Abstract
Though it is well established that gamma interferon (IFN-γ) is crucial to the early innate defense of murine listeriosis, its sources remain controversial. In this study, intracellular cytokine staining of IFN-γ-expressing splenocytes early after Listeria monocytogenes infection revealed that NK1.1+, CD11c+, CD8+ T, and CD4+ T cells expressed IFN-γ 24 h after infection. Contrary to the previous report, most IFN-γ+ dendritic cells (DC) were CD8α− DC. Unexpectedly, almost all CD11c+ IFN-γ-expressing cells also expressed NK1.1. These NK1.1+ CD11c+ cells represented primary IFN-γ-expressing cells after infection. In situ studies showed these NK1.1+ CD11c+ cells were recruited to the borders of infectious foci and expressed IFN-γ. A significant NK1.1+ CD11c+ population was found in uninfected spleen, lymph node, blood, and bone marrow cells. And its number increased significantly in spleen, lymph node, and bone marrow after L. monocytogenes infection. Using interleukin-12 (IL-12) p40−/− mice, IFN-γ expression was found to be largely IL-12 p40 dependent, and the number of IFN-γ-expressing cells was only about one-third of that of wild-type mice. Moreover, the IFN-γ expression was absolutely dependent on live L. monocytogenes infection, as no IFN-γ was detected after inoculation of heat-killed L. monocytogenes. Our findings not only provide an insight into IFN-γ expression after in vivo infection but may also change the current perceptions of DC and natural killer cells.
Listeria monocytogenes infection in mice has long been used as an experimental model to study the mechanisms involved in host defense against pathogens. Initially as a model to study cell-mediated immunity at the macrophage level (20), it appears that the host defense mechanisms against L. monocytogenes infection are composed of many components of innate and adaptive immune responses and their complicated interactions (3, 15). Macrophage activation through gamma interferon (IFN-γ) and tumor necrosis factor alpha is a paradigm for controlling intracellular pathogens like L. monocytogenes (3, 4). Indeed, IFN-γ−/− or IFN-γ receptor−/− mice are highly susceptible to even very low doses of L. monocytogenes, succumbing to infection within the first week (9, 11).
The early cellular sources of IFN-γ during L. monocytogenes infection are of great interest and a point of controversy. In 1989, Bancroft et al. found that anti-asialo-GM1 antibodies blocked IFN-γ expression of splenocyte from severe combined immune deficiency mice after in vitro stimulation with heat-killed L. monocytogenes (4). Since that time, the NK cells have been thought of as the primary source of IFN-γ, and this NK-derived IFN-γ was dependent on interleukin-12 (IL-12) and tumor necrosis factor alpha production by macrophages (31). However, Ohteki et al. later said that NK-depleted RAG2−/− mice had levels of IFN-γ in their sera comparable to those of nondepleted mice 48 h postinfection (hpi) and that incubation of NK-depleted splenocytes with L. monocytogenes still resulted in production of IFN-γ. After incubating a sorted dendritic cell (DC) population with IL-12 in vitro, they concluded that CD8α+ DCs produced IFN-γ in an IL-12-dependent way (24). Andersson et al. also showed that γc−/− mice, which completely lacked NK cells, could produce IFN-γ, while γc−/− RAG1−/− mice could not, as indicated from serum IFN-γ levels 48 hpi (1). Therefore, the authors concluded that T cells were another major source of early IFN-γ besides NK cells after L. monocytogenes infection. In addition, CD8+ T cells with memory markers were found to express IFN-γ after overnight culture with L. monocytogenes-infected macrophages, and this IFN-γ expression was thought to respond to combinatory stimulation of IL-12 and IL-18 (5). Lastly, Kirby et al. claimed that neutrophils and macrophages were the dominant sources of IFN-γ during Salmonella infection, a facultative intracellular bacterium like L. monocytogenes (17). Therefore, in spite of all those studies, the cellular source of early IFN-γ expression L. monocytogenes infection is unclear.
Due to the crucial role of IFN-γ in the defense of L. monocytogenes infection, it is important to unequivocally identify the sources of IFN-γ. We believe the conflicting data in previous reports probably resulted from the use of different mice and different in vivo and in vitro manipulations, for example, in vivo depletion or prolonged in vitro culture. In this study, we applied the technique of intracellular cytokine staining to visualize IFN-γ-expressing cells early after L. monocytogenes infection. This approach minimizes all in vitro and in vivo manipulations. Herein, we report that NK1.1+ CD11c+ cells are the primary source of early IFN-γ after L. monocytogenes infection.
MATERIALS AND METHODS
Animals and bacteria.
Normal 6- to 8-week-old female C57BL/6 mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan). IL-12 p40−/− breeding pairs were purchased from Jackson Laboratory (Bar Harbor, ME). Animals were housed at a specific-pathogen-free facility with an individual ventilation cage system at the Laboratory Animal Center, Tzu-Chi University, and cared for in accordance with the Institutional Animal Care and Use Committee of Tzu-Chi University. L. monocytogenes strain 10403S was provided by Eric Pamer (Sloan-Kettering Cancer Center) and grown in brain heart infusion broth (Difco) to log phase. L. monocytogenes was administrated intravenously through tail veins. In our hands, the 50% lethal dose (LD50) of L. monocytogenes was approximately 3 × 104 to 4 × 104 CFU in C57BL/6 mice. The preparation of heat-killed (HK) L. monocytogenes was as described previously (19).
Cell preparation.
Spleens were removed from naive mice and mice infected with L. monocytogenes 24 h and 48 h earlier. Splenocytes were harvested by dissociation through a wire mesh, followed by lysis of erythrocytes with ammonium chloride, and were subsequently resuspended in RP10, which consists of RPMI 1640 (Biowest) supplemented with 10% fetal calf serum, l-glutamine, HEPES, 2-mercaptoethanol, penicillin, and streptomycin. Lymph node (LN) cells were prepared from popliteal and inguinal LNs. Bone marrow cells were flushed from femurs. Blood samples were obtained via cardiac puncture. Red blood cells were lysed before staining. Cell numbers were determined by exclusion of 0.45% trypan blue. To purify NK1.1+ cells, two-step magnetic activated cell sorting (MACS) was performed. Briefly, total splenocytes were incubated with 0.7 μg/ml anti-NK1.1-biotin (e-bioscience) for 10 min at 4°C. After washing with MACS buffer (0.5% bovine serum albumin-phosphate-buffered saline [PBS] with 5 mM EDTA), anti-biotin magnetic beads (Miltenyi, Germany) were applied as per the manufacturer's instructions and incubated for 15 min at 4°C. After washing with MACS buffer, NK1.1+ cells were eluted through an LS column (Miltenyi Biotec, Bergisch Gladbach, Germany). The purified NK1.1+ cells were cytospun for immunostaining or were prepared for flow cytometric sorting.
Flow cytometry.
