Abstract
Here we report that Salmonella enterica serovar Typhimurium pathogenicity island 4 carries a type I secretion system (siiCDF) which secretes an ∼600-kDa protein (encoded by siiE). SiiE is surface expressed, and its production is regulated by HilA. SiiE and SiiF influence colonization in cattle and the invasion of bovine enterocytes.
Serovars of Salmonella enterica infect a broad range of animal species, resulting in a spectrum of outcomes ranging from asymptomatic carriage to severe systemic disease. For instance, oral infection of chickens with serovar Typhimurium results in asymptomatic colonization of the gastrointestinal tract, while oral dosing of calves results in fever and diarrhea with high mortality in the absence of antibiotic treatment. Salmonellosis in livestock results in both animal suffering and economic losses, and the bacteria can contaminate the food chain and environment from these animal reservoirs.
The clinical outcomes associated with infection with S. enterica depend on both the infecting serovar and the host species. It is, however, becoming increasingly apparent that serovar Typhimurium uses different repertoires of genes to infect different hosts and that the expression of various virulence determinants may contribute both to host specificity and to the clinical outcome of infection.
Many of the virulence determinants used by serovar Typhimurium to cause infections in cattle are well characterized, and most of these are carried on horizontally acquired regions of DNA called Salmonella pathogenicity islands. Genes carried on Salmonella pathogenicity island 1 (SPI-1) have been shown to be essential in calves for the invasion of the intestinal barrier and the induction of enteritis (22, 26, 28). SPI-1 encodes a type III secretion system (T3SS-1) and secreted translocator proteins which together mediate the delivery of effector proteins into enterocytes. A subset of the effector proteins rearranges the actin cytoskeleton, resulting in membrane ruffling and, consequently, the internalization of Salmonella into epithelial cells. Another subset induces the enteropathogenic response to infection. Many of the effector proteins are encoded on loci outside SPI-1, including other pathogenicity islands and bacteriophages (23, 32).
Genes on SPI-2 influence both enteric and systemic virulence in calves following the initial invasion into the epithelial cells of the intestine (2, 22). SPI-2 encodes a second type III secretion system (T3SS-2) that secretes effector proteins across the membrane of the Salmonella-containing vacuole and enables the persistence of Salmonella inside host cells by modulating vesicular trafficking (reviewed in reference 25).
We recently screened a bank of 1,045 signature-tagged mutagenesis (STM) mutants of serovar Typhimurium in order to identify genes required for intestinal colonization in cattle, chickens (17), and pigs (S. C. Carnell, A. J. Bowen, E. Morgan, D. J. Maskell, T. S. Wallis, and M. P. Stevens, submitted for publication). Of the mutants attenuated in intestinal colonization in these host species, 98 had transposon insertions in genes required for colonization in cattle. As expected, a significant proportion of these genes were on SPI-1 and SPI-2, but we also isolated 11 mutants with transposon insertions within SPI-4 which were attenuated only in the colonization of the intestines of calves. SPI-4 was initially identified using a hybridization-based approach to search for large segments of DNA which were present in serovar Typhimurium but not in Escherichia coli K-12 and could therefore potentially constitute pathogenicity islands (30). A 27-kb Salmonella-specific region of DNA was sequenced and predicted to carry 18 open reading frames (spi4_A to spi4_R). A previously identified locus required for survival in murine macrophages was located at spi4_L and was the basis for the definition of this segment of DNA as a pathogenicity island (30). The publication of the complete genome sequence of serovar Typhimurium strain LT2 refined the identified sequence of SPI-4 into six open reading frames numbered STM4257 to STM4262 (15). We have since confirmed that the sequence is almost identical in the strain used in our studies (ST4/74 Nalr) and have renamed the genes siiA to siiF (17). We have also predicted through the sequence analysis of siiA to siiF that siiE encodes a 595-kDa secreted protein and that siiC, siiD, and siiF encode components of a type I secretion system (17). The remaining genes, siiA and siiB, do not have homologs in the protein or DNA databases, and their functions may be unrelated to those of the remainder of the island as transposon insertion mutations producing strains deficient in the colonization of calves were identified only in siiD, siiE, and siiF (17). However, upstream of siiA we identified a highly conserved operon polarity suppressor (ops) motif which is required for transcription elongation under the control of the RfaH protein and is associated with several other virulence gene clusters in the Enterobacteriaceae (17). Although this finding may indicate that the siiA to siiF genes are transcribed as a single unit, a recent study suggested that only the siiABCD genes are regulated by RfaH (18).
The aim of this study was to characterize the genes carried on SPI-4 and to explore their role in colonization in cattle. We chose to approach this issue through the in vitro and in vivo characterization of mutants of serovar Typhimurium strain ST4/74 Nalr, which is a virulent bovine isolate (17). Defined nonpolar deletions of siiE and siiF were created since the mutant strains were to be characterized in vivo and in vitro, and strains with nonpolar transposon insertion mutations in siiC and siiD were isolated from the signature-tagged mutagenesis transposon mutant bank (17) for characterization in vitro.
A mutant with a miniTn5Km2 transposon insertion in siiD was described previously (17). An siiC::miniTn5Km2 mutant was located within the STM mutant bank (17) by hierarchical screening of pools of mutants by PCR and was a kind gift from Gillian Pullinger, Institute for Animal Health, Compton, United Kingdom. The transposon insertion site for the siiC::miniTn5Km2 mutant was verified through sequencing of the specific PCR product by using a siiC-specific primer (5′-CCTGACCATGAACCACTG-3′) and a transposon-specific primer (5′-CCTAGGCGGCCAGATCTG-3′).
