Abstract
The ability of 111 Streptococcus agalactiae strains to bind to human fibrinogen was quantified. We correlated the percentages of bacteria that bound to immobilized fibrinogen with fibrinogen-binding (fbs) gene characteristics of strains and with clinical origin, serotypes, and phylogenetic positions of strains. Percentages varied from 0.4 to 29.9%. Fifty-five strains (49.5%) had the fbsB gene sensu stricto described by Gutekunst et al. (Infect. Immun., 72:3495-3504, 2004), allowing adhesion to human fibrinogen, and all of the other strains had an fgag variant gene. Ninety strains (81.1%) had a fbsA gene and 55 of them also had the fbsB gene. The other 21 strains (18.9%) had a truncated form of fbsA without the fbsB gene sensu stricto. The numbers of 48-nucleotide repeat sequences (rs) in the fbsA gene varied from 2 to 26. The population of strains with the highest ability to bind to human fibrinogen significantly more frequently had the fbsB gene sensu stricto and 4 to 7 rs in the fbsA gene (P < 0.05). However, the single strain that carried the highest number of rs (26 rs) in the fbsA gene showed high fibrinogen-binding activity (24.3%). Strains exhibiting significantly higher levels of binding to human fibrinogen belonged to a phylogenetic group of strains associated with neonatal meningitis, currently known as the ST-17 clone, that is mostly composed of serotype III strains. These findings indicate that S. agalactiae strains possess a wide variety of fbs gene content that markedly influences the ability of strains to bind to human fibrinogen. Variations in the configuration and the expression of the Fbs proteins may therefore partly explain the variability of virulence in S. agalactiae species.
Streptococcus agalactiae is the leading cause of invasive neonatal infections in many industrialized countries. Vertical transmission of the bacteria to the neonate follows asymptomatic colonization of the genital and/or intestinal tract of the mother, which occurs in 15 to 35% of pregnant women, and approximately 1 to 2% of colonized neonates develop a severe infection, including pneumonia, sepsis, and/or meningitis (26, 27). Phenotypic differences have been observed between the S. agalactiae strains causing invasive infections in neonates and those colonizing healthy infants, including capsular polysaccharides and surface protein antigens, but no single factor can account for the pathogenicity of the S. agalactiae species (9, 19, 27). Multilocus enzyme electrophoresis (MLEE) was the first method that made possible the distinction of two strongly differentiated lineages in S. agalactiae species (20, 21), confirmed by multilocus sequence typing (14, 16, 18). Most of the strains responsible for neonatal meningitis are clustered in particular genogroups of these two phylogenetic divisions, indicating differences in the virulence of strains (20, 21).
Several studies have shown the contribution of S. agalactiae adhesins and invasins to virulence. The three surface-exposed bacterial proteins, FbsA, ScpB, and Lmb, interact with fibrinogen, fibronectin, and laminin, respectively, and the excreted FbsB protein interacts with fibrinogen. Bacterial binding to these proteins, which are components of the extracellular eucaryotic matrix and/or of the host cell surface, thus allows the bacteria to adhere to or invade tissues (1, 7, 13, 19, 24, 28-30). FbsA and FbsB are proteins with no structural homology which both bind to human fibrinogen, mediate the bacterial adhesion to or invasion of host cells, and contribute to the escape from the immune system. Using fusion proteins and deleted and recomplemented strains for the fbsA and fbsB genes has shown that FbsA is the major receptor for fibrinogen and promotes adhesion to epithelial and endothelial cells, whereas FbsB more particularly mediates bacterial invasion into host cells (13, 24, 25, 29).
FbsA, a surface-exposed cell wall-anchored protein, includes a repeat region very close to the N-terminal extremity, which forms the major part of this protein and is responsible for binding to fibrinogen. The protein size varies from 186 to 618 amino acids and is related to the number of repeat units, each 16 amino acids in length (mostly GNVLERRQRDAENRSQ), which varies from 3 to 30, depending on the strain. The fibrinogen-binding capacity of a strain varies with the number of repeats, but the use of synthetic peptides demonstrated that a single repeat is sufficient to bind human fibrinogen (25). Some S. agalactiae strains exhibit a truncated fbsA gene, with a deletion in the 5′ end and a single repeat, and are inactivated by a frameshift mutation (5, 32).
FbsB, a 635-amino-acid protein encoded by the fbsB gene (13), which is the gbs0850 gene of NEM316, an entirely sequenced S. agalactiae strain (12), is also named FgagV2 (i.e., fibrinogen-binding protein variant 2 in S. agalactiae) (15) or FbsB2 (5). It is devoid of a cell wall-anchoring domain and transmembrane regions, suggesting that it is a secreted rather than a covalently surface-exposed protein, and binds to human fibrinogen with its 388 N-terminal amino acids (13). An fbsB variant, named fbsB2b, showing 98.7% identity with fbsB has recently been described (5). Various strains from human and bovine specimens lack the fbsB gene sensu stricto and carry a fgag gene variant. The various Fgag proteins lack the 388 N-terminal residues involved in the binding of FbsB to human fibrinogen and contain 223 almost identical C-terminal residues that are similar to those found in the FbsB protein and are involved in binding to bovine but not human fibrinogen (15). A recent study called these fgag variant genes fbsB1 and fbsB3 and identified two other variants of fgag genes that were named fbsB4 and fbsB5, without demonstrating that these genes were involved in binding to fibrinogen (5).