For intracellular staining of IFN-γ, splenocytes were treated with 2.5 μg/ml brefeldin A (BFA; Sigma) for 5 h at 37°C. Next, after blocking with 0.25 μg Fc block (ebioscience), 1 × 106 to 2 × 106 splenocytes were incubated in staining buffer (PBS-0.5% bovine serum albumin-0.02% sodium azide) for 1 h on ice in the presence of various monoclonal antibodies (MAbs) for surface staining. Dead cells were excluded by staining with 1.25 μg/ml ethidium monoazide bromide (Sigma) and exposure to light during the last 10 min of staining to cross-link DNA-bound ethidium monoazide bromide (16). Cells were subsequently washed three times in staining buffer, fixed in 1% paraformaldehyde-PBS for 10 min, and permeabilized with 0.1% saponin (Sigma) for 10 min before applying anti-IFN-γ-fluorescein isothiocyanate (FITC) MAb (clone XMG1.2; Caltag). For staining of CD68, anti-CD68-biotin MAb (clone FA-11; Serotec) was mixed with anti-IFN-γ-FITC MAb. After washing with staining buffer, streptavidin-allophycocyanin (ALPC; Caltag) was applied. Flow cytometric acquisition was performed on a FACSCalibur. We usually collected 2.5 × 105 of total splenocytes, and data were further analyzed with CellQuest Pro software (Becton Dickinson). The following MAbs were from Caltag: anti-CD4-phycoerythrin (PE, clone RM4-5), anti-CD8α-PE (clone 5H10), anti-Gr-1-PE (clone RB68C5), anti-T-cell receptor γδ (TCRγδ)-PE (clone GL3), and anti-CD8α-ALPC (clone 5H10). The following were from Biolegend: anti-NK1.1-PE (clone PK136), normal mouse immunoglobulin G2a (IgG2a)-PE (clone MOPC-173), anti-CD11c-PE (clone N418), anti-CD44-PE (clone IM7), anti-CD11c-ALPC (clone N418), normal hamster IgG-ALPC (clone HTK888), and anti-TCRβ-ALPC (clone H57-597).
Immunofluorescence microscopy.
For immunofluorescence studies, spleen tissue was embedded in OCT (Sakura Finetechnical, Japan), snap-frozen in liquid nitrogen, cryosectioned, and fixed for 30 s in cold acetone. Sections or cytospun slides were first incubated in 1% newborn calf serum-PBS for 30 min at room temperature. For simultaneous detection of L. monocytogenes, Ly49G2, and CD11c, the following antibodies were applied sequentially: rabbit-anti-L. monocytogenes serum (Difco), donkey anti-rabbit-FITC, rat anti-Ly49G2 MAb (clone 4D11; BD), donkey anti-rat-rhodamine, hamster anti-CD11c-biotin (clone HL3; BD), goat anti-biotin (Sigma), and donkey anti-goat-aminomethylcoumarin acetate. After application of antibodies, sections were incubated for 40 min at room temperature in a wet chamber. Between each step, the sections were washed for 10 min in PBS-0.1% Tween 20. A control staining with appropriate primary antibodies, namely normal rabbit serum, normal rat IgG, and normal hamster IgG-biotin (all from Jackson ImmunoResearch), was also prepared and showed no background staining of secondary antibodies used. For simultaneous detection of IFN-γ, Ly49G2, and CD11c, the following antibodies were applied: rabbit anti-IFN-γ (biosource), donkey anti-rabbit-FITC, rat anti-Ly49G2, donkey anti-rat-rhodamine, hamster anti-CD11c-biotin, goat anti-biotin, and donkey anti-goat-aminomethylcoumarin acetate. All fluorochrome-conjugated antibodies were from Jackson ImmunoResearch. The photographs were taken using a Nikon E400 fluorescent microscope with a Coolpix 9500 digital camera.
RESULTS
NK, DC, and T cells express IFN-γ after L. monocytogenes infection.
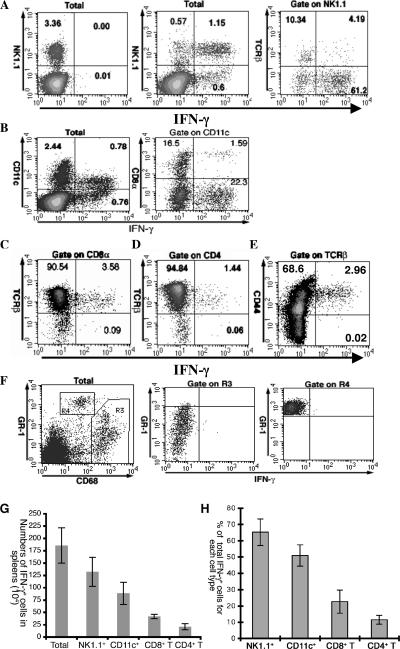
To test whether we could detect IFN-γ-expressing cells, we first infected mice with 105 L. monocytogenes organisms, a lethal dose that would kill the mice at around 72 hpi (Fig. 1). Approximately 1.57% ± 0.09% (average ± standard deviation, n = 4) of splenocytes expressed IFN-γ at 24 hpi (Fig. 1A, middle plot). The expression of IFN-γ was dependent on L. monocytogenes stimulation, since naive splenocytes did not express IFN-γ (0.02% ± 0.01% of splenocytes) (Fig. 1A, left plot). The staining of IFN-γ was highly specific. To rule out that the detected IFN-γ was induced by endogenous IL-12 during in vitro BFA incubation prior to staining, anti-IL-12 antibodies were added during incubation, but the results were not affected (data not shown). Costaining with various cell surface markers revealed that NK1.1+ cells, CD11c+ cells, and T cells were primary cell types expressing IFN-γ (Fig. 1A to D). Costaining of NK1.1 and TCRβ revealed that, though both NK cells and NK T cells expressed IFN-γ, the majority of IFN-γ+ NK1.1+ cells were not NK T cells (Fig. 1A, right plot).
FIG. 1.
NK1.1+, CD11c+, and T cells express IFN-γ 24 hpi. Splenocytes from naive mice (A, left plot) and C57BL/6 mice infected with 105 CFU L. monocytogenes intravenously 24 h earlier (all other plots) were stained for intracellular IFN-γ after staining of NK1.1 and TCRβ (A), CD11c and CD8α (B), CD8 and TCRβ (C), CD4 and TCRβ (D), CD44 and TCRβ (E), and CD68 and Gr-1 (F). Plots were gated on live splenocytes (label “Total”) or surface markers as indicated above each plot. The numbers in the quadrants indicate the percentage of cells in each quadrant. All plots except the naive one (A, left plot) are from the same representative mouse of four mice. (G) The numbers of IFN-γ+ cells are enumerated for each cell type. (H) The contribution of each IFN-γ-expressing cell type to the total number of IFN-γ+ cells is presented. The data represent the averages of results for four mice and one of three independent experiments. Error bars represent the standard deviations.
In agreement with the results of Ohteki et al., we demonstrated that CD11c+ cells did express IFN-γ (Fig. 1B, left plot). But contrary to their report (24), we found that the majority of IFN-γ+ DC were CD8α− DC (Fig. 1B, right plot). They constituted 93.3% of IFN-γ+ DC.