Mutant strains carrying deletions of siiE and siiF were created by overlapping PCR followed by allelic exchange with the positive-selection suicide vector pDM4 (16). Sequences flanking the serovar Typhimurium siiE gene were separately amplified by PCR from ST4/74 Nalr genomic DNA with PfuTurbo DNA polymerase (Stratagene, La Jolla, California) using the primer pairs siiE1 (5′-TATATAGAGCTCGGATATCTATTCACCGGTTGACG-3′) and siiE2 (5′-TTCTTGATTATCCTTTTGTATGCTTTTATTTCCC-3′) and siiE3 (5′-AGCATACAAAAGGATAATCAAGAAGAACACGC-3′) and siiE4 (5′-TATATAGAGCTCAATCTTGTTCATCGATAAAGAG-3′). The primary PCR products were gel purified and combined in a mixture for overlapping PCR (10) by using the flanking primers siiE1 and siiE4. The secondary PCR product was then cloned into pDM4 by using SstI sites incorporated into the primers. The resulting plasmid, pDM4ΔsiiE, was maintained in E. coli PIR1 cells (Invitrogen Life Technologies, Paisley, United Kingdom) and introduced into the ST4/74 Nalr strain by conjugation from E. coli S17-1λpir (21). Merodiploids were isolated on Luria-Bertani (LB) agar containing nalidixic acid (Nal; 20 μg ml−1) and chloramphenicol (25 μg ml−1). Double recombinants were selected by growing merodiploids to late logarithmic phase in LB broth lacking chloramphenicol and plating onto LB agar (without NaCl) containing 6% (wt/vol) sucrose at 30°C. Sucrose-resistant colonies were screened for deletions by colony PCR, and recombinants were verified by sequencing using flanking primers siiE5 (5′-CTTCAGGAACTTGAGGTTGTTAAC-3′) and siiE6 (5′-CTAAATACTGGGATAGTTAACG-3′). The deletion resulted in the juxtaposition of the first and last eight predicted codons of siiE, theoretically permitting the translational coupling of the mutated siiE gene and the downstream siiF gene.
A mutant with a deletion of the siiF gene was constructed in the same way except that sequences flanking siiF were amplified by PCR using the primer pairs siiF1 (5′-TATATAGAGCTCGCGTGGTGAAGGTGACAGC-3′) and siiF2 (5′-TTTATCCGGAGAATAAGGTTCTAGTTTTTTATC C-3′) and siiF3 (5′-CTAGAACCTTATTCTCCGGATAAATTATTAATG-3′) and siiF4 (5′-TATATAGAGCTCGCAACTGCTGAGCGCCG-3′). Recombinants were verified by sequencing using flanking primers siiF5 (5′-CTACGCATCTTCGTACAGAGC-3′) and siiF6 (5′-CATAACCACTGGTGTCG-3′). The deletion resulted in the juxtaposition of the first and last eight predicted codons of siiF.
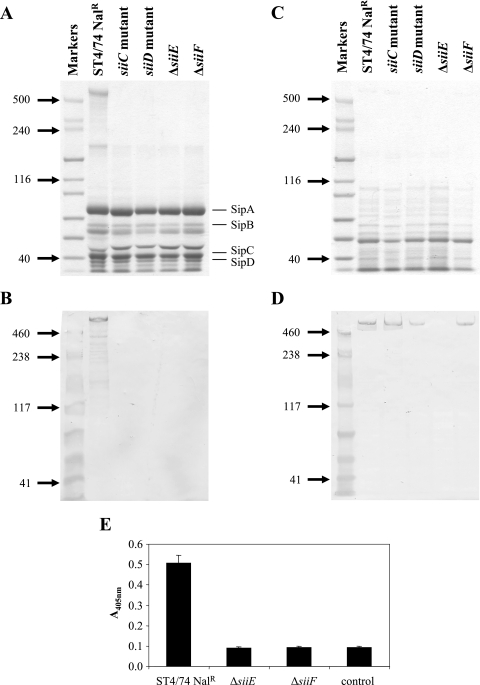
As we had predicted that siiE encoded a 595-kDa secreted protein, the secreted proteomes of the ST4/74 Nalr, siiC::miniTn5Km2 mutant, siiD::miniTn5Km2 mutant, ΔsiiE, and ΔsiiF strains were compared. As we had demonstrated by reverse transcription-PCR that siiE transcripts could be detected in the ST4/74 Nalr strain during aerated growth in LB broth from early to late log phase (data not shown), secreted proteins were precipitated by using trichloroacetic acid (26) from culture supernatants of bacteria grown overnight at 25°C in LB broth and then diluted 1:10 into 40 ml of fresh medium and grown with shaking for 4 h to an optical density at 600 nm of ∼1.0. The proteins were resuspended in a total volume of 40 μl, and 2 μl was separated on NuPAGE 3 to 8% Tris-acetate gels (Invitrogen, Paisley, United Kingdom) and stained with Coomassie brilliant blue. A large protein, which was present above the 500-kDa protein standard, was visible in the secreted proteome of the ST4/74 Nalr wild-type strain but absent from the secreted proteomes of the siiC::miniTn5Km2, siiD::miniTn5Km2, ΔsiiE, and ΔsiiF mutant strains (Fig. 1A). In order to confirm that the >500-kDa protein was SiiE, we cloned, expressed, and purified the C-terminal region of SiiE and used this protein fragment to raise monoclonal antibodies against SiiE.
FIG. 1.