We report here the ability of 111 clinically and phylogenetically well-documented S. agalactiae strains isolated from mothers and neonates to bind to human fibrinogen and correlate the results with the genetic variability of fbsA and fbsB genes. We observed that the group of strains that had the highest percentage of binding to fibrinogen carried the fbsB gene sensu stricto and 4 to 7 repeats of 48 nucleotides in the fbsA gene. These properties were examined according to the clinical origin, the capsular serotype, and the phylogenetic position of the strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 111 human strains of Streptococcus agalactiae of genital and neonatal origin were included in the present study: 54 strains from the cerebrospinal fluid (CSF) of neonates, 21 strains from the gastric fluid of colonized asymptomatic neonates, and 36 strains from vaginal swabs of colonized asymptomatic pregnant women. All strains had previously been serotyped on the basis of capsular polysaccharides (21). Six serotypes were identified, i.e., serotypes Ia (16 strains), Ib (12 strains), II (17 strains), III (55 strains), IV (3 strains), and V (3 strains), and five strains were not typeable. The strains were also analyzed by MLEE, which identified two major phylogenetic divisions, MLEE I and II (21). Forty isolates, including 27 from CSF, were from MLEE I, a genetically homogeneous group posing a high risk of central nervous system (CNS) infection in neonates. In this division, 29 strains were from two electrophoretic types (ETs 49 and 58) that included 26 strains from the CSF. Seventy-one strains were from MLEE II, which is genetically more heterogeneous, but the 27 CSF isolates in this group were mainly clustered in ET 12 (14 strains) and ET 11 (5 strains). The strains were stored at −80°C in Schaedler-vitamin K3 broth (bioMérieux, Marcy l'Etoile, France) with 10% glycerol. The bacteria were grown for 24 h on 5% horse blood Trypticase soja (TS) agar plates (bioMérieux) at 37°C and then grown in 10 ml of brain heart infusion broth (bioMérieux) overnight at 37°C.
Binding of S. agalactiae to immobilized human fibrinogen.
Bacterial cells were harvested by centrifugation (10 min, 3,000 rpm), suspended in sterile phosphate-buffered saline (PBS) (150 mM NaCl, 10 mM sodium phosphate, pH 7.2), and adjusted to 5 × 107 to 1 × 108 bacteria per ml by spectrophotometry. The bacterial inocula were checked by viable count on TS agar plates.
All binding assays were performed in triplicate. Flat-bottom 96-well polystyrene plates (Ciblex, Ivry sur Seine, France) were coated for 18 h at 4°C with 21 nM human fibrinogen (Diagnostica Stago, Asnières, France) diluted in PBS (13). Fibrinogen-coated wells were washed three times with PBS, and then 100 μl of PBS containing 5 × 107 to 1 × 108 CFU per ml was added to each well. After incubation for 90 min at 37°C, nonbinding bacteria were removed by five washings with PBS. Bound bacteria were subsequently unbound by the addition of 100 μl of a 0.01% (weight/vol) solution of protease-serine protease mix (Sigma) to each well, and 15-min incubation of the plates at 37°C. The contents of each well were harvested by 10 vigorous pipettings, and then the viable bacteria were quantified by plating serial dilutions onto TS agar plates. It was checked that protease did not affect bacterial viability in these experimental conditions.
For each preparation, the percentage of binding to human fibrinogen was obtained by the ratio between the number of bound bacteria and the number of bacteria present in the inoculum. After fixation for 10 min with methanol and staining for 15 min with 10% Giemsa (Sigma), the plates were observed by light microscopy (×1,000) to check that the protease treatment released the totality of bound bacteria.
Investigation of fbsA, fbsB, and fgag genes.
All primers used (Table 1) were purchased from Eurogentec (Seraing, Belgium). (i) Selection of fbsA primers. Primers were designed from the conserved sequences of the fbsA genes of S. agalactiae strains 176H4A, 090R, SS169, 6313, 706S2 (25), NEM316 (12), and A909 (31) and from sag1052, the fbsA truncated form of the 2603V/R strain (32). The number in each primer corresponds to the position of the first nucleotide in the fbsA gene of strain 176H4A (accession number AJ437621) which presents 3 48-nucleotide repeat sequences (rs), and “rc” indicates that the primer was identical to the lagging strand. The fbsA86 and fbsA555rc primers were designed in the 3′ and the 5′ part of the fbsA gene, respectively, flanking the rs. The fbsA282 and fbsA405rc primers were designed in the 3′ part of the gene, downstream from the rs. Primer set fbsA86/fbsA555rc was used to amplify the fbsA gene, and primer set fbsA282/fbsA405rc was used to amplify a sequence present in the fbsA gene and in the fbsA truncated form.
TABLE 1.
Oligonucleotide primers used for amplification
| S. agalactiae reference strain (reference[s]) | Gene in reference strain | Name of primer | Sequence (5′-3′) | Amplified gene(s) |
|---|---|---|---|---|
| 176H4A (25) | fbsA | fbsA86 | ATCAAGTCCTGTATCTGCTAT | fbsA |
| fbsA555rc | TTCATTGCGTCTCAAACCG | |||
| fbsA282 | CAACTTATAGGGAAAAATCCAC | fbsA and truncated fbsA | ||
| fbsA405rc | AGTTAACATCGGTCTATTAGC | |||
| NEM316 (12, 13) | gbs0850 | fbsB349 | ATGAAGCGATTGTGAATAGAA | fbsBa |
| fbsB1926rc | TCGCCTTGATAGCAGTGTC | |||
| fbsB143 | TCGGTCATAAAATAGCGTATGG | fbsBa | ||
| fbsB1710rc | AAGAATTCAACGGTCGGCTTCGT | |||
| 4:74 (15) | fgag | fgag | ACGATGCTATTTCGGCGTAT | fbsBa and all fgag variants |
| fgagrc | AAGAATTCAACGGTCGGCTTCGT | |||
| 090R (28) | lmb | lmb130 | GTTGTGAGTTTAGTAATGATAGCGA | lmb |
| lmb970rc | TATGTCTTGTTCCGCTTG | |||
| BM110 (17, 20) | gbs2018 | ST-17S | ATACAAATTCTGCTGACTACCG | gbs2018-ST-17b |
| ST-17AS | TTAAATCCTTCCTGACCATTCC | |||
| NEM316 (12, 17) | dltR | dltRS | TTGACAGGTCTCTATGATTTAGTC | dltR |
| dltRAS | GTCTGGTTCTCAGCCTAATTC |
(ii) Selection of fbsB and fgag primers.