To verify whether T cells express IFN-γ postinfection, we costained the splenocytes for CD8 and TCRβ (Fig. 1C) as well as CD4 and TCRβ (Fig. 1D). As shown in cytometric plots, significant numbers of both CD8+ T cells and CD4+ T cells expressed IFN-γ. The population of IFN-γ+ CD8+ T cells was equivalent to 3.8% of the total CD8+ T cells, while IFN-γ+ CD4+ T cells were about 1.5% of the total CD4+ T cells. In addition, these IFN-γ+ T cells had a CD44high phenotype (Fig. 1E), indicating that they were recently activated or memory cells. No significant IFN-γ expression was found on macrophages and neutrophils (Fig. 1F, middle and right plots). The macrophages were defined as CD68+ cells (R3 in Fig. 1F, left plot), while neutrophils were defined as Gr-1highCD68− (R4) as defined by Kirby et al. (17). γδ T cells were also negative for IFN-γ staining (data not shown).
The presence of NK1.1+ CD11+ cells in spleen.
We identified NK1.1+, CD11c+, CD8+ T, and CD4+ T cells as four IFN-γ-expressing cell types after L. monocytogenes infection. To determine their relative contribution to total IFN-γ production, we enumerated the numbers of IFN-γ+ cells for these four cell types (Fig. 1G) and calculated their percentage relative to the total number of IFN-γ-expressing cells (Fig. 1H). We found that NK1.1+ cells were the major IFN-γ-expressing cells (65.3% ± 3.1%), followed by CD11c+ cells (51% ± 3%), CD8+ T cells (22.7% ± 2.1%), and CD4+ T cells (11.6% ± 0.25%) at 24 hpi. Unexpectedly, the sum (150% ± 8.45%) of these four IFN-γ-expressing cells far exceeded the total number of IFN-γ+ cells. These results were unlikely an outcome of nonspecific staining, since the staining of IFN-γ was highly specific as described above. Second, all surface marker antibodies were common clones and widely used. Therefore, we reasoned that there might exist bimarker cells.
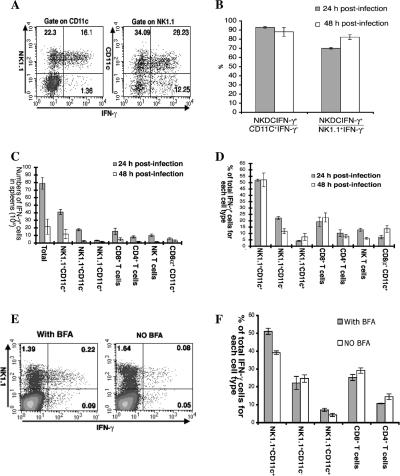
Since NK.1.1+ cells and CD11c+ cells were relatively the most abundant IFN-γ-expressing cells, it prompted us to examine whether some cells expressed both NK1.1 and CD11c. Indeed, costaining of naive splenocytes for NK1.1 and CD11c (Fig. 2A, left plot) revealed a population of cells expressing both NK1.1 and CD11c markers. Surprisingly, these NK1.1+ CD11c+ cells constituted about 50% of NK.1.1+ cells. To rule out that trace amounts of residual Ca2+ and Mg2+ in the cell culture medium used in the splenocyte preparations might lead to cell-cell adhesion, 10 mM EDTA was added to the staining buffer during staining. However, the population of NK1.1+ CD11c+ cells did not significantly change (Fig. 2A, right plot). Also, applying isotype control antibodies for anti-CD11c-ALPC (Fig. 2B) and anti-NK1.1-PE (Fig. 2C) confirmed that the staining was specific and that the double-positive cells were not a result of incorrect flow cytometric compensation.
FIG. 2.
The presence of NK1.1+ CD11c+ cells in the spleen. (A to C) Splenocytes from a naive C57BL/6 mouse were stained with anti-NK1.1-PE and anti-CD11c-ALPC in the presence or absence of 10 mM EDTA (A), anti-NK1.1-PE and normal hamster IgG-ALPC (isotype control for anti-CD11c-ALPC) (B), and normal mouse IgG2a-PE and anti-CD11c-ALPC (C). Plots were gated on live splenocytes, and the numbers in each quadrant indicate the percentages of cells in each quadrant. (D to F) The percentage of CD11c+ cells from sorted NK1.1+ cells. Enriched NK1.1+ cells (by MACS) were stained with anti-NK1.1-PE and then sorted on the NK1.1+ population. The purity of sorted cells is presented in panel D. (E) The sorted NK1.1+ cells were then cytospun and stained for Ly49G2 (red) and CD11c (green). Arrows in the plots indicate cells stained both green and red, while symbols *, #, and & denote cells stained green or red or without any staining, respectively. (F) From stained slides, the percentages of Ly49G2+, CD11c+, and Ly49G2+ CD11c+ cells were determined. Twelve random photographs were taken from each of four slides, and the results of the four slides were averaged and presented. The presented data are representative of two independent experiments. Error bars represent the standard deviations.
To further verify that splenocytes really contained a significant population of NK1.1+ CD11c+ cells, we determined the percentage of NK1.1+ CD11c+ cells of sorted NK1.1+ cells on cytospun slides. After sorting, we achieved a purity of over 95% (Fig. 2D). Sorted NK1.1+ cells were cytospun on slides and stained for Ly49G2 in red and CD11c in green (Fig. 2E). Ly49G2 was chosen as a marker for NK cells because Dokun et al., having previously screened a panel of antibodies, found only MAb 4D11 (anti-Ly49G2) was suitable for in situ staining of NK cells though only half of NK cells expressed Ly49G2 (10). The result showed that CD11c+ cells constituted 44% ± 7.26% of sorted NK1.1+ cells, while Ly49G2+ CD11c+ cells represented 26.7% ± 6% of sorted NK1.1+ cells (Fig. 2F). Consistent with the results of Dokun et al., only about half of sorted NK1.1+ cells were Ly49G2+. Thus, we concluded that splenocytes really contained a significant population of NK1.1+ CD11c+ cells, which might constitute up to 50% of NK1.1+ cells, and that the cytometrically stained NK1.1+ CD11c+ cells were not doublets consisting of NK1.1+ cells associated with CD11c+ cells.
NK1.1+ CD11c+ cells are primary IFN-γ-expressing cells after L. monocytogenes infection.
With the identification of NK1.1+ CD11c+ cells, we then reevaluated IFN-γ expression on NK1.1+ and CD11c+ cells. When gated on CD11c+ cells, the majority of IFN-γ+ CD11c+ cells actually coexpressed NK1.1 (Fig. 3A, left plot), and they represented 93% ± 0.89% of total IFN-γ+ CD11c+ cells (Fig. 3B, 24 hpi). When gated on NK1.1+ cells, NK1.1+ CD11c+ cells still constituted the major proportion of IFN-γ+ cells, but a significant number of NK.1.1+ CD11c− cells also expressed IFN-γ (Fig. 3A, right plot). IFN-γ+ NK1.1+ CD11c+ cells made up 70.13% ± 0.81% of total IFN-γ+ NK1.1+ cells (Fig. 3B, 24 hpi).