SiiE is secreted in a siiC-, siiD-, and siiF-dependent manner and is surface associated. (A) Proteins isolated from culture supernatants were separated by SDS-PAGE and stained with Coomassie blue. Molecular masses (kilodaltons) of standard proteins are shown on the left. The locations of T3SS-1-secreted proteins (SipA to SipD) are indicated on the right. (B) Proteins isolated from culture supernatants were transferred onto membrane by Western blotting and probed with the anti-SiiE monoclonal antibody. Molecular masses (kilodaltons) of standard proteins are shown on the left. Lanes are as shown for panel A. (C) Total cellular proteins were separated by SDS-PAGE and stained with Coomassie blue. Molecular masses (kilodaltons) of standard proteins are shown on the left. (D) Total cellular proteins were transferred onto membrane by Western blotting and probed with the anti-SiiE monoclonal antibody. Molecular masses (kilodaltons) of standard proteins are shown on the left. Lanes are as shown for panel C. (E) Fixed bacteria were probed by ELISA with the anti-SiiE monoclonal antibody. The control wells were coated with ST4/74 Nalr bacteria but incubated without the monoclonal antibody.
A DNA fragment encoding residues 5065 to 5559 of the predicted SiiE protein (SiiE5065-5559) was amplified by PCR from ST4/74 Nalr genomic DNA by using KOD Hot Start DNA polymerase (Novagen; Merck Biosciences Ltd., Nottingham, United Kingdom) with the primer pair siiE-LIC1 (5′-GACGACGACAAGATGAATCGTTGGGAAGATGT G-3′) and siiE-LIC2 (5′-GAGGAGAAGCCCGGTTTATGCGTGTTCTTCTTG-3′). The resulting DNA fragment was inserted by ligation-independent cloning into pET-30 Ek/LIC (Novagen; Merck Biosciences Ltd., Nottingham, United Kingdom) with N-terminal His6 and S tags to produce pET-SiiE. For purification of the His6-SiiE5065-5559 protein, E. coli BL21 Star (DE3) cells (Invitrogen Life Technologies, Paisley, United Kingdom) transformed with the pET-SiiE plasmid were grown at 37°C in LB broth supplemented with kanamycin (30 μg ml−1) and 1% (wt/vol) glucose. At an optical density at 600 nm of 0·6 to 0.8, the transcription of the SiiE5065-5559-encoding fragment was induced for 3 h with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by centrifugation (8,000 × g for 10 min at 4°C) and resuspended in 20 ml of phosphate buffer (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4) with 60 mM imidazole and lysozyme (200 μg ml−1). Following lysis by ultrasonic disruption, cell debris was removed by centrifugation as described above and the supernatant was applied to a 1-ml HisTrap HP affinity chromatography column (GE Healthcare UK Ltd., Chalfont St. Giles, United Kingdom). Proteins were eluted in phosphate buffer with 300 mM imidazole. The purification was monitored by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by Western blotting using a monoclonal antibody against the His6 tag (Novagen; Merck Biosciences Ltd., Nottingham, United Kingdom). The protein was purified further by electroelution of the protein from zinc-stained SDS-PAGE gels and was dialyzed overnight against water by using Slide-A-Lyzer dialysis cassettes (Perbio Science UK Ltd., Cramlington, Northumberland, United Kingdom). For production of a monoclonal antibody against SiiE, a six-week-old female BALB/c mouse was immunized twice at a 14-day interval with 20 μg of the purified His6-SiiE5065-5559 protein in Titermax Gold adjuvant (Sigma-Aldrich Company Ltd, Gillingham, United Kingdom) via the subcutaneous route. Forty-five days after the second immunization, a further 50 μg of purified His6-SiiE5065-5559 was given without adjuvant via the intraperitoneal route. Four days after the final booster dose, the spleen was removed and splenocytes prepared and fused with SP2/0 mouse myeloma cells by using polyethylene glycol 1500 according to standard procedures. The fused cells were diluted in RPMI-hypoxanthine-aminopterin-thymidine medium with 10% (vol/vol) fetal calf serum and plated onto 96-well plates seeded 48 h previously with mouse peritoneal macrophages. The plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 10 days. The hybridomas were screened for the production of SiiE-specific monoclonal antibodies by enzyme-linked immunosorbent assay (ELISA) and immunoblotting against purified His6-SiiE5065-5559. Clones secreting antibody reactive with SiiE5065-5559 were expanded, and SiiE-specific antibody was purified from the culture supernatant by using a protein G-Sepharose column (GE Healthcare UK Ltd., Chalfont St. Giles, United Kingdom) according to standard techniques.
In order to confirm that the >500-kDa protein present in the secreted proteome of ST4/74 Nalr was SiiE, 0.2 μl of the secreted protein preparations from ST4/74 Nalr, siiC::miniTn5Km2 mutant, siiD::miniTn5Km2 mutant, ΔsiiE, and ΔsiiF strains was transferred to Hybond ECL membrane by standard Western blotting techniques and SiiE was detected using the anti-SiiE monoclonal antibody at 1 μg ml−1. A band corresponding to the >500-kDa protein was visible on the blot in the lane carrying proteins from the ST4/74 Nalr strain but not visible in the lanes carrying proteins derived from the siiC::miniTn5Km2, siiD::miniTn5Km2, ΔsiiE, and ΔsiiF mutant strains (Fig. 1B), thus confirming that SiiE is secreted into the culture supernatant. As the SiiE protein was absent from the secreted proteins isolated from the siiC::miniTn5Km2, siiD::miniTn5Km2, and ΔsiiF mutant strains, we investigated whether SiiE was produced, but not secreted, by the siiC::miniTn5Km2, siiD::miniTn5Km2, and ΔsiiF mutant strains by isolation and comparison of the total cellular proteins of the ST4/74 Nalr, siiC mutant, siiD mutant, ΔsiiE, and ΔsiiF strains. The bacteria were grown in LB broth as described above and harvested by centrifugation (8,000 × g at 4°C for 10 min), and total cellular proteins were prepared from the bacterial cells by using the ReadyPreps protein preparation kit (Epicenter, Madison, WI).