Two primer sets (fbsB349/fbsB1926rc and fbsB143/fbsB1710rc) were used to amplify the fbsB gene sensu stricto described by Gutekunst et al. (13). Primers were designed from the sequence of gbs0850, the fbsB gene of the S. agalactiae human NEM316 strain (accession number AL766847) (12). Primer set fgag/fgagrc was used to amplify a 240-nucleotide 3′-terminal sequence conserved in all variants of fgag genes (5, 15). This 240-nucleotide sequence is also conserved in the fbsB gene.
The lmb gene, used as a control, was amplified with primer set lmb130/lmb970rc determined from the AF062533 sequence of S. agalactiae R268 strain (28).
(iii) PCR assay.
Bacterial genomic DNA (20 ng), extracted and purified by conventional methods (23), was used as the template for PCR assays. The mixture (20 μl) contained primers (0.2 μM each), deoxynucleoside triphosphates (200 μM each), Taq DNA polymerase (0.5 U) (Roche Diagnostics, Mannheim, Germany), and 1.5 mM MgCl2, in 1× buffer. The PCR consisted of an initial 5-min hold at 94°C, followed by 30 cycles, each of 1 min denaturation at 94°C, 0.5 min annealing at 55°C, and 1 min elongation at 72°C, followed by a final 10-min elongation step at 72°C (GeneAmp PCR System 2700; Applied Biosystems, Foster City, CA).
PCR products were analyzed by agarose gel electrophoresis for 1 h (for fbsB and lmb) and 2 h (for fbsA and fgag) at a constant voltage (110 V). Gels contained 1% (for fbsB and lmb) or 2% (for fbsA and fgag) agarose (Eurogentec) in 89 mM Tris, 89 mM borate, and 2.5 mM EDTA buffer (pH 8.0). A 1-kb DNA ladder (Invitrogen, Cergy-Pontoise, France) and a 100-bp DNA ladder (Amersham Biosciences, Buckinghamshire, England) were used as the molecular size standards. Gels were stained with ethidium bromide (1 μg/ml) (Sigma, Lyon, France) for 20 min and visualized by UV transillumination.
DNA sequencing of PCR products.
DNA sequencing of fbsA and fbsB amplicons was performed using the BigDye Terminator chemistry and the Applied BioSystem 3730 sequencer (Genome Express, Meylan, France).
Real-time PCR screening of ST-17 clone.
Detection of the ST-17 clone was performed as recently described (17) by a real-time PCR assay with primer set ST-17S/ST17AS characterizing the gbs2018 gene variant encoding a surface protein that is specific for the ST-17 clone. The additional primer set dltR/dltRAS characterizing the dltR regulation gene present in all S. agalactiae strains (17) was used as an amplification control (Table 1). The PCR assay was performed on a Chromo 4 System Instrument (Bio-Rad, Hercules, CA) in a final volume of 25 μl containing 5 μl of extracted DNA, 0.5 μM concentrations of each primer, and iQ SYBR green Supermix 1× (Bio-Rad) including 3 mM MgCl2. Amplification was performed using 40 cycles, each of 10 s at 95°C, 5 s at 55°C, and 10 s at 72°C. The reaction product was then cooled to 35°C and subjected to a post-PCR melting cycle by elevating the temperature by 0.2°C at each 10-s cycle, up to 95°C.
Data expression and statistical analysis.
For binding assays to fibrinogen, all results were expressed as mean values ± standard deviations (SD). Data were analyzed using a paired Student's t test (two samples) or one-way analysis of variance with Fisher's post hoc test (group comparisons). When conditions of normality were not met to use parametric analysis, a nonparametric analysis of variance test (Kruskal-Wallis) with pairwise multiple-comparison procedures (Dunn's method) was used to find statistical differences between results. Statistically significant differences were determined at a 95% confidence level. Parametric tests were carried out with Minitab version 10.51 (MINITAB Inc., State College, PA), and nonparametric tests were carried out with SigmaStat version 2.03 (Systat Software, Inc., Richmond, CA).
RESULTS
fbsB and fgag genes in S. agalactiae strains.
By using an initial set of primers (fbsB349/fbsB1926rc) to amplify the fbsB gene sensu stricto described by Gutekunst et al. (13), a fragment of the predicted size of 1,578 bp was obtained for 55 of the 111 S. agalactiae strains (49.5%) (Table 2). PCR using a second set of primers (fbsB143/fbsB1710rc) amplified the predicted fragment of 1,568 bp in the fbsB gene for the same 55 strains. DNA sequencing of the 1,578-bp fragment obtained by PCR with one strain showed 100% identity with the fbsB2b gene and 98.7% identity with gbs0850, the fbsB gene of the entirely sequenced S. agalactiae NEM316 strain (5, 12). For all the strains of our collection, including the 56 strains lacking the fbsB gene, a 240-bp amplicon was obtained using the primer set fgag/fgagrc annealing in the 3′ end of the fbsB gene in a region that is conserved in all variants of fgag genes. Thus, all of our strains harbored the 3′ part which is common to fbsB and fgag genes, but only 49.5% of our strains had the fbsB gene sensu stricto described by Gutekunst et al. (13).