FIG. 3.
NK1.1+ CD11+ cells are primary IFN-γ-expressing cells in spleens after L. monocytogenes infection. (A and B) Analysis of IFN-γ expression on NK1.1+ and CD11c+ cells. Splenocytes from C57BL/6 mice infected with 104 L. monocytogenes 24 h (A and B) or 48 h (B) earlier were stained for intracellular IFN-γ after surface staining of NK1.1 and CD11c. (A) The representative flow plots display NK1.1 staining (left plot) or CD11c staining (right plot) on the y axis and IFN-γ staining on the x axis after gating on CD11c+ cells (left plot) or NK1.1+ cells (right plot). The numbers in the quadrants are the percentages of cells in each quadrant. (B) The ratios of IFN-γ+ NK1.1+ CD11+ cells to total IFN-γ+ CD11c+ or to IFN-γ+ NK1.1+ cells are presented. (C and D) Identification of NK1.1+ CD11+ cells as primary IFN-γ-expressing cells in spleens after L. monocytogenes infection. Splenocytes from mice 24 and 48 hpi were stained for intracellular IFN-γ after surface staining of NK1.1 and CD11c and various combinations of surface markers as for Fig. 1. (C) The numbers of IFN-γ+ cells were calculated for each cell type. The P value (t test) for the number of total IFN-γ-expressing cells at 24 versus 48 hpi is 1 × 10−4. (D) The contribution of each IFN-γ-expressing cell type to the total number of IFN-γ+ cells is presented. The P value (t test) for NK1.1+ CD11c+ cells versus NK1.1+ CD11c− cells is 1 × 10−8. (E and F) Comparison of IFN-γ staining with or without BFA incubation. Splenocytes from mice infected with 104 L. monocytogenes 24 h earlier were stained for intracellular IFN-γ after surface staining of NK1.1 and CD11c, CD8 and TCRβ, and CD4 and TCRβ. (E) The representative flow plots with (left plot) or without (right plot) BFA incubation display NK1.1 staining on the y axis and IFN-γ staining on the x axis. (F) Comparison of the contribution of each IFN-γ-expressing cell type to total number of IFN-γ-expressing cells using the staining with or without BFA incubation protocols. All data shown represent the averages of results for four mice and one of two independent experiments. Error bars represent the standard deviations.
Next, to evaluate the relative contribution of NK1.1+ CD11c+ cells to total IFN-γ expression, we reanalyzed the IFN-γ-expressing cells with a panel of various surface antibodies (Fig. 3C and D). In addition, in our previous experiments, we infected mice with 105 CFU L. monocytogenes, a lethal dose (Fig. 1), and identified large numbers of IFN-γ-expressing cells 24 hpi. But we also found a dramatic decrease in IFN-γ+ cells 48 hpi (data not shown). Nonetheless, the mice were very sick when given such a high dose of L. monocytogenes, and they usually died within the next 24 h. To test whether such a decrease in IFN-γ-expressing cells also occurred at sublethal doses, mice were given 104 CFU L. monocytogenes. As shown in Fig. 3C and D, NK1.1+ CD11c+ cells were the primary IFN-γ-expressing cells 24 hpi (51.96% ± 0.8%), followed by NK1.1+ CD11c− cells (22.14% ± 1.14%), CD8+ T cells (19.25% ± 3.61%), CD4+ T cells (10.28% ± 2.59%), and finally, NK1.1− CD11c+ cells (4.14% ± 0.5%). The sum of these five populations was 107.87% ± 8.64% of total IFN-γ+ cells. We thought it was within a reasonable range of experimental error. The P value (t test) for NK1.1+ CD11c+ cells versus NK1.1+ CD11c− cells is 1 × 10−8. Thus, we concluded that NK1.1+ CD11c+ cells represented the major cell type expressing IFN-γ after L. monocytogenes infection and that the majority of IFN-γ+ CD11c+ cells were actually NK1.1+ CD11c+ cells (Fig. 3A, left plot, and Fig. 3B).
Comparing IFN-γ expression at 24 and 48 hpi, we found that IFN-γ expression peaked at about 24 hpi and diminished rapidly thereafter. There was a dramatic reduction in the number of IFN-γ+ cells 48 hpi (about 27.2% of that 24 hpi) (Fig. 3C). The P value for the number of total IFN-γ-expressing cells at 24 versus 48 hpi is 1 × 10−4. This reduction was synchronous among all cell populations, suggesting a common mechanism.
To rule out that our result might be biased by in vitro BFA incubation, we performed intracellular cytokine staining without BFA incubation. In the absence of BFA treatment, the number of IFN-γ-expressing cells was only about 40% of that with BFA treatment (Fig. 3E). However, NK1.1+ CD11c+ cells were still the primary IFN-γ-expressing cells (Fig. 3F). Therefore, we believe that our results faithfully reflect the situation in vivo.
Identification of IFN-γ-expressing NK1.1+ CD11c+ cells in situ.
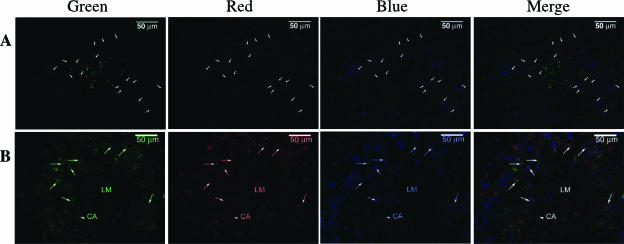
To provide further evidence that NK1.1+ CD11c+ cells expressed IFN-γ, we sought to identify IFN-γ-expressing NK1.1+ CD11c+ cells in situ. To elucidate the localization of NK cells relative to infectious foci, splenic sections from naive (data not shown) and 24-hpi mice were stained for L. monocytogenes, Ly49G2, and CD11c in green, red, and blue, respectively (Fig. 4A). In agreement with other's findings (23), we found L. monocytogenes usually formed infectious foci in the white pulps of spleens. Consistent with the finding of Dokun et al., in naive spleens, Ly49G2+ cells were primarily located at red pulp (10). In contrast, numerous Ly49G2+ cells and CD11c+ cells were found at the border of infectious foci. Many Ly49G2+ cells actually coexpressed CD11c, as indicated in Fig. 4A. Since MAb 4D11 only stained half of NK1.1+ cells, many Ly49G2− CD11c+ cells around infectious foci might actually be NK1.1+ CD11c+ cells, too. Though not precisely, we estimated that Ly49G2+ CD11c+ cells at least constituted 30% of the total Ly49G2+ cells. Thus, NK1.1+ CD11c+ cells did represent a very significant proportion of NK1.1+ cells or CD11c+ cells. Next, we wanted to see whether Ly49G2+ CD11c+ cells expressed IFN-γ. The splenic sections were stained for IFN-γ, Ly49G2, and CD11c in green, red, and blue, respectively (Fig. 4B). As indicated by arrows, a lot of IFN-γ+ Ly49G2+ CD11c+ cells were found around a putative “infectious focus” (an empty space without any staining), indicated by an “LM” label. No IFN-γ was detected in the naive spleen (data not shown).