The proteins were separated by electrophoresis as described above (Fig. 1C), and the presence of SiiE in the protein preparations was investigated by Western blotting of the proteins and probing with the anti-SiiE monoclonal antibody. The SiiE protein was present in equal amounts in the preparations of total cellular proteins from the ST4/74 Nalr, siiC::miniTn5Km2 mutant, siiD::miniTn5Km2 mutant, and ΔsiiF strains (Fig. 1D) but was not present in the preparation of total cellular protein from the ΔsiiE mutant, indicating that the SiiE gene was translated in the siiC::miniTn5Km2, siiD::miniTn5Km2, and ΔsiiF mutants but that the protein was not secreted. Although the siiC::miniTn5Km2 and siiD::miniTn5Km2 mutant strains are transposon insertion mutants, the presence of SiiE in the total cellular protein fractions from these strains indicates that the mutations do not have polar effects on the siiE gene. However, we cannot preclude the possibility that the mutation of siiC has polar effects on siiD.
We also sought to establish whether SiiE was associated with the bacterial surface through ELISA using fixed bacteria. ST4/74 Nalr, ΔsiiE, and ΔsiiF strains were grown as described above, harvested by centrifugation (8,000 × g at 4°C for 10 min), washed in phosphate-buffered saline (PBS), and resuspended in 5 ml of 4% (wt/vol) paraformaldehyde in PBS for 30 min. The bacteria were harvested by centrifugation as described above to remove any SiiE protein that may have been released by fixation and resuspended in 4 ml of 0.05 M carbonate buffer, pH 9.6. Microtiter plates (Maxisorp; Nunc, Rochester, NY) were coated with 50 μl of the prepared bacteria at 4°C overnight. The wells were washed and blocked with 3% (wt/vol) bovine serum albumin in PBS-0.05% (vol/vol) Tween for 1 h and then incubated with the anti-SiiE monoclonal antibody at 3 μg ml−1 in the blocking buffer for 2 h. A control in which wells coated with the ST4/74 Nalr strain were incubated without the anti-SiiE monoclonal antibody was included in order to obtain a background reading. The plates were washed again with PBS-Tween and incubated with alkaline phosphatase-conjugated goat anti-mouse antibody (Sigma) at 1/1,000 in the blocking buffer for 2 h. After washing, the reaction was developed with p-nitrophenyl phosphate and the absorbance was read at 405 nm. Each strain was tested in triplicate wells on three occasions. The ELISA showed that SiiE was present on the surfaces of ST4/74 Nalr cells, but the signal from the ΔsiiE and ΔsiiF mutant bacteria was reduced to background levels (Fig. 1E), indicating that SiiE was not present on the bacterial surface in these strains and confirming the specificity of the anti-SiiE antibody. The inability to detect SiiE by ELISA in the ΔsiiF mutant bacteria confirmed that at least a subset of the SiiE detected in the parent strain was surface associated.
Several previous studies have suggested that genes carried on SPI-4 are expressed under conditions similar to those under which genes carried on SPI-1 are expressed (3, 6, 8, 14, 24) and that the transcription of siiE is influenced by the SPI-1-encoded transcriptional regulator HilA (1, 3). A recent study identified a putative HilA box at +3,279 bp from the start codon of siiE (3), but it is unclear how this internal HilA box would allow the transcriptional regulation of siiE. The authors concluded that the HilA box is in the promoter region of the spi4_H gene which was predicted as part of the original incorrect annotation of SPI-4 (30). Since we have now shown that siiE encodes a secreted protein of ca. 595 kDa, it is unlikely that the spi4_H gene exists. We have, however, identified a motif which matches the consensus sequence of the HilA box predicted by De Keersmaecker et al. (3) at −52 to −40 bp upstream of the predicted translational start of siiC (data not shown). It is therefore possible that a HilA-regulated promoter lies upstream of siiC which may control the expression of the type I secretion system and SiiE.
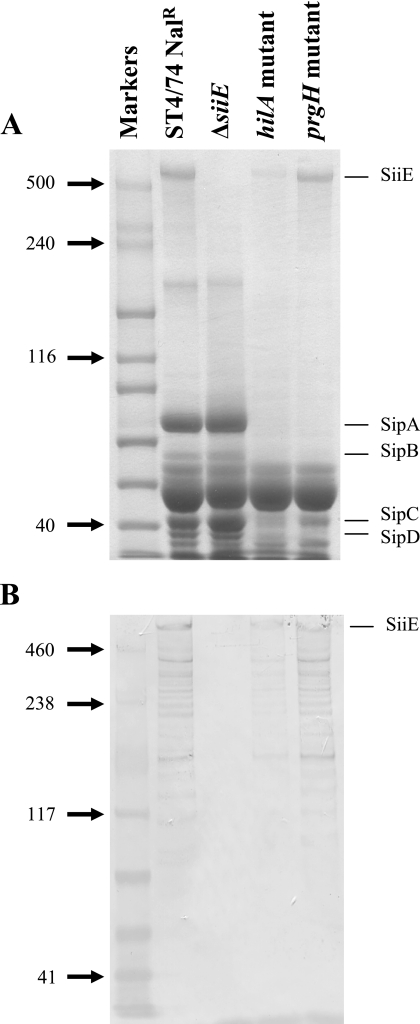
As genes carried on SPI-1 and SPI-4 may be coregulated, we investigated whether hilA influenced the secretion of SiiE through the examination of the secreted proteome of an ST4/74 Nalr hilA::miniTn5Km2 mutant (17). The level of SiiE present in the culture supernatant of the hilA mutant strain was significantly lower than that in the supernatant of the parent strain (Fig. 2), indicating that hilA influences the transcription or secretion of SiiE. It is likely that this occurs independently of the need for a functional T3SS-1 since a prgH mutation did not affect the secretion of SiiE (Fig. 2).