TABLE 2.
Binding to fibrinogen in relation to the distribution of fbsA and fbsB genes in 111 strains of S. agalactiae
| Presence of fbsAa | Presence of fbsBb | No. (%) of strains | % Binding to fibrinogen (SD) |
|---|---|---|---|
| + | + | 55 (49.5) | 11.3 (7.2) |
| + | − | 35 (31.6) | 4.6 (4.1) |
| − | − | 21 (18.9) | 4.3 (3.3) |
fbsA genes of various sizes, excluding the truncated form of fbsA.
fbsB gene sensu stricto (13).
fbsA gene in S. agalactiae strains.
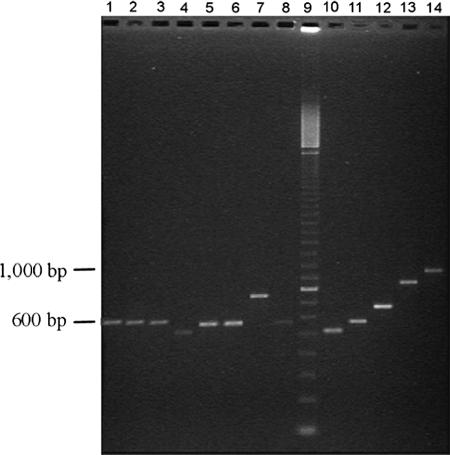
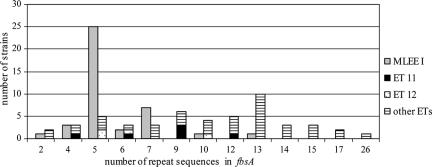
To detect the fbsA gene, a set of primers (fbsA86/fbsA555rc) annealing outside the repeat units of the fbsA gene was used for PCR. The fbsA gene was found for 90 of the 111 strains (81.1%) (Table 2). The amplicon size varied (Fig. 1), and there were 13 different PCR products from 0.4 to 1.6 kb in size. From the individual sizes of the 13 different fragments and DNA sequencing of each of them, we determined that the differences in size were related to different numbers of the 48-nucleotide rs described by Schubert et al. in the fbsA gene (25). The numbers of rs varied from 2 to 26 in our collection of strains (Fig. 2), and the most frequent configurations were of 5 rs (found in 30 strains), 13 rs (found in 11 strains), and 7 rs (found in 10 strains). The other numbers of rs each occurred in one to six strains. The deduced amino acid sequences of the rs obtained from 21 strains showed five types of 16-amino-acid units. The most frequent pattern was GNVLERRQRDVENKSQ. The other sequences carried one or two of the following substitutions: V11A, E12D, and K14R (Table 3). Four of these sequences had previously been described, and the different substitutions that we observed were all shown not to affect fibrinogen binding ability (25).
FIG. 1.
PCR products obtained with the fbsA86/fbsA555rc primer set and analyzed by agarose gel electrophoresis with chromosomal DNA from different clinical S. agalactiae isolates belonging to the MLEE I (lanes 1 to 8) and to the MLEE II (lanes 10 to 14) phylogenetic divisions. Lane 9, 100-bp ladder.
FIG. 2.
Distribution of the 48-nucleotide repeat sequences in the fbsA gene. MLEE I phylogenetic division, ET 11, and ET 12 are three high-risk groups for neonatal meningitis.
TABLE 3.
Characteristics of the 16-amino-acid rs in FbsA
| Amino acid sequencea | No. (%) of rs:
|
||
|---|---|---|---|
| 4-7 | 2 or 9-17 | 26 | |
| GNVLERRQRDVENKSQ | 16 (35.6) | 30 (31.9) | 9 (45.0) |
| ..........A..... | 21 (46.7) | 21 (22.3) | 2 (10.0) |
| ..........AD.... | 7 (15.6) | 32 (34.0) | 7 (35.0) |
| ..........A..R.. | 0 | 7 (7.4) | 2 (10.0) |
| .............R.. | 1 (2.2) | 4 (4.3) | 0 |
| Total no. of rs analyzed | 45 | 94 | 20 |
Dots indicate identical residues.
The 21 strains (18.9%) that did not harbor the fbsA gene carried the truncated fbsA gene with a deletion in the 5′ end (5, 32), as shown by amplification with the fbsA282/fbsA405rc primer set.
Association of fbsA and fbsB genes in S. agalactiae strains.
Twenty-one of the 111 strains (18.9%) were devoid of the fbsA gene and fbsB gene sensu stricto, 35 strains (31.6%) had only the fbsA gene, and 55 strains (49.5%) had both genes. The fbsB gene was never present when the fbsA gene was absent (Table 2). The distribution of the number of rs in the fbsA gene was different according to the presence of the fbsB gene. The 55 strains with the fbsB gene presented 2 to 12 rs; 28 of these strains (50.9%) had 5 rs, and 9 strains (16.4%) had 7 rs. The 35 strains without the fbsB gene had 2 to 26 rs, and 11 strains (31.4%) had 13 rs (Fig. 3).
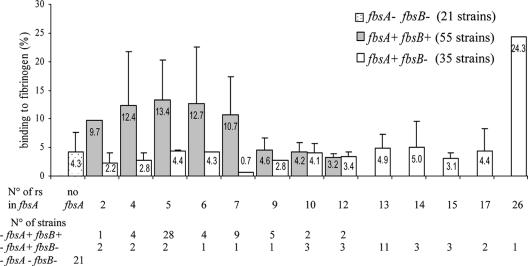
FIG. 3.
Binding to fibrinogen of 111 strains of S. agalactiae in relation to the presence of the fbsA and the fbsB genes and to the numbers of 48-nucleotide repeat sequences in the fbsA gene. Boxes are means and bars are standard deviations of the means of the fibrinogen-binding percentages for the different strains in each category.
Detection of the ST-17 clone.
Real-time PCR confirmed that the 111 strains belonged to S. agalactiae species, in which 41 strains belonged to the ST-17 clone: 37 of the 40 MLEE I strains (92.5%) and 4 of the 71 MLEE II strains (5.6%), 3 of which were ET 12 CSF strains. The ST-17 clone included 29 of the 54 CSF strains (53.7%) and 37 of the 55 serotype III strains (67.3%).
Binding of S. agalactiae strains to immobilized human fibrinogen.
The percentages of bacteria that bound to fibrinogen varied considerably according to strain (0.4 to 29.9% in our collection of strains). Statistical comparisons of the ability of strains to bind to fibrinogen showed that the group of 55 strains with the simultaneous presence of fbsA and fbsB genes was significantly associated with a stronger ability to bind to fibrinogen (11.3% ± 7.2) than the group of 21 strains with no fbs genes (4.3% ± 3.3%) and than the group of 35 strains with the fbsA gene but without the fbsB gene (4.6% ± 4.1%) (P < 0.001) (Table 2). The binding ability of these last two groups was not statistically different.
Statistical analysis was performed for each of the rs groups shown in Fig. 3. Five rs groups showed a significantly higher ability to bind to fibrinogen than the group of 21 strains lacking both the fbsA and fbsB genes (P < 0.05). The first four groups were the 4-rs, 5-rs, 6-rs, and 7-rs groups of the population of strains with both the fbsA and fbsB genes. We named this population the“4- to 7-rs fbsA/fbsB group.” The last group was represented by a single strain that had the fbsA gene without the fbsB gene and a high number of rs (26 rs) in the fbsA gene. This strain demonstrated a very strong fibrinogen binding capacity (24.3% ± 1.9%, with the SD value of 1.9% corresponding to the SD obtained for the triplicate assays of this strain).
Fibrinogen binding, fbsA and fbsB genes, and clinical origin, serotype, and phylogenetic position of strains.
The ability of strains to bind to fibrinogen according to the genetic diversity of fbs genes is reported in Table 4 for each clinical origin, capsular serotype, and phylogenetic position of strains. Neonatal CSF strains bound significantly more strongly to fibrinogen (9.6% ± 7.6%) than vaginal strains (6.8% ± 5.7%) or neonatal gastric fluid strains (5.0% ± 4.2%) (P = 0.01) due to the fact that CSF strains more frequently contained the fbsB gene and the fbsA gene (35/54 strains, 64.8%) with 4 to 7 rs in the fbsA gene (30/54 strains, 55.5%). Accordingly, the strains isolated from the gastric fluid were those that had the lowest fibrinogen-binding ability in relation to the lowest number of strains that possessed the fbsB gene with the fbsA gene (4/21 strains, 19.0%) and the lowest number of strains in the 4- to 7-rs fbsA/fbsB group (3/21 strains, 14.3%) (Table 4).
TABLE 4.
Binding of S. agalactiae strains to fibrinogen and presence of fbsA and fbsB genes in isolates correlated with clinical origin, serotype, and phylogenetic position of strains
| Strain group | No. (% ± SD) of strains in groupa:
|
|||||
|---|---|---|---|---|---|---|
| All strains (n = 111) |
fbsA+ fbsB+ (n = 55)
|
fbsA+ fbsB− (n = 35)
|
fbsA− fbsB− (n = 21) | |||
| 4-7 rs | Other no. of rs | 26 rs | Other no. of rs | |||
| Clinical origins | ||||||
| Vaginal | 36 (6.8 ± 5.7) | 12 (9.4 ± 6.7) | 4 (3.6 ± 1.6) | 1 (24.3) | 14 (4.4 ± 1.9) | 5 (6.0 ± 4.5) |
| Gastric | 21 (5.0 ± 4.2) | 3 (11.8 ± 6.9) | 1 (3.7) | 0 | 16 (4.0 ± 2.6) | 1 (2.8) |
| CSF | 54 (9.6 ± 7.6) | 30 (14.1 ± 7.0) | 5 (5.9 ± 2.7) | 0 | 4 (2.6 ± 1.6) | 15 (3.8 ± 2.9) |
| Polysaccharide serotypes | ||||||
| Ia | 16 (4.1 ± 1.6) | 6 (5.5 ± 1.4) | 7 (3.6 ± 1.2) | 0 | 3 (2.7 ± 0.7) | 0 |
| Ib | 12 (4.4 ± 2.2) | 0 | 0 | 0 | 12 (4.4 ± 2.2) | 0 |
| II | 17 (3.8 ± 3.2) | 0 | 0 | 0 | 9 (3.1 ± 2.0) | 8 (4.6 ± 4.0) |
| III | 55 (11.1 ± 7.3) | 36 (14.5 ± 6.7) | 3 (7.5 ± 2.1) | 0 | 4 (6.3 ± 3.1) | 12 (3.5 ± 2.3) |
| IV | 3 (6.4 ± 4.5) | 0 | 0 | 0 | 2 (4.3 ± 3.8) | 1 (10.5) |
| V | 3 (3.1 ± 1.0) | 0 | 0 | 0 | 3 (3.1 ± 1.0) | 0 |
| NT | 5 (9.2 ± 9.3) | 3 (5.8 ± 5.6) | 0 | 1 (24.3) | 1 (4.5) | 0 |
| Phylogenetic positions | ||||||
| MLEE Ib | 40 (13.3 ± 7.2) | 36 (14.2 ± 7.1) | 2 (7.5 ± 3.0) | 0 | 2 (4.1 ± 3.0) | 0 |
| ET 49 | 10 (14.0 ± 7.7) | 8 (15.6 ± 7.8) | 2 (7.5 ± 3.0) | 0 | 0 | 0 |
| ET 58 | 19 (15.1 ± 7.2) | 19 (15.1 ± 7.2) | 0 | 0 | 0 | 0 |
| Other ETs | 11 (9.8 ± 6.1) | 9 (11.0 ± 6.0) | 0 | 0 | 2 (4.1 ± 3.0) | 0 |
| MLEE IIc | 71 (4.7 ± 3.6) | 9 (6.7 ± 2.4) | 8 (4.1 ± 1.7) | 1 (24.3) | 32 (4.0 ± 2.3) | 21 (4.3 ± 3.3) |
| ET 11 | 6 (5.5 ± 1.7) | 2 (6.3 ± 0.2) | 4 (5.1 ± 2.0) | 0 | 0 | 0 |
| ET 12 | 15 (4.6 ± 2.6) | 3 (7.6 ± 0.9) | 0 | 0 | 1 (5.9) | 11 (3.6 ± 2.4) |
| Other ETs | 50 (4.6 ± 4.0) | 4 (6.3 ± 3.7) | 4 (3.1 ± 0.5) | 1 (24.3) | 31 (3.9 ± 2.3) | 10 (5.0 ± 4.1) |
| Sequence type | ||||||
| ST-17 | 41 (13.6 ± 6.9) | 40 (13.6 ± 7.0) | 1 (9.7) | 0 | 0 | 0 |
| Non-ST-17 | 70 (4.5 ± 3.4) | 5 (5.2 ± 1.4) | 9 (4.2 ± 1.7) | 1 (24.3) | 34 (4.0 ± 2.2) | 21 (4.3 ± 3.3) |
%, mean percentage of binding to fibrinogen; +, present; −, absent.
In MLEE division I, no significant difference was observed between the mean percentages of binding to fibrinogen of strains from ET 49 and ET 58 (mostly composed of isolates from CSF) and strains from other ETs.
In MLEE division II, no significant difference was observed between the mean percentages of binding to fibrinogen of strains from ET 11 and ET 12 (mostly composed of isolates from CSF) and strains from other ETs.
The 55 serotype III strains bound significantly more strongly to fibrinogen (11.1% ± 7.3%) than the 45 strains belonging to the other main I and II serotypes (3.8% to 4.4%) (P < 0.01) due to the fact that 36 serotype III strains belonged to the 4- to 7-rs fbsA/fbsB group with significantly higher fibrinogen-binding ability (14.5% ± 6.7%) (P < 0.01) (Table 4). Nevertheless, the serotype III strains belonged to the two MLEE phylogenetic divisions (37 were in MLEE I and 18 were in MLEE II). The fibrinogen-binding ability of the 18 MLEE II serotype III strains was low (4.7% ± 2.8%) and significantly lower than the binding ability of the 37 MLEE I serotype III strains (14.2% ± 6.8%) (P < 0.01). This difference was related to their fbs gene characteristics. Indeed, 34 of the 37 MLEE I serotype III strains (91.9%) belonged to the 4- to 7-rs fbsA/fbsB group, while only 2 of the 18 MLEE II serotype III strains (11.1%) belonged to this group. It is notable that 14 of the 18 MLEE II serotype III strains belonged to ET 12, a group of strains associated with neonatal meningitis despite its low ability to bind to fibrinogen and the frequent lack of both fbs genes (11/15 strains) (Table 4).
According to their fbs gene status, serotype Ib and serotype II strains, which are rarely isolated in meningitis, bound weakly to fibrinogen (Table 4). By contrast, serotype Ia strains that possessed the fbsB gene in 81.2% of isolates (13/16 strains) and belonged to the 4- to 7-rs fbsA/fbsB group in 37.5% of isolates (6/16 strains), were weakly able to bind to fibrinogen (4.1% ± 1.6%). Nevertheless, serotype Ia strains can be found in neonatal meningitis (5 of our 16 serotype Ia strains), and these five CSF strains had significantly higher fibrinogen-binding ability (5.6% ± 1.1%) than the other serotype Ia strains (3.4% ± 1.3%) (P < 0.05).
The five NT strains appeared to have a high average fibrinogen-binding activity (9.2% ± 9.3%). In fact, this was mainly related to the presence in the NT group of the only strain with 26 rs in the fbsA gene with 24.3% binding to fibrinogen (Fig. 3 and Table 4).
Each phylogenetic MLEE division was marked by a different distribution of rs in the fbsA gene (Fig. 1 and 2 and Table 4). The numbers of rs varied from 2 to 13 in the MLEE I strains, and 36 of these 40 strains (90%) belonged to the 4- to 7-rs fbsA/fbsB group, with 25/36 strains (69.4%) having five repeats. By contrast, the numbers of rs varied from 2 to 26 for the MLEE II strains, and only 9 strains of this group (12.7%) belonged to the 4- to 7-rs fbsA/fbsB group. This difference in distribution in the 4- to 7-rs fbsA/fbsB group for strains originating from the two phylogenetic divisions that compose S. agalactiae species was significant (P < 0.01). According to this fbs gene status, the mean percentage of the fibrinogen binding of strains from MLEE I (13.3% ± 7.2%) was significantly higher than that of MLEE II (4.7% ± 3.6%) (P < 0.01). Rapid detection of the ST-17 clone by real-time PCR showed a high level of concordance between MLEE I phylogenetic division and the ST-17 clone. ST-17 strains were strongly associated with the 4- to 7-rs fbsA/fbsB group, since 40 of the 41 ST-17 strains belonged to this group (Table 4). Therefore, strains from the ST-17 clone were significantly more able to bind to fibrinogen (13.6% ± 6.9%) than non-ST-17 strains (4.5% ± 3.4%) (P < 0.01).
DISCUSSION
Recent studies have characterized two proteins, FbsA and FbsB, involved in the adhesion and/or invasion of host cells by human strains of S. agalactiae by allowing bacterial binding to human fibrinogen (13, 24, 25). By studying a collection of 111 human strains of S. agalactiae of genital and neonatal origin previously typed by several techniques (6, 21, 22), we quantified the ability of strains to bind to immobilized human fibrinogen and correlated these properties with the fbsA and fbsB gene characteristics of strains at a population level.
The bacterial properties of binding to fibrinogen were correlated with the genetic status of fbs genes (Table 2 and Fig. 3). Indeed, strains devoid of both genes were weakly adherent (4.3% ± 3.3%), while high binding properties (11.6% ± 7.3%) were significantly associated with the presence of both the fbsB gene and the fbsA gene with moderate numbers of rs (4 to 7). Schubert et al., who studied the binding ability of five strains to soluble fibrinogen, found that one strain with 19 rs in the fbsA gene bound 3- to 10-fold more than the other four strains (24). This strain may thus be compared with one of our strains that had unusual features. Lacking the fbsB gene sensu stricto, and with a high number of rs in the fbsA gene (26 rs), it showed a high fibrinogen-binding ability (24.3%). Such high-binding strains may thus form a small part of the S. agalactiae bacteria population.
In S. agalactiae species, the prevalence of the fbsB gene sensu stricto was lower (49.5%) than that of the fbsA gene (81.1%). Similarly, the fbsA gene was found in 25/27 strains (92.6%) by Schubert et al., and in 55/75 strains (70.7%) by Brochet et al. (5, 25). The fbsB gene sensu stricto, also described as the fbsB2 or fbsB2b variant gene by Brochet et al., was found in 35/75 strains (46.6%) (5). Thus, in contrast to many virulence genes involved in S. agalactiae adhesion and invasion, such as the scpB, lmb, and bca genes which have been found in almost all human strains tested (2-4, 10, 11, 28), these findings show that S. agalactiae strains may lack one (31.6% of strains in this study) or both (18.9% of strains in this study) fbs genes. In addition, a strain may have the fbsA gene without the fbsB gene (34 of our 111 strains). In contrast, we never observed a strain with the fbsB gene without fbsA. Among the 75 strains studied by Brochet et al., only two strains from humans had the fbsB gene without the fbsA gene (5).
The presence of the fbsA gene was not sufficient to result in strong binding to fibrinogen, since fbsA alone allowed weak binding, similar to that observed when both genes were absent. It therefore appears that fbsB plays an important role in binding to fibrinogen. Using a deleted mutant for fbsB, Gutekunst et al. demonstrated that the FbsB protein was more weakly involved in the adhesion of S. agalactiae to host cells and in binding to fibrinogen than FbsA (13). However, the strain they studied was that described by Schubert et al. which exhibited a very strong ability to bind to fibrinogen, related to its fbsA gene with 19 rs (24). Our findings show that strains that have a large number of rs have a high ability to bind to human fibrinogen, even in the absence of the fbsB gene, but these strains are poorly represented in the species. Indeed, only one of our 111 strains corresponded to this profile. We therefore conclude that the ability to bind to fibrinogen requires the fbsB gene sensu stricto associated with the fbsA gene in most S. agalactiae strains.
All 111 strains studied had either the fbsB gene sensu stricto (49.5%) or a fgag variant gene (50.5%), and strains without the fbsB gene sensu stricto bound significantly more weakly to human fibrinogen (4.4% ± 3.8% versus 11.3% ± 7.2%). Jacobsson suggested that the almost identical 223 C terminal amino acids found in all Fgag and FbsB proteins are involved in binding to bovine but not to human fibrinogen (15). Our results confirm that fgag variant genes probably play a minor role in the ability to bind to fibrinogen in human neonatal infections. On the other hand, the FbsB protein sensu stricto allows binding to human fibrinogen, and the binding specificity is located within the 388 N-terminal residues in a region excluding the 223 C-terminal amino acids (13). Therefore, the recent denomination “fbsB variants” used to name all fgag variant genes (5) is somewhat confusing, and the ability of strains that possess these variant genes to bind to human fibrinogen remains to be explored before renaming them fgag or fbsB genes.
S. agalactiae strains that are able to invade the CNS of neonates originate from the maternal vaginal flora and then from amniotic/gastric fluid. Surprisingly, strains isolated from the gastric fluid of asymptomatic neonates (i.e., the amniotic fluid at birth) exhibited the lowest ability to bind to fibrinogen (5.0% ± 4.2%). These results suggest that amniotic fluid could constitute a valuable barrier to the most pathogenic strains, limiting their transfer from the mother's genital tract to the fetus. By contrast, strains from meningitis had the highest ability to bind to human fibrinogen (9.6% ± 7.6%), suggesting that this property could play a significant role in the diffusion of S. agalactiae strains to the CNS of colonized neonates.
Strains isolated from neonatal meningitis are frequently of serotype III (26, 27). Our findings indicated that the ability of serotype III strains to bind to human fibrinogen varied markedly according to the fbs gene status and to the phylogenetic position of strains (14.2% ± 6.8% for MLEE I serotype III strains versus 4.7% ± 2.8% for MLEE II serotype III strains). The MLEE I division was shown to be a high-virulence group of strains (21), corresponding to the high-virulence clone first identified by MLEE by Musser et al. in 1989 (20). Our findings show that the MLEE I division corresponds strongly to the ST-17 clone recently defined by multilocus sequence typing (16, 18). Our findings therefore support the hypothesis that the virulence of serotype III isolates is related to the phylogenetic position of strains and that differences in the ability to bind to human fibrinogen may be one of the factors explaining differences in virulence within serotype III strains.
Serotype Ia strains, which are often responsible for neonatal CNS infections, seem to be particular. Despite the high prevalence of the fbsB gene (81.3%), their fibrinogen-binding ability was low. Thus, as suggested by Chhatwal et al., capsular antigens may interfere with the surface exposure of Fbs proteins in some strains (8).
In conclusion, binding properties to human fibrinogen in S. agalactiae vary according to the fbs gene status, which itself correlates with the phylogenetic distribution of strains. These various genetic configurations probably influence the level of transcription of fbsA and fbsB genes, transport of FbsA and FbsB proteins across the cytoplasmic membrane, and the cell wall anchoring or excretion of these proteins. Quantification of fbsA and fbsB gene transcripts, use of isogenic mutants, and the use of animal models of bacteremia with strains representing each of these genetic configurations may help to clarify the mechanisms explaining the variations in ability of strains to bind to human fibrinogen.
Acknowledgments
We thank Mazen Salloum for his technical assistance.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 11 December 2006.
REFERENCES
- 1.Beckmann, C., J. D. Waggoner, T. O. Harris, G. S. Tamura, and C. E. Rubens. 2002. Identification of novel adhesins from group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidet, P., N. Brahimi, C. Chalas, Y. Aujard, and E. Bingen. 2003. Molecular characterization of serotype III group B-Streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 188:1132-1137. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack, J. F., S. Takahashi, L. Hammitt, D. V. Miller, A. A. Aly, and E. E. Adderson. 2000. Genetic polymorphisms of group B Streptococcus scpB alter functional activity of a cell-associated peptidase that inactivates C5a. Infect. Immun. 68:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolduc, G. R., M. J. Baron, C. Gravekamp, C. S. Lachenauer, and L. C. Madoff. 2002. The alpha C protein mediates internalization of group B Streptococcus within human cervical epithelial cells. Cell. Microbiol. 4:751-758. [DOI] [PubMed] [Google Scholar]
- 5.Brochet, M., E. Couvé, M. Zouine, T. Vallaeys, C. Rusniok, M.-C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227-1243. [DOI] [PubMed] [Google Scholar]
- 6.Chatellier, S., C. Ramanantsoa, P. Harriau, K. Rolland, A. Rosenau, and R. Quentin. 1997. Characterization of Streptococcus agalactiae strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 35:2573-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhatwal, G. S., C. Lämmler, and H. Blöbel. 1984. Guanine extraction enhances the binding of human fibrinogen to group-B streptococci. Med. Microbiol. Immunol. 173:19-27. [DOI] [PubMed] [Google Scholar]
- 9.Doran, K. S., and V. Nizet. 2004. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 54:23-31. [DOI] [PubMed] [Google Scholar]
- 10.Duarte, R. S., B. C. Bellei, O. P. Miranda, M. A. V. P. Brito, and L. M. Teixeira. 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franken, C., G. Haase, C. Brandt, J. Weber-Heynemann, S. Martin, C. Lämmler, A. Podbielski, R. Lütticken, and B. Spellerberg. 2001. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 41:925-935. [DOI] [PubMed] [Google Scholar]
- 12.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couvé, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 13.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2004. The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72:3495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hery-Arnaud, G., G. Bruant, P. Lanotte, S. Brun, A. Rosenau, N. van der Mee-Marquet, R. Quentin, and L. Mereghetti. 2005. Acquisition of insertion sequences and the GBSi1 intron by Streptococcus agalactiae isolates correlates with the evolution of the species. J. Bacteriol. 187:6248-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobsson, K. 2003. A novel family of fibrinogen-binding proteins in Streptococcus agalactiae. Vet. Microbiol. 96:103-113. [DOI] [PubMed] [Google Scholar]
- 16.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. M. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamy, M.-C., S. Dramsi, A. Billoët, H. Réglier-Poupet, A. Tazi, J. Raymond, F. Guérin, E. Couvé, F. Kunst, P. Glaser, P. Trieu-Cuot, and C. Poyart. 2006. Rapid detection of the “highly virulent” group B streptococcus ST-17 clone. Microbes Infect. 8:1714-1722. [DOI] [PubMed] [Google Scholar]
- 18.Lin, F.-Y. C., A. Whiting, E. Adderson, S. Takahashi, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, J. Regan, P. Clark, G. G. Rhoads, C. E. Fraasch, J. Troendle, and J. F. Bohnsack. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl, G., M. Stålhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Schubert, A., K. Zakikhany, G. Pietrocola, A. Meinke, P. Speziale, B. J. Eikmanns, and D. J. Reinscheid. 2004. The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72:6197-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert, A., K. Zakikhany, M. Schreiner, R. Frank, B. Spellerberg, B. J. Eikmanns, and D. J. Reinscheid. 2002. A fibrinogen receptor from group B streptococcus interacts with fibrinogen by repetitive units with novel ligand binding sites. Mol. Microbiol. 46:557-569. [DOI] [PubMed] [Google Scholar]
- 26.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting parardigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spellerberg, B. 2000. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2:1733-1742. [DOI] [PubMed] [Google Scholar]
- 28.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura, G. S., and C. E. Rubens. 1995. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol. Microbiol. 15:581-589. [DOI] [PubMed] [Google Scholar]
- 30.Tenenbaum, T., C. Bloier, R. Adam, D. J. Reinscheid, and H. Schroten. 2005. Adherence to and invasion of human brain microvascular endothelial cells are promoted by fibrinogen-binding protein FbsA of Streptococcus agalactiae. Infect. Immun. 73:4404-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]