FIG. 4.
NK1.1+ CD11+ cells express IFN-γ in situ. Splenic frozen sections from mouse infected with 4,000 L. monocytogenes organisms 24 h earlier were stained for L. monocytogenes (green), Ly49G2 (red), and CD11c (blue) (A) or IFN-γ (green), Ly49G2 (red), and CD11c (blue) (B). (A) The arrows indicate Ly49G2+ CD11c+ cells. (B) An arrowhead indicates a central arteriole (CA), and the arrows denote the Ly49G2+ CD11c+ IFN-γ+ cells. “LM” indicates the putative infectious focus.
IFN-γ expression is mainly dependent on IL-12 p40 and live L. monocytogenes infection.
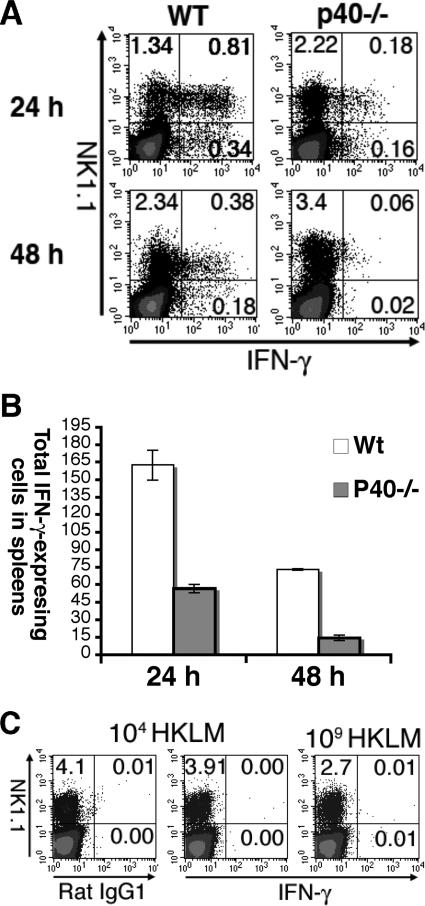
IL-12/IL-23 has been regarded as the primary IFN-γ inducer (32). However, IL-12/IL-23-independent IFN-γ expression has also been shown in other infectious models (25). To examine whether IFN-γ expression after L. monocytogenes infection is dependent on IL-12/IL-23, we analyzed IFN-γ expression in IL-12 p40−/− mice. IFN-γ-expressing cells in IL-12 p40−/− mice 24 hpi were significantly decreased, indicating that IFN-γ expression 24 hpi was largely IL-12/IL-23 dependent (Fig. 5A and B). The P value for the number of IFN-γ-expressing cells between wild-type and IL-12 p40−/− mice is 1 × 10−4. For two independent experiments, the IFN-γ-expressing cells in IL-12 p40−/− mice were only about 34.9% and 35.1%, respectively, of those of wild-type mice. Nonetheless, our result also clearly demonstrated the existence of IL-12 p40-independent IFN-γ expression after L. monocytogenes infection. Consistent with earlier findings, IFN-γ expression was rapidly down-regulated by 48 hpi.
FIG. 5.
IFN-γ expression is largely dependent on IL-12 p40 and live L. monocytogenes infection. (A and B) Wild-type C57BL/6 and IL-12 p40−/− mice were infected with 104 L. monocytogenes organisms. Splenocytes from 24 and 48 hpi were stained for NK1.1 and intracellular IFN-γ. (A) The representative flow plots display NK1.1 staining on the y axis and IFN-γ staining on the x axis. The plots are gated on live cells. The numbers in the quadrants indicate the percentages of cells in each quadrant. (B) The numbers of total IFN-γ+ cells are presented. The data are the averages of results for three mice and represent one of two independent experiments. Error bars represent the standard deviations. The P value for the number of IFN-γ-expressing cells between wild-type and IL-12 p40−/− mice is 1 × 10−4. (C) Wild-type C57BL/6 mice were inoculated with 104 (left and middle plots) or 109 (right plot) heat-killed L. monocytogenes organisms intravenously. IFN-γ expression 24 h later was analyzed and displayed as above. The left plot displays the staining of isotype control.
Next, since HK L. monocytogenes inoculation was shown not to induce IL-12 expression (34), we would like to know whether the IFN-γ expression after HK L. monocytogenes inoculation was different from live infection. To our surprise, we were unable to detect any IFN-γ expression with either low (104) or high (109) doses of HK L. monocytogenes inoculation (Fig. 5C). Thus, IFN-γ expression is absolutely dependent on live infection.
NK1.1+ CD11c+ cells are present in naive spleen, LN, blood, and bone marrow cells, and their amount increases after L. monocytogenes infection.
The presence of NK1.1+ CD11c+ cells in the naive spleen (Fig. 2) suggested that they are not induced by L. monocytogenes infection. So it was interesting to know whether they were present in other lymph organs. Costaining for NK1.1 and CD11c in LN, blood, and bone marrow cells revealed significant populations of NK1.1+CD11c+ cells (Fig. 6A). They represented 41.4% ± 2.95%, 44.1% ± 4.27%, 42% ± 4.34%, and 32.2% ± 3.21% of total NK1.1+ cells within splenocytes, LN cells, blood cells (lysed red blood cells), and bone marrow cells, respectively. On the contrary, fewer NK1.1+ CD11c+ cells could be found in the thymus, constituting only 8.4% ± 1.47% of the total numbers of NK1.1+ cells within the thymus (Fig. 6B). Comparing the naive and 72 hpi mice revealed a dramatic increase in the number of NK1.1+ CD11c+ cells in the spleen, LN, and bone marrow (Fig. 6C to F) (P < 0.01 for all organs). This increase was largely due to a relative increase in the percentage of NK1.1+ CD11c+ cells in each organ, as the ratio of NK1.1+ CD11c+ cells over total NK1.1+ cells was significantly increased, too (Fig. 6D) (P < 0.01). The total numbers of cells in the spleen, LN, and bone marrow were only slightly increased or unchanged (data not shown).
FIG. 6.