FIG. 2.
The secretion of SiiE is influenced by hilA but not T3SS-1. (A) Proteins isolated from culture supernatants were separated by SDS-PAGE and stained with Coomassie blue. Molecular masses (kilodaltons) of standard proteins are shown on the left. The locations of T3SS-1-secreted proteins (SipA to SipD) are indicated on the right. (B) Proteins isolated from culture supernatants were transferred onto membrane by Western blotting and probed with the anti-SiiE monoclonal antibody. Molecular masses (kilodaltons) of standard proteins are shown on the left. Lanes are as shown for panel A.
We previously found through STM that mutants with transposon insertions in siiD, siiE, and siiF were absent from the ileal walls of calves 4 days postinfection (17). In order to further explore the role of SPI-4 in intestinal colonization and to define the basis of the attenuation seen in the STM screens, we compared the progressions of infections following oral inoculation of 28-day-old calves with the parent strain or the defined ΔsiiE and ΔsiiF mutants. All animal experiments were conducted according to the requirements of the Animal (Scientific Procedures) Act of 1986. Twenty-eight-day-old calves from the Institute for Animal Health farm were used as described previously (2). Bacterial cultures were prepared by inoculating LB broth with bacterial colonies from a fresh agar plate and incubated at 37°C with shaking for 18 h. Calves were separately infected with approximately 1 × 108 CFU of each bacterial strain mixed with reconstituted powdered milk and fed by syringe immediately before the morning feed. Following inoculation, calves were monitored for signs of disease at least twice daily as described previously (2). The calves reached predetermined end points at 5 to 6 days postinfection, at which point all calves were humanely killed. At postmortem examination, samples of contents, walls, and mesenteric lymph nodes were taken from the mid ileum, cecum, and spiral colon and also the liver and spleen, and the bacteria associated with these sites were enumerated by counting viable cells. All samples were taken in triplicate, and the tissue samples were washed gently to remove nonadherent bacteria from the surfaces. One gram of sample was homogenized in 9 ml of 0.9% (wt/vol) saline. Serial dilutions were performed, and 20 μl of each dilution was spread in triplicate onto brilliant green agar plates containing nalidixic acid. Fecal shedding and pyrexial response data were statistically analyzed for the effect of mutation by means of an F test, with data taken as repeated measurements (Proc GLM, Statistical Analysis System [SAS]; SAS Institute, NC). Postmortem data were analyzed by means of Student's t test (SAS). P values of <0.05 were taken to be significant.
For testing of the ΔsiiE strain, four calves were inoculated orally with 1.3 × 108 CFU of the ST4/74 Nalr ΔsiiE mutant strain and three calves were inoculated orally with 1.0 × 108 CFU of the ST4/74 Nalr parent strain. All calves rapidly exhibited clinical signs of salmonellosis, a strong pyrexial response accompanied by the shedding of large numbers of bacteria in the feces and diarrhea. The experiment was terminated on day 6 as one calf in each group had reached the end point threshold of a scour score of 20 or above. Scour (diarrhea) was scored by using a cumulative daily scoring scheme based on the consistency and the contents of the feces. Scores for consistency were as follows: 0, normal; +1, semisolid; +2, liquid; and +3, watery. Scores for the fecal contents were as follows: +1, fresh blood; +2, sloughed mucosa (23a).
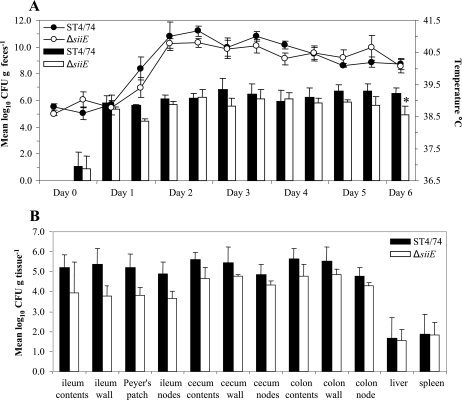
At most time intervals during the experiment, the mean numbers of bacteria recovered from the feces of calves challenged with the ST4/74 Nalr ΔsiiE mutant strain were lower than those recovered from calves challenged with the parent strain, but the difference was statistically significant only at day 6 postinfection (P < 0.05) (Fig. 3A). No statistically significant differences in the magnitude or duration of the pyrexial responses were detected at any time point. However, at 32 h, at which time point a significant difference in the magnitude of the response between the groups of calves infected with the ST4/74 Nalr and ΔsiiF strains was seen (see below), there was a tendency towards a statistically significant difference in the magnitude of the response (P < 0.07).
FIG. 3.
SiiE contributes to intestinal colonization in cattle. (A) Mean rectal temperature responses and mean numbers of bacteria recovered per gram of feces following infection with either the parental strain or the ΔsiiE mutant strain for 6 days. Each bar or datum point is presented with the standard error of the mean *, P of <0.05. (B) Mean numbers of bacteria of the parental strain or the ΔsiiE mutant strain recovered per gram of tissue. Each bar is presented with the standard error of the mean.
At postmortem examination, the numbers of bacteria associated with intestinal and systemic sites were determined. Although the mean number of ST4/74 Nalr ΔsiiE bacteria recovered from intestinal sites was lower than the mean number of ST4/74 Nalr bacteria recovered, none of the data were statistically significant (Fig. 3B).
In order to determine whether the mutation of the type I secretion apparatus had an effect on intestinal colonization similar to or greater than that of the deletion of siiE, calves were inoculated orally with 1.8 × 108 CFU of either the ST4/74 Nalr ΔsiiF mutant strain (four calves) or the ST4/74 Nalr parent strain (three calves). Again, all calves rapidly exhibited signs of salmonellosis, and the experiment was terminated 5 days postinfection as two of the three calves in the group challenged with the ST4/74 Nalr strain were unable to stand unaided and were displaying symptoms of anorexia. In contrast, the four calves challenged with the ST4/74 Nalr ΔsiiF mutant appeared to be relatively healthy.
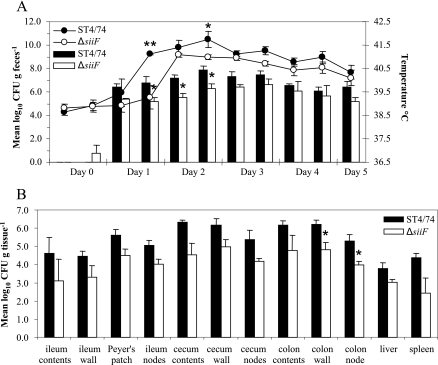
At all time points, the mean numbers of bacteria recovered per gram of feces from calves infected with the ST4/74 Nalr ΔsiiF mutant were lower than those from the calves infected with the parent strain. However, the data were statistically significant only at 32, 48, and 56 h postinfection (P < 0.05) (Fig. 4A). The mean rectal temperatures of the calves infected with the ST4/74 Nalr ΔsiiF mutant were lower than those of the calves infected with the ST4/74 Nalr strain at all time points and significantly so at 32 h (P < 0.001) and 56 h (P < 0.05) (Fig. 4A).
FIG. 4.
SiiF contributes to intestinal colonization in cattle. (A) Mean rectal temperature responses and mean numbers of bacteria recovered per gram of feces following infection with either the parental strain or the ΔsiiF mutant strain for 6 days. Each bar or datum point is presented with the standard error of the mean. *, P of <0.05; **, P of <0.001. (B) Mean numbers of bacteria of the parental strain or the ΔsiiF mutant strain recovered per gram of tissue. Each bar is presented with the standard error of the mean. *, P of <0.05.
At postmortem examination, the mean numbers of ST4/74 Nalr ΔsiiF bacteria recovered from the calves were lower than those of the parent strain for all sites examined, but the difference between the means was statistically significant only for the tissues taken from the colon walls and colon nodes (P < 0.05) (Fig. 4B).
As siiE and siiF contribute to the infection of calves following oral inoculation, we chose to investigate further their role in infection through the examination of the ability of the ΔsiiE and ΔsiiF mutants to invade the bovine epithelium and to persist in primary macrophages.
To investigate whether siiE and siiF are required for the invasion of the bovine epithelium by serovar Typhimurium, the bovine ligated ileal loop model to quantify intestinal invasion was used as described by Paulin et al. (20). At 2 h postinfection, gentamicin-protected tissue-associated bacteria were enumerated by the direct plating of serial dilutions of tissue homogenates as described above.
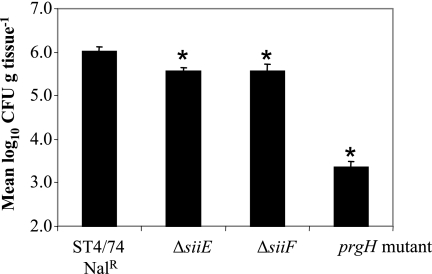
Fewer of the ΔsiiE and ΔsiiF mutant bacteria than of the parent strain bacteria were recovered from the tissues 2 h postinfection (P < 0.0001) (Fig. 5), indicating that siiE and siiF contribute to the intestinal invasion of bovine enterocytes. The degree of attenuation of the ΔsiiE and ΔsiiF mutants, however, was not as severe as that of a prgH::miniTn5Km2 mutant strain (17). The mutation of siiE and siiF did not affect the secretion of T3SS-1-secreted effector proteins as assessed by the analysis of the secreted proteomes of the ΔsiiE and ΔsiiF mutants (Fig. 1A).
FIG. 5.
SiiE and SiiF contribute to the invasion of bovine enterocytes. The mean numbers of gentamicin-protected bacteria of the ST4/74 Nalr, ΔsiiE, ΔsiiF, and prgH::miniTn5Km2 mutant strains recovered per gram of tissue are shown. Each bar is presented with the standard error of the mean. *, P of <0.0001.
Several studies with conflicting results have reported the requirement for genes carried on SPI-4 for survival in macrophages. Fields et al. (7) found that a mutant with a transposon insertion in a locus mapped to SPI-4 by Wong et al. (30) was deficient in its ability to survive in murine peritoneal macrophages. A microarray study also suggested that SPI-4 genes are up-regulated under in vitro conditions which mimic the phagocytic vacuole (4), although other microarray studies of serovar Typhimurium gene expression in macrophages have suggested that the reverse is true (6, 31).
We previously demonstrated that four mutants with transposon insertions in siiD, siiE, and siiF were not attenuated in persistence in porcine alveolar macrophages (17). Since that study was conducted, however, we have shown that mutants with transposon insertions in genes within SPI-4 are not attenuated in intestinal colonization in pigs (Carnell et al., submitted). We therefore chose to test the ΔsiiE and ΔsiiF mutants for invasion and persistence in bovine alveolar macrophages.
Alveolar macrophages were isolated from healthy Friesian cattle by bronchoalveolar lavage (9) and used essentially as described previously (29) except that the cells were inoculated with the bacteria in Iscove's modified Dulbecco's medium (without phenol red and l-glutamine) supplemented with 5% (vol/vol) fetal calf serum and 2 mM l-glutamine. In each case, the strains were tested in triplicate on two occasions. The survival rates of the strains at 6 h and 24 h were calculated as follows: percentage of surviving bacteria = (number of bacteria recovered at 6 h or 24 h/number of bacteria recovered at 2 h) × 100. The data were statistically analyzed across two biological replicates for the effect of the strain by using a two-way analysis of variance (Proc Mixed; SAS).
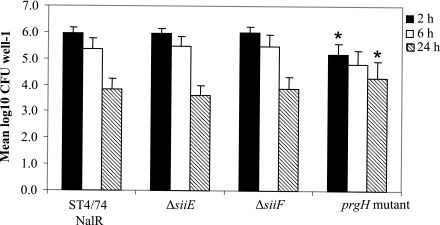
The means and standard errors of the means of results from the two experiments are shown in Fig. 6. The ΔsiiE and ΔsiiF mutants were taken up by the macrophages at levels equivalent to that of the parent strain as assessed by the recovery of gentamicin-protected bacteria at 2 h postinfection, although the mean numbers of prgH::miniTn5Km2 mutant bacteria recovered were significantly lower than those of the ΔsiiF strain (P < 0.05) and showed a statistical tendency towards being reduced compared to those of the parent strain and the ΔsiiE strain (P < 0.056). Neither of the ΔsiiE and ΔsiiF mutants were attenuated in intracellular survival over 6 h or 24 h, although the numbers of gentamicin-protected prgH::miniTn5Km2 mutant bacteria recovered were greater than those of the other strains at 24 h (P < 0.05), presumably due to reduced macrophage lysis (27).
FIG. 6.
SiiE and SiiF do not influence persistence in bovine alveolar macrophages. The mean numbers of gentamicin-protected bacteria of the ST4/74 Nalr, ΔsiiE, ΔsiiF, and prgH::miniTn5Km2 mutant strains recovered per well at 2 h, 6 h, and 24 h postinfection are shown. Each bar is presented with the standard error of the mean. *, P <0.05.
In summary, we report that SPI-4 encodes a ca. 600-kDa protein (SiiE) which is secreted into culture supernatants through an associated putative type I secretion system (encoded by siiC, siiD, and siiF) and have identified a minor role for siiE and siiF in the invasion of bovine enterocytes and intestinal colonization in calves. Although the data gathered following the oral infection of calves with ΔsiiE were statistically significant only for fecal shedding at 6 days postinfection, the data show a clear trend indicating that siiE contributes to infection in calves given that at most time points the mean rectal temperature responses and mean numbers of bacteria recovered from fecal samples were lower for the calves infected with the ΔsiiE strain than for those infected with the parent strain. Similarly, for each site examined postmortem, the mean numbers of bacteria recovered from the infected tissues were lower for the calves infected with the ΔsiiE strain than for those infected with the parent strain (Fig. 3).
A similar trend was seen following the oral challenge of calves with the ΔsiiF strain or the parent strain, although the data obtained from these animals suggested that the ΔsiiF mutant was slightly more attenuated than the ΔsiiE mutant (Fig. 4). We cannot preclude the possibility that the type I secretion system, of which SiiF is a component, is required for the secretion of proteins other than SiiE and that these secreted substrates also contribute to infection in calves. As yet, we have no data to support this hypothesis as only SiiE appears to be absent from the secreted proteome of the ΔsiiF mutant (Fig. 1A). In addition, the ΔsiiE and ΔsiiF mutants were attenuated to comparable levels in the invasion of bovine enterocytes as assessed by the bovine ligated ileal loop model (Fig. 5), even though the levels of attenuation were not equal following oral challenge. This apparent discrepancy may reflect the complex interactions that take place in vivo over the 5 to 6 days of examination following oral challenge compared to those over a 4-h time course in the bovine ligated ileal loop model and may indicate a role for SPI-4 at early time points postinfection.
That the mutants were clearly attenuated in the bovine ligated ileal loop model, but not following oral challenge, also highlights the difficulties of experimental procedures performed with large animals, including the variability among animals which arises from the use of outbred animals and the limits on experimental group size due to practicability and cost. However, the findings are supported by the independent isolation of eight siiE and two siiF transposon mutants of serovar Typhimurium in a signature-tagged mutagenesis screen for mutants defective in persistence in the bovine ileal mucosa 4 days post-oral inoculation (17). This finding emphasizes the sensitivity of the technique of signature-tagged mutagenesis in identifying genes which contribute a minor role in infections but also reinforces a requirement to substantiate the results of such screens. Although siiE and siiF appear to contribute in a minor role to the pathogenesis of salmonellosis in cattle, it is likely that a large number of genes contribute in a small, but significant, way to infections in cattle as suggested by the large numbers of mutants identified by signature-tagged mutagenesis as being attenuated in intestinal colonization (17).
Although we have demonstrated here that siiE and siiF contribute to infections in calves at the intestinal level, it is unclear what role the SPI-4 genes play in virulence in other animal species. We previously demonstrated by signature-tagged mutagenesis that mutants with transposon insertions in SPI-4 genes were not attenuated in colonization in chickens (17) or pigs (Carnell et al., submitted) but that transposon mutants with insertions in siiE and siiF were attenuated in systemic virulence following the oral challenge of BALB/c mice but not following the intraperitoneal infection of Nrampr mice (17). Similarly, Detweiler et al. found that an siiF mutant was not attenuated in bacterial persistence at 3 weeks following the intraperitoneal infection of Nramp1+/+ mice (4). However, a recent report contradicted this finding and suggested that SPI-4 genes do contribute to long-term systemic infections in mice (13). Through the use of a microarray-based negative selection screen for mutants unable to persist over 28 days in Nramp1+/+ mice, siiC and siiF mutants were shown to be lost from the livers and spleens of mice 21 days following intraperitoneal infection. The role of SPI-4 genes in long-term systemic virulence in mice was confirmed in a competitive-index experiment using a ΔsiiABCD::Km deletion mutant. This study, however, also showed that SPI-1-carried genes are also required for long-term systemic virulence in mice, although previously SPI-1 was thought to be required primarily for the gastrointestinal phase of infection. These conflicting results underline the importance of studying bacterial gene functions in target animal hosts wherever possible.
Although the mode of action of SiiE has not been established, few other type I secretion systems are known to influence virulence. These secretion systems secrete a wide range of proteins in both prokaryotic and eukaryotic cells, and the proteins are secreted from the cytoplasm to the extracellular medium without a periplasmic step. SiiC is predicted to be an outer membrane component, and SiiD is a putative membrane fusion protein which links the inner and outer membranes. SiiF is predicted to be the ATP-binding cassette component which in other systems is localized in the cytoplasmic membrane and is thought to be responsible for the specificity of the secretion process. Further studies are required to establish the mode of translocation of SiiE across the periplasm and outer membrane and to determine the exact roles of SiiC, SiiD, and SiiF in this process.
The best-characterized type I secretion system is that required for the secretion of the HlyA alpha-hemolysin of uropathogenic E. coli. HlyA belongs to the repeats-in-toxin protein family, and strains carrying alpha-hemolysin are mainly associated with extraintestinal infections (11), although some have been linked with intestinal infections (5). Few type I secretion systems are known to contribute to virulence in Salmonella. MacAB, which is a drug efflux system, is required for virulence in mice and is regulated by the global virulence regulator PhoP (19). We previously showed that siiCDEF genes share 40% overall nucleotide identity with four genes on Salmonella pathogenicity island 9 (17). These genes also encode a type I secretion system and a predicted 365-kDa protein, BapA, which is required for biofilm formation and virulence in mice (12). Despite the similarities between SiiE and BapA, no evidence has been found for a role for SiiE in biofilm formation (12) and further studies are required to establish the mode of action of SiiE in vivo.
Acknowledgments
This work was supported by BBSRC grant number 201/D19269 and DEFRA grant number OZ0319.
We thank Microbiological Services (Institute for Animal Health, Compton, United Kingdom) for anti-SiiE monoclonal antibody production, Andrew Green for assistance with the animal work, and Pauline van Diemen for assistance with statistical analysis.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Bispham, J., B. N. Tripathi, P. R. Watson, and T. S. Wallis. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun. 69:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Keersmaecker, S. C. J., K. Marchal, T. L. A. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, S. J., S. Srinivas, M. J. Albert, K. Alam, R. M. Robins-Browne, S. T. Gunzburg, B. J. Mee, and B. J. Chang. 1998. Characterization of the roles of hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect. Immun. 66:2040-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 7.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, I. Hautefort, A. Thompson, J. C. Hinton, and F. Van Immerseel. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72:946-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilloteau, L. A., T. S. Wallis, A. V. Gautier, S. MacIntyre, D. J. Platt, and A. J. Lax. 1996. The Salmonella virulence plasmid enhances Salmonella-induced lysis of macrophages and influences inflammatory responses. Infect. Immun. 64:3385-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penadés, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322-1339. [DOI] [PubMed] [Google Scholar]
- 13.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathogens 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangan, M. W., S. Lucchini, V. Danino, T. Ó. Cróinín, J. C. D. Hinton, and C. J. Dorman. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:1831-1847. [DOI] [PubMed] [Google Scholar]
- 15.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 16.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 18.Nagy, G., V. Danino, U. Dobrindt, M. J. Pallen, R. Chaudhuri, L. Emody, J. C. Hinton, and J. Hacker. 2006. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74:5914-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, K., T. Latifi, and E. A. Groisman. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126-141. [DOI] [PubMed] [Google Scholar]
- 20.Paulin, S. M., P. R. Watson, A. R. Benmore, M. P. Stevens, P. W. Jones, B. Villarreal-Ramos, and T. S. Wallis. 2002. Analysis of Salmonella enterica serotype-host specificity in calves: avirulence of S. enterica serotype Gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infect. Immun. 70:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon, R., U. Preifer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 22.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 23a.Wallis, T. S., S. M. Paulin, J. S. Plested, P. R. Watson and P. W. Jones. 1995. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect. Immun. 63:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 25.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 26.Watson, P. R., E. E. Galyov, S. M. Paulin, P. W. Jones, and T. S. