with sterile medium were used as a control. WT, wild type.
The third virulence assay performed was an analysis of the dissemination of each strain individually over time compared to the dissemination of the wild type. Dissemination to the heart, brain, and spleen was measured at 15, 30, and 60 min and 24 h after intramuscular injection of 1 × 105 CFU into the dorsal muscle. For these assays, we use the term dissemination to refer to the number of bacteria found in a particular organ at a specific time, although we could not distinguish actual dissemination as opposed to multiplication of the strain once it was in the organ. In addition, it must be noted that the bacteria passed through several host tissues to reach the organs, which could greatly affect the final number of CFU. The first observation of note is the large number of wild-type bacteria that were found to be disseminated systemically in the first 15 min after intramuscular injection; approximately 103 CFU was found in both the spleen and heart at this early time (Table 3). By 60 min the amount had quadrupled, and by 24 h there were ∼1 × 105 CFU of wild-type bacteria in the spleen alone resulting from the initial inoculation into the dorsal muscle (Table 3 and Fig. 5). The 24-h dissemination analysis can be compared to the competitive indices for the heart and brain since these indices were also analyzed at 24 h; the only differences were the presence of the wild-type strain in the competitive index analysis and, therefore, the host immune factors that may have been elicited by the presence of the wild-type strain.
TABLE 3.
S. iniae dissemination to organs
| Organ | Strain | No. of CFU at:
|
||
|---|---|---|---|---|
| 15 min postinjection | 30 min postinjection | 60 min postinjection | ||
| Spleen | Wild type | 791 ± 148 | 4,167 ± 642 | 4,097 ± 941 |
| ΔcpsY | 950 ± 196 | 2,461 ± 443a | 2,771 ± 361 | |
| cpsA | 385 ± 88b | 297 ± 100b | 832 ± 162b | |
| F12-1 | 621 ± 104 | 1,050 ± 170b | 747 ± 261b | |
| E6-9 | 4,400 ± 940a | 7,200 ± 1,616 | 12,325 ± 3,185 | |
| Heart | Wild type | 858 ± 195 | 796 ± 178 | 594 ± 252 |
| ΔcpsY | 792 ± 96 | 553 ± 222 | 523 ± 130 | |
| cpsA | 170 ± 86a | 380 ± 166 | 132 ± 52 | |
| F12-1 | 840 ± 407 | 636 ± 206 | 443 ± 110 | |
| E6-9 | 481 ± 130 | 312 ± 69 | 1,178 ± 288a | |
| Brain | Wild type | 336 ± 69 | 270 ± 55 | 267 ± 53 |
| ΔcpsY | 44 ± 9b | 21 ± 6b | 14 ± 3b | |
| cpsA | 30 ± 12b | 25 ± 6b | 13 ± 4b | |
| F12-1 | 153 ± 41 | 217 ± 74 | 109 ± 31 | |
| E6-9 | 190 ± 40 | 54 ± 9a | 95 ± 7 | |
P < 0.05.
P < 0.01.
FIG. 5.
Dissemination of S. iniae wild-type strain 9117 (9117 WT) and four capsule mutants at 24 h postinfection. Zebrafish were infected separately with each strain as described in Materials and Methods. The numbers of CFU of each strain were determined at 24 h postinfection in spleens (solid bars), hearts (striped bars), and brains (dotted bars) using a minimum of eight zebrafish. The number above each bar is the average number of CFU for that organ and that strain. The error bars indicate standard errors of the means. An asterisk indicates that the P value is <0.05.
At all times, the dissemination of the cpsA mutant to the spleen and brain was significantly attenuated compared to the dissemination of the wild type, results which correlated with the results of the other analyses. The cpsY mutant was also significantly deficient for dissemination to the spleen and brain at 24 h (P < 0.05) but only to the brain (P < 0.01) at early times. The results for cpsY mutant dissemination to the brain are similar to the competitive index data for the brain. In contrast, the competitive index for the cpsY mutant in the heart revealed almost no attenuation, while the dissemination to the heart at 24 h was 12-fold less than the dissemination of the wild-type strain, consistent with the hypothesis that the wild-type strain complements the cpsY mutant in the heart. Dissemination of the F12-1 mutant was significantly decreased in the spleen at 30 and 60 min (P < 0.01) and at 24 h (P < 0.05). Surprisingly, the values for F12-1 dissemination to the heart and brain are actually higher than the values for the wild type at 24 h (Fig. 5). This is in direct contrast to the competitive indices for F12-1 in the heart and brain at 24 h in the presence of the wild-type strain, which showed that there was at least a 1,000-fold decrease (P < 0.0001) in F12-1 CFU in these organs (compare Fig. 3A and B and Fig. 5). The number of E6-9 CFU was also greater than the number of wild-type CFU in all organs at 24 h and in the spleen at early times. However, the competitive indices in the brain showed that there was significant attenuation, 0.377 (P < 0.005), suggesting that in the presence of the inflammation elicited by the wild-type strain, the E6-9 mutant strain was less able to compete.
DISCUSSION
Like most systemic pathogens, S. iniae produces a capsule that is required for its pathogenesis and survival in the host (5, 17). To determine the composition of the S. iniae polysaccharide operon, we sequenced and annotated the entire locus and compared it to other streptococcal capsule operons and the operon of a known commensal S. iniae strain, strain 9066 (Fig. 1). We assigned names to the genes and putative functions to the protein products based on homologies to genes found in other gram-positive pathogens. Our analysis revealed that the 9066 commensal strain contains similar capsule genes at both the 5′ and 3′ ends of the operon but lacks a large central region of the capsule locus. Of note, the central region that is not present in 9066 is where the majority of the mutations of the virulent 9117 strain that were found to play a role in pathogenesis were located (17).
As mentioned above, the S. iniae capsule operon exhibits homology to the capsule operon of S. thermophilus, an organism commonly used in the dairy industry due to the texturizing properties that it confers to yogurt products. The exopolysaccharide produced by this organism leads to yogurt cultures with the desired smooth “ropy” or viscous texture. Nonropy strains of S. thermophilus have been identified, and when the exopolysaccharide operons of ropy and nonropy strains were compared, it was discovered that genes epsE through epsM were missing in the nonropy strains (33). This scenario is very similar to what we observed when we compared the operons of the capsule-producing S. iniae 9117 strain and the non-capsule-producing 9066 strain (Fig. 1A and B), except that 9066 also lacks some additional putative open reading frames not found in S. thermophilus and has the cpsE gene of 9117. The S. iniae 9117 capsule locus contains an insertion sequence with 98% homology to the IS981 insertion sequence from L. lactis (23, 24). IS981 has been found in the capsular operons of many strains of S. thermophilus and has been postulated to be an agent of genetic rearrangement between capsular operons of several species of bacteria over time (3, 4, 11). The presence of the IS981 insertion sequence element upstream of the cpsM gene in the S. iniae 9117 capsule operon suggests that horizontal transfer of genetic elements may occur between capsule-producing and non-capsule-producing strains.
Analysis of the S. iniae 9117 genetic regions sequenced in a previous study (17), as well as sequences listed in the GenBank database, revealed a G+C content of ∼33%. The entire capsule operon has an average G+C content of 32.2%, but the G+C contents of the individual genes within the operon vary from 22.8% to 40%. Most striking is the center region of the operon encompassing the sequences from cpsF through cpsL that are present in 9117 but have been shown to be absent in 9066. This region has an overall G+C content of only 27%, which is much lower than the G+C contents of S. iniae sequences reported to date, further supporting the hypothesis that this region was acquired by horizontal transfer from another low-G+C-content bacterial genome.
Virulence analysis of strains with mutated capsule genes located at the beginning, middle, and end of the capsule operon revealed multiple phenotypes. By comparing the results of virulence analyses using three different criteria, we were able to examine how the mutants fared in several host environments against factors elicited both in the presence and in the absence of the wild-type strain. Furthermore, the results provide insight into the different host responses to individual mutants. Table 4 summarizes the results of the three analyses used to compare the mutants. The most attenuated mutant overall, the cpsA insertion mutant, exhibited a similar level of attenuation in all analyses. The S. iniae CpsA protein is 65% homologous to the CpsA (previously called CpsX) protein of GBS, which has been suggested to be an attenuator of transcription as it exhibits homology with the lytR gene in Bacillus subtilis (13). However, a nonpolar deletion in the type Ia GBS cpsA gene resulted in two- to threefold less transcript from the cps operon than from the wild-type strain operon, suggesting that, in contrast to B. subtilis LytR, the GBS CpsA most likely functions as an activator of capsule expression (7). If, as in GBS, CpsA is an activator of capsule expression, then loss of this protein would predictably result in an attenuation of virulence and would most likely show the same loss of capsule both in vitro and in vivo. Moreover, if CpsA regulates transcription of additional factors other than the capsule promoter, then multiple factors may be missing from the wild-type survival mechanism.
TABLE 4.
Summary of infection assays
| Strain | CI at 24 ha
|
CFU at 24 h
|
% Survival after 4 days | |||
|---|---|---|---|---|---|---|
| Heart | Brain | Spleen | Heart | Brain | ||
| 9117 wild type | 1.0000 | 1.0000 | 1.0 × 105 | 5.7 × 103 | 1.0 × 104 | 8 |
| cpsA | 0.0002b | 0.0003b | 7.9 × 101b | 1.1 × 102 | 3.5 × 101b | 59 |
| F12-1 | 0.0002b | 0.0014b | 5.7 × 103b | 2.1 × 104 | 3.3 × 104 | 24 |
| ΔcpsY | 0.8790 | 0.0230b | 9.7 × 102b | 4.7 × 102 | 2.9 × 102b | 42 |
| E6-9 | 0.3770b | 0.5390b | 1.1 × 105 | 4.0 × 104 | 3.6 × 104 | 17 |
A CI of <1 indicates attenuation.
P ≤ 0.05.
Similar to the cpsA mutant, the F12-1 mutant has a very high buoyant density, suggesting loss of polysaccharide on the cell surface. The F12-1 insertion occurred in the cpsH gene (Fig. 1A and Table 2), which encodes a product that exhibits homology to a protein having an unknown function encoded in the exopolysaccharide locus of S. thermophilus. However, this gene occurs in a region of the operon encoding other glycosyl transferases, suggesting that it may play a similar role. Unlike the similarly unencapsulated cpsA mutant, the F12-1 mutant did not exhibit a consistently attenuated phenotype in all three analyses. The F12-1 competitive indices showed that there was a significantly attenuated profile in the presence of the wild-type strain; in sharp contrast, the dissemination results for the F12-1 mutant at 24 h showed that there was an ∼3-fold increase in the number of CFU in both the heart and the brain compared to the number of wild-type CFU (Table 4). These results are consistent with the hypothesis that the capsule differences between F12-1 and the wild-type strain cause different responses in the host. While the presence of the encapsulated wild-type strain causes the F12-1 strain to be cleared, when F12-1 is inoculated alone, the host is not able to clear the strain, suggesting that the immune responses of the host most likely differ in the presence and in the absence of the wild-type strain. This is in contrast to the level of dissemination to the spleen at 24 h, which was significantly less (19-fold; P < 0.05) than that of the wild type. Therefore, although the levels of dissemination to the heart and brain at 24 h are higher than the levels of dissemination for the wild type, the spleen environment can still efficiently clear the mutant strain in the absence of the wild-type strain, suggesting that there are differences in how the organs or environments react to a particular strain. An alternative explanation is that there may be an increased requirement for capsule in this environment. Although the cpsA mutant and the F12-1 mutant had almost the same level of capsule as determined by buoyant density, F12-1 exhibited significantly higher levels of dissemination to the spleen, heart, and brain at 24 h than the cpsA mutant exhibited, suggesting either that loss of capsule alone is not the entire reason for the high level of attenuation of the cpsA mutant or that the F12-1 mutant still has some capsule regulation in these environments.
The cpsY gene is directly upstream of the cpsA gene and the putative promoter of the S. iniae capsule operon, and the cpsY product exhibits over 78% identity and 89% similarity at the amino acid level to the CpsY transcriptional regulators encoded in the genomes of S. pyogenes and S. agalactiae (also called MtaR [30]). Initially, CpsY was proposed to be the regulator of capsule synthesis based on its homology to proteins in the LysR family of transcriptional regulators (13, 27). However, a recent report on inactivation of the cpsY gene of the type III COHI strain of S. agalactiae revealed no change in capsule production, although the strain was attenuated for virulence in a neonatal rat model (30). The analysis showed that the GBS CpsY mutant strain was deficient for growth in media with low concentrations of methionine and was also deficient for methionine transport, and subsequently the gene designation was changed from cpsY to mtaR (30). The growth of the S. iniae cpsY in-frame deletion mutant was analyzed in media deficient in methionine, as well as other amino acids. However, no difference was observed between the growth of the S. iniae wild-type strain and the growth of the cpsY deletion strain regardless of the amino acid content of the media (data not shown).
When the results for the cpsY mutant in the three virulence analyses were compared, the picture was very different than the picture obtained for the cpsA mutant. While the results for the ΔcpsY strain obtained in the heart competitive index analyses (in the presence of the wild-type strain) were similar to the results for the wild type (Table 4), the numbers of ΔcpsY CFU that disseminated to the spleen and the brain by 24 h (the same time that the CI was determined) were significantly less than the numbers of wild-type CFU. This correlates with the less virulent phenotype observed in the survival-over-time analysis (Fig. 4). These results suggest that the presence of the wild-type strain in competitive index analyses can complement the deficiency in the ΔcpsY mutant. Usually, this scenario occurs when a secreted factor is missing from the mutant that can be supplemented by having the mutant in close contact with a strain that is capable of secreting the factor. The function of cpsY in S. iniae is still unknown, but its high level of homology to LysR transcriptional regulators is consistent with the possibility that it regulates other virulence genes, some of which may be secreted. While both the cpsA and ΔcpsY strains were highly attenuated in the dissemination and survival analyses, the ΔcpsY strain, as demonstrated by the density gradient analysis, has just slightly less capsule than the wild-type strain, suggesting that the presence of capsule alone is not enough to rescue a strain from clearance by the host.
The E6-9 insertion occurred in the cpsM gene, which encodes a protein with homology to polysaccharide transport proteins. Comparison of the levels of dissemination to the heart and the brain of the E6-9 and F12-1 mutants at 24 h revealed a very high level of similarity between these two strains. However, in the Percoll gradient analysis the E6-9 mutant had the lowest buoyant density of any strain (most capsule), while the F12-1 mutant had one of the highest buoyant densities (smallest amount of capsule). Survival in the bloodstream (in this case, the heart) almost always correlates with strains that possess an antiphagocytic capsule, but the F12-1 mutant survives in this environment in the absence of this property. Furthermore, strains of S. pneumoniae and S. agalactiae that have less capsule have been shown to invade across the blood-brain barrier more efficiently (21, 25), but the number of CFU of the E6-9 mutant in the brain was the same as the number of CFU of the F12-1 mutant. Clearly, additional factors play a role in this process.
In summary, the results obtained using three different criteria for measuring virulence provide a more complete picture of the role of capsule in virulence, with each individual factor contributing to the overall survival of the pathogen. Furthermore, the multiple analyses provided insights into possible differences in the immune responses of the host to particular phenotypes. While an insertion in the putative capsule regulator gene, cpsA, should predictably result in an attenuated phenotype, the results for the F12-1 insertion mutant, which shows a loss of capsule similar to that of the cpsA mutant, are not as easily explained. The current dogma for how a systemic pathogen survives during dissemination and in the bloodstream is that a capsule is required for prevention of phagocytosis and subsequent clearance. Our results with F12-1 argue against the need for capsule during these steps in the infection process. On the other hand, even a small amount of excess capsule, as in the E6-9 insertion mutant, resulted in about 0.5-log attenuation in the CI analysis, and even though this is a small amount, it was found to be significant (P < 0.005). The data presented here illustrate that although too little capsule can be tolerated in some environments, it is less satisfactory in other tissues, and an excess of capsule can be advantageous in some environments but is also not optimal. Therefore, for the overall “ideal” infection, capsule needs to be regulated during dissemination, with expression of different amounts of capsule in various tissues. We are currently exploring the potential of this modulation and its implications for virulence in the host. Clearly, the role of polysaccharide capsule in virulence is very complex, and more work is needed to understand the dynamic interaction with the host immune system.
Acknowledgments
The technical assistance provided by Donna Runft in this project is greatly appreciated.
This work was supported by Public Health Service grant AI52141 from the NIAID, National Institutes of Health.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Adamou, J. E., T. M. Wizemann, P. Barren, and S. Langermann. 1998. Adherence of Streptococcus pneumoniae to human bronchial epithelial cells (BEAS-2B). Infect. Immun. 66:820-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bourgoin, F., G. Guedon, M. Pebay, Y. Roussel, C. Panis, and B. Decaris. 1996. Characterization of a mosaic ISS1 element and evidence for the recent horizontal transfer of two different types of ISS1 between Streptococcus thermophilus and Lactococcus lactis. Gene 178:15-23. [DOI] [PubMed] [Google Scholar]
- 4.Bourgoin, F., A. Pluvinet, B. Gintz, B. Decaris, and G. Guedon. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151-161. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan, J. T., J. A. Stannard, X. Lauth, V. E. Ostland, H. C. Powell, M. E. Westerman, and V. Nizet. 2005. Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development. Infect. Immun. 73:6935-6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 8.Farley, M. M. 2001. Group B streptococcal disease in nonpregant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 9.Fuller, J. D., D. J. Bast, V. Nizet, D. E. Low, and J. C. de Azavedo. 2001. Streptococcus iniae virulence is associated with a distinct genetic profile. Infect. Immun. 69:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller, J. D., A. C. Camus, C. L. Duncan, V. Nizet, D. J. Bast, R. L. Thune, D. A. Low, and J. C. S. de Azavedo. 2002. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect. Immun. 70:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guedon, G., F. Bourgoin, M. Pebay, Y. Roussel, C. Colmin, J. M. Simonet, and B. Decaris. 1995. Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol. Microbiol. 16:69-78. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt, S., S. Wolff, A. Hocke, S. Rosseau, E. Muller, and M. Rohde. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koskiniemi, S., M. Sellin, and M. Norgren. 1998. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression in group B streptococci. FEMS Immunol. Med. Microbiol. 21:159-168. [DOI] [PubMed] [Google Scholar]
- 14.Llull, D., R. Lopez, and E. Garcia. 2001. Genetic bases and medical relevance of capsular polysaccharide biosynthesis in pathogenic streptococci. Curr. Mol. Med. 1:475-491. [DOI] [PubMed] [Google Scholar]
- 15.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 60:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. D., and M. N. Neely. 2005. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect. Immun. 73:921-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. D., and M. N. Neely. 2004. Zebrafish as a model host for streptococcal pathogenesis. Acta Trop. 91:53-68. [DOI] [PubMed] [Google Scholar]
- 19.Morona, J. K., D. C. Miller, R. Morona, and J. C. Paton. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189:1905-1913. [DOI] [PubMed] [Google Scholar]
- 20.Neely, M., J. Pfeifer, and M. G. Caparon. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 70:3904-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nizet, V., K. S. Kim, M. Stins, M. Jonas, E. Y. Chi, D. Nguyen, and C. E. Rubens. 1997. Invasion of brain microvascular endothelial cells by group B streptococci. Infect. Immun. 65:5074-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Casal, J., J. A. Price, E. Maugin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 23.Polzin, K. M., and L. L. McKay. 1991. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl. Environ Microbiol. 57:734-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polzin, K. M., D. Romero, M. Shimizu-Kadota, T. R. Klaenhammer, and L. L. McKay. 1993. Copy number and location of insertion sequences ISS1 and IS981 in lactococci and several other lactic acid bacteria. J. Dairy Sci. 76:1243-1252. [DOI] [PubMed] [Google Scholar]
- 25.Ring, A., J. Weiser, and E. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz, N., B. Wang, A. Pentland, and M. G. Caparon. 1998. Streptolysin O and adherence synergistically modulate proimflammatory responses of kerationocytes to group A streptococci. Mol. Microbiol. 27:337-346. [DOI] [PubMed] [Google Scholar]
- 27.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 28.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellin, M., S. Hakansson, and M. Norgren. 1995. Phase-shift of polysaccharide capsule expression in group B streptococci, type III. Microb. Pathog. 18:401-415. [DOI] [PubMed] [Google Scholar]
- 30.Shelver, D., L. Rajagopal, T. O. Harris, and C. E. Rubens. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J. Bacteriol. 185:6592-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 32.Spellerberg, B. 2000. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2:1733-1742. [DOI] [PubMed] [Google Scholar]
- 33.Stingele, F., J. R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstein, M. R., M. Litt, D. A. Kertesz, P. Wyper, D. Rose, M. Coulter, A. McGeer, R. Facklam, C. Ostach, B. Willey, A. Borczyk, and D. E. Low. 1997. Invasive infections due to a fish pathogen, Streptococcus iniae. N. Engl. J. Med. 337:589-594. [DOI] [PubMed] [Google Scholar]
- 35.Zlotkin, A., H. Hershko, and A. Eldar. 1998. Possible transmission of Streptococcus iniae from wild fish to cultured marine fish. Appl. Environ Microbiol. 64:4065-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]