NK1.1+ CD11+ cells are present in naive spleen, LN, blood, and bone marrow, and their amount increases after infection. (A and B) Splenocytes, blood, LN, and bone marrow cells and thymocytes from naive C57BL/6 mice were stained for NK1.1 and CD11c. (A) The representative flow plots display NK1.1 staining on the y axis and CD11c staining on the x axis. The plots are gated on total live cells. The numbers in the quadrants indicate the percentages of cells in each quadrant. (B) Percentages of NK1.1+ CD11+ cells of total NK1.1+ cells from each indicated organ are presented. The results are the averages from four mice from one of two independent experiments. Error bars represent the standard deviations. (C to F) Splenocytes, blood, LN, and bone marrow cells from naive and 72 hpi C57BL/6 mice were stained for NK1.1 and CD11c. (C) The representative flow plots display NK1.1 staining on the y axis and CD11c staining on the x axis as described above. (D) Percentages of NK1.1+ CD11+ cells of total NK1.1+ cells from each indicated organ are presented. (E) The total number of NK1.1+ CD11c+ cells in spleen is presented. (F) The total number of NK1.1+ CD11c+ cells in LN and femur is presented. The P value between naive mice and 72 hpi mice is <0.01 for each indicated organ. The results are the averages from three mice per group from one of two independent experiments. Error bars represent the standard deviations.
DISCUSSION
In this study, we used intracellular cytokine staining to define the early source of IFN-γ after L. monocytogenes infection. In our approach, the only manipulation was a 5-h in vitro BFA incubation prior to the staining of splenocytes. By minimizing in vivo and in vitro manipulation, we believe our data faithfully reflect what happens in vivo during L. monocytogenes infection. Indeed, the hierarchy in the numbers of IFN-γ-expressing cells was the same in the presence or absence of an in vitro BFA incubation (Fig. 3G). Our analysis not only recapitulates several previous findings but also provides new insights into IFN-γ expression in vivo.
We found that NK1.1+, CD11c+, and T cells expressed IFN-γ early after L. monocytogenes infection. Surprisingly, almost all IFN-γ+ CD11c+ cells were in fact NK1.1+ CD11+ cells (Fig. 3). Several authors had reported that DC (defined as CD11c+ cells) expressed IFN-γ after in vitro stimulation with IL-12 (24, 29). It is our belief that if these DC were costained by an antibody against NK1.1, many of these cells would actually be NK1.1+ CD11+ cells. As our study indicated, at least in an in vivo infection setting, CD11c+ NK1.1− cells only had a very limited capacity to produce IFN-γ (Fig. 3A, left plot). Our result is consistent with a recent report that DX5+ CD11clo cells, but not DX5− CD11chigh cells, express IFN-γ after stimulation with IL-12 and IL-18 (18). In addition, we found that the majority of IFN-γ+ CD11c+ cells were CD8α−. This is in stark contrast to the report of Ohteki et al. that sorted CD8α+ DCs produce IFN-γ in vitro in an IL-12-dependent way (24). It is difficult to reconcile this discrepancy. One explanation might be that CD8α+ DCs are “restrained” to receive IL-12 in vivo, even though they can respond to IL-12 in vitro. Whatever the explanation, caution is needed when extrapolating an in vitro observation into an in vivo scenario.
T cells (both CD4+ and CD8+) are another source of early innate IFN-γ after L. monocytogenes infection. They represent about 30% of the total IFN-γ-expressing cells. And those IFN-γ-expressing T cells have a CD44high phenotype. Our results are consistent with an earlier report that T cells are one of the major sources of early IFN-γ after L. monocytogenes infection (1). Indeed, memory T cells have been shown to rapidly induce IFN-γ expression in response to non-TCR stimuli both in vitro and in vivo (5, 6).
Using IL-12 p40−/− mice, we found that IFN-γ expression was largely IL-12/IL-23 dependent, but IL-12/IL-23-independent IFN-γ expression also contributed to about one-third of total IFN-γ (Fig. 5). Our result fits well with previous findings that IL-12 p35−/− mice, unlike IFN-γ-deficient mice, could resist low doses of L. monocytogenes infection (7). Their data implied that there was an IL-12-independent source of IFN-γ, and here we provided direct evidence for both IL-12/IL-23-dependent and -independent IFN-γ expression.
HK L. monocytogenes inoculation is notorious for its failure to induce effective Th1 and CD8+ T-cell responses. Here, we showed that the expression of early innate IFN-γ was also dependent on live infection. Our result is consistent with an earlier report that no IFN-γ could be detected by PCR and enzyme-linked immunosorbent assay after HK L. monocytogenes administration (33). One possibility for lack of IFN-γ expression after HK L. monocytogenes administration might be that IFN-γ induction requires a cytosolic invasion by live L. monocytogenes. Indeed, it has been shown that induction of IL-12 in macrophage needs the invasion of L. monocytogenes into the cytosol (21). Further investigation with listeriolysin O-deficient L. monocytogenes will clarify this important issue.
We found a very tight regulation of expression of IFN-γ after L. monocytogenes infection. When mice were infected with a sublethal dose of L. monocytogenes, the amount of IFN-γ-expressing cells peaked at about 24 hpi and then rapidly declined thereafter. This finding is significant in that the numbers of L. monocytogenes in infected spleens peak around day 3 after infection. It means that IFN-γ expression is turned down even before its original inducers reach their peak, probably as a mechanism to avoid the detrimental effect of uncontrolled production of IFN-γ. The mechanisms for this rapid down-regulation require further investigation. Moreover, the early down-regulation of IFN-γ expression implies that IFN-γ acts within a very early window of innate response. Indeed, this is consistent with the report of Harty and Bevan that IFN-γ is essential for defense of primary L. monocytogenes infection but is dispensable in secondary L. monocytogenes infection once the T cells responses are induced by anattenuated strain of L. monocytogenes (11). Their data suggest that IFN-γ primarily acts during the innate phase and that T-cell-mediated immunity can overcome the requirement of IFN-γ in the innate phase.
We unexpectedly identified NK1.1+ CD11+ cells as the primary IFN-γ-expressing cell in vivo. Since conventional CD4+ and CD8+ T cells have been shown to induce NK1.1 expression after activation (28) and since activated T cells might also express CD11c (13), some NK1.1+ CD11c+ cells might actually be activated T cells. However, from our analysis, we knew that the majority of IFN-γ-expressing NK1.1+ cells did not express TCRβ (Fig. 1A, right plot). Therefore, they are not activated T cells.
Although NK1.1+ CD11+ cells appeared as a novel cell type to us, we searched scientific literature for the existence of this population. Joisen et al.'s report that depicted a subset of cytolytic DCs in rats was probably the first to describe this population (14). Later, Homann et al. described DX5+ CD11c+ cells as regulatory cells and their role in preventing autoimmune diabetes after CD40L blockade (12). More recently, Pillarisetty et al. described their discovery of NK1.1+ CD11+ cells and referred to these cells as natural killer dendritic cells (NKDC). They showed that NKDC produced IFN-γ in response to CpG via autocrine IL-12 in vitro and had antigen-presenting function in vitro (27). Consistent with their data, we found that only a small fraction of NK1.1+ CD11c+ cells expressed major histocompatibility class II (data not shown). Nonetheless, we are not sure whether the NK1.1+ CD11+ cells we defined are identical to NKDC, since their abilities for antigen presentation are still under investigation. Besides, we want to emphasize one significant difference between our data and that of Pillarisetty et al., which is that, in this study, NK1.1+ CD11+ cells were never found to be a minor population in spleen, LN, blood, or bone marrow cells, unlike in their report. For example, in the spleen, we usually found that NK1.1+ CD11+ cells represented 40 to 65% of total NK1.1+ cells or 20 to 60% of total CD11c+ cells by flow cytometric analysis. But in their report, NKDC (also defined as NK1.1+ CD11+ cells) only constituted 2.75% of NK1.1+ or 4% of CD11c+ cells (see Table 1 in reference 27). In addition to cytometric analysis, staining of both cytospun slides and splenic sections indicated that NK1.1+ CD11+ cells did represent a very significant proportion of NK1.1+ cells. In agreement with our result, Homann et al. also found that, in uninfected mice, DX5+ CD11c+ cells constituted 30% of CD11c+ cells (see page 408 of reference 12). Lastly, during our preparation of the manuscript, two articles reported that a population of NK1.1+ CD11cint B220+ cells or DX5+ CD11cint B220+ cells, designated as interferon-producing killer dendritic cells, were major IFN-γ-producing cells after stimulation and they also possessed the cytolytic capacity as well as antigen-presenting cell activity (8, 30). These reports are consistent with our findings and point out that NK1.1+ CD11c+ cells may be heterogeneous.
Our finding that NK1.1+ CD11c+ cells are primary IFN-γ-expressing cells has significant meaning in the defense of L. monocytogenes infection. Since IFN-γ is essential for the defense of L. monocytogenes infection, that means NK1.1+ CD11c+ cells contribute significantly to the control of infection, as they represent 50% of total IFN-γ-expressing cells. Besides the contribution of IFN-γ, NK1.1+ CD11c+ cells may help clear the infection indirectly through participating in T-cell priming, a function of NK1.1+ CD11c+ cells, as suggested by other investigators. It is well established that sterile clearance of L. monocytogenes infection requires effective T-cell responses. Moreover, since NK1.1+ CD11c+ cells may possess both cytolytic and antigen-presenting abilities, they may play a special role in the defense of L. monocytogenes infection by killing the infected cells and then facilitating an effective T-cell response.
Regardless of whether NK1.1+ CD11c+ cells are truly bifunctional, the fact that they may constitute a very significant proportion of NK1.1+ or CD11c+ cells will have an impact on the phenomenon of NK-DC interaction, a field with increasing attention (22). Traditionally, in the murine system, we define NK1.1+ cells as “NK” cells and CD11c+ cells as “DC.” If “NK” cells and “DC” could be the same cells, special attention should be paid when describing the interaction between these “two” populations. For instance, several authors have used MAb to deplete NK1.1+ cells in vivo, observed a decrease in the numbers of DC or subset of DC, and then inferred from their results that there were interactions between “NK” cells and “DC” in vivo (2, 26). Could this conclusion be misled, at least partially, by the depletion of NK1.1+ CD11+ cells? This issue will be critical when NK1.1+ CD11+ cells may represent up to 50% of NK1.1+ cells or CD11c+ cells, as our data showed.
The development and origin of NK1.1+ CD11+ cells are other interesting questions. Are they an activated status or developmental stage of NK cells or DC? However, we found numerous NK1.1+ CD11+ cells in the bone marrow, blood, LN, and spleen of naive mice. It implies that there are “endogenous” stimuli if they are developed from NK cells or CD11c+ cells. Alternatively, they may represent a unique cell lineage distinct from NK and DC. In this case, it will be interesting to know whether NK1.1+ CD11+ cells share the same progenitor and developmental pathway with NK or DC.
We found a dramatic increase in the population of NK1.1+ CD11c+ cells in the spleen and LN after L. monocytogenes infection. Even more surprising to us, a similar increase was found in the blood and bone marrow. This suggests that the expanded populations in the spleen and LN are mainly recruited from blood, not expanded in situ. Moreover, our result suggested that L. monocytogenes infection somehow altered the hematopoiesis in bone marrow. Since we administrated L. monocytogenes intravenously, it will be interesting to know whether L. monocytogenes establishes an infectious focus in the bone marrow. If not, it implies that immune responses elicited by L. monocytogenes in other organs regulate hematopoiesis remotely.
In summary, we have identified NK1.1+ CD11+ cells as primary IFN-γ-expressing cells in vivo after L. monocytogenes infection. A significant proportion of NK1.1+ and CD11c+ cells were actually NK1.1+ CD11+ cells even in the naive spleen, LN, blood, and bone marrow. The functional similarities and differences, as well as developmental relationships among NK1.1+ CD11+, NK1.1+ CD11c−, and NK1.1− CD11c+ cells require further investigation.
Acknowledgments
We thank M. Chang for critical review of the manuscript.
This work was supported by grant 92-2320-B-320-032 and grant 93-2320-B-018 from the National Science Council, Taiwan, to S.-L.W.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 11 December 2006.
REFERENCES
- 1.Andersson, A., W. J. Dai, J. P. Di Santo, and F. Brombacher. 1998. Early IFN-gamma production and innate immunity during Listeria monocytogenes infection in the absence of NK cells. J. Immunol. 161:5600-5606. [PubMed] [Google Scholar]
- 2.Andrews, D. M., A. A. Scalzo, W. M. Yokoyama, M. J. Smyth, and M. A. Degli-Esposti. 2003. Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 4:175-181. [DOI] [PubMed] [Google Scholar]
- 3.Bancroft, G. J., R. D. Schreiber, G. C. Bosma, M. J. Bosma, and E. R. Unanue. 1987. A T cell-independent mechanism of macrophage activation by interferon-gamma. J. Immunol. 139:1104-1107. [PubMed] [Google Scholar]
- 4.Bancroft, G. J., K. C. Sheehan, R. D. Schreiber, and E. R. Unanue. 1989. Tumor necrosis factor is involved in the T cell-independent pathway of macrophage activation in scid mice. J. Immunol. 143:127-130. [PubMed] [Google Scholar]
- 5.Berg, R. E., C. J. Cordes, and J. Forman. 2002. Contribution of CD8+ T cells to innate immunity: IFN-gamma secretion induced by IL-12 and IL-18. Eur. J. Immunol. 32:2807-2816. [DOI] [PubMed] [Google Scholar]
- 6.Berg, R. E., E. Crossley, S. Murray, and J. Forman. 2003. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J. Exp. Med. 198:1583-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brombacher, F., A. Dorfmuller, J. Magram, W. J. Dai, G. Kohler, A. Wunderlin, K. Palmer-Lehmann, M. K. Gately, and G. Alber. 1999. IL-12 is dispensable for innate and adaptive immunity against low doses of Listeria monocytogenes. Int. Immunol. 11:325-332. [DOI] [PubMed] [Google Scholar]
- 8.Chan, C. W., E. Crafton, H. N. Fan, J. Flook, K. Yoshimura, M. Skarica, D. Brockstedt, T. W. Dubensky, M. F. Stins, L. L. Lanier, D. M. Pardoll, and F. Housseau. 2006. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat. Med. 12:207-213. [DOI] [PubMed] [Google Scholar]
- 9.Dai, W. J., W. Bartens, G. Kohler, M. Hufnagel, M. Kopf, and F. Brombacher. 1997. Impaired macrophage listericidal and cytokine activities are responsible for the rapid death of Listeria monocytogenes-infected IFN-gamma receptor-deficient mice. J. Immunol. 158:5297-5304. [PubMed] [Google Scholar]
- 10.Dokun, A. O., D. T. Chu, L. Yang, A. S. Bendelac, and W. M. Yokoyama. 2001. Analysis of in situ NK cell responses during viral infection. J. Immunol. 167:5286-5293. [DOI] [PubMed] [Google Scholar]
- 11.Harty, J. T., and M. J. Bevan. 1995. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity 3:109-117. [DOI] [PubMed] [Google Scholar]
- 12.Homann, D., A. Jahreis, T. Wolfe, A. Hughes, B. Coon, M. J. van Stipdonk, K. R. Prilliman, S. P. Schoenberger, and M. G. von Herrath. 2002. CD40L blockade prevents autoimmune diabetes by induction of bitypic NK/DC regulatory cells. Immunity 16:403-415. [DOI] [PubMed] [Google Scholar]
- 13.Huleatt, J. W., and L. Lefrancois. 1995. Antigen-driven induction of CD11c on intestinal intraepithelial lymphocytes and CD8+ T cells in vivo. J. Immunol. 154:5684-5693. [PubMed] [Google Scholar]
- 14.Josien, R., M. Heslan, J. P. Soulillou, and M. C. Cuturi. 1997. Rat spleen dendritic cells express natural killer cell receptor protein 1 (NKR-P1) and have cytotoxic activity to select targets via a Ca2+-dependent mechanism. J. Exp. Med. 186:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerksiek, K. M., and E. G. Pamer. 1999. T cell responses to bacterial infection. Curr. Opin. Immunol. 11:400-405. [DOI] [PubMed] [Google Scholar]
- 16.Kerksiek, K. M., A. Ploss, I. Leiner, D. H. Busch, and E. G. Pamer. 2003. H2-M3-restricted memory T cells: persistence and activation without expansion. J. Immunol. 170:1862-1869. [DOI] [PubMed] [Google Scholar]
- 17.Kirby, A. C., U. Yrlid, and M. J. Wick. 2002. The innate immune response differs in primary and secondary Salmonella infection. J. Immunol. 169:4450-4459. [DOI] [PubMed] [Google Scholar]
- 18.Laouar, Y., F. S. Sutterwala, L. Gorelik, and R. A. Flavell. 2005. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 6:600-607. [DOI] [PubMed] [Google Scholar]
- 19.Lauvau, G., S. Vijh, P. Kong, T. Horng, K. Kerksiek, N. Serbina, R. A. Tuma, and E. G. Pamer. 2001. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science 294:1735-1739. [DOI] [PubMed] [Google Scholar]
- 20.Mackaness, G. B. 1969. The influence of immunologically committed lymphoid cells on macrophage activity in vivo. J. Exp. Med. 129:973-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaffrey, R. L., P. Fawcett, M. O'Riordan, K. D. Lee, E. A. Havell, P. O. Brown, and D. A. Portnoy. 2004. A specific gene expression program triggered by gram-positive bacteria in the cytosol. Proc. Natl. Acad. Sci. USA 101:11386-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moretta, A. 2005. The dialogue between human natural killer cells and dendritic cells. Curr. Opin. Immunol. 17:306-311. [DOI] [PubMed] [Google Scholar]
- 23.Muraille, E., R. Giannino, P. Guirnalda, I. Leiner, S. Jung, E. G. Pamer, and G. Lauvau. 2005. Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur. J. Immunol. 35:1463-1471. [DOI] [PubMed] [Google Scholar]
- 24.Ohteki, T., T. Fukao, K. Suzue, C. Maki, M. Ito, M. Nakamura, and S. Koyasu. 1999. Interleukin 12-dependent interferon gamma production by CD8alpha+ lymphoid dendritic cells. J. Exp. Med. 189:1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oxenius, A., U. Karrer, R. M. Zinkernagel, and H. Hengartner. 1999. IL-12 is not required for induction of type 1 cytokine responses in viral infections. J. Immunol. 162:965-973. [PubMed] [Google Scholar]
- 26.Pan, P. Y., P. Gu, Q. Li, D. Xu, K. Weber, and S. H. Chen. 2004. Regulation of dendritic cell function by NK cells: mechanisms underlying the synergism in the combination therapy of IL-12 and 4-1BB activation. J. Immunol. 172:4779-4789. [DOI] [PubMed] [Google Scholar]
- 27.Pillarisetty, V. G., S. C. Katz, J. I. Bleier, A. B. Shah, and R. P. Dematteo. 2005. Natural killer dendritic cells have both antigen presenting and lytic function and in response to CpG produce IFN-gamma via autocrine IL-12. J. Immunol. 174:2612-2618. [DOI] [PubMed] [Google Scholar]
- 28.Slifka, M. K., R. R. Pagarigan, and J. L. Whitton. 2000. NK markers are expressed on a high percentage of virus-specific CD8+ and CD4+ T cells. J. Immunol. 164:2009-2015. [DOI] [PubMed] [Google Scholar]
- 29.Stober, D., R. Schirmbeck, and J. Reimann. 2001. IL-12/IL-18-dependent IFN-gamma release by murine dendritic cells. J. Immunol. 167:957-965. [DOI] [PubMed] [Google Scholar]
- 30.Taieb, J., N. Chaput, C. Menard, L. Apetoh, E. Ullrich, M. Bonmort, M. Pequignot, N. Casares, M. Terme, C. Flament, P. Opolon, Y. Lecluse, D. Metivier, E. Tomasello, E. Vivier, F. Ghiringhelli, F. Martin, D. Klatzmann, T. Poynard, T. Tursz, G. Raposo, H. Yagita, B. Ryffel, G. Kroemer, and L. Zitvogel. 2006. A novel dendritic cell subset involved in tumor immunosurveillance. Nat. Med. 12:214-219. [DOI] [PubMed] [Google Scholar]
- 31.Tripp, C. S., S. F. Wolf, and E. R. Unanue. 1993. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc. Natl. Acad. Sci. USA 90:3725-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watford, W. T., M. Moriguchi, A. Morinobu, and J. J. O'Shea. 2003. The biology of IL-12: coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 14:361-368. [DOI] [PubMed] [Google Scholar]
- 33.Yang, J., I. Kawamura, and M. Mitsuyama. 1997. Requirement of the initial production of gamma interferon in the generation of protective immunity of mice against Listeria monocytogenes. Infect. Immun. 65:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan, Y., and C. Cheers. 1998. Control of IL-12 and IFN-gamma production in response to live or dead bacteria by TNF and other factors. J. Immunol. 161:1447-1453. [PubMed] [Google Scholar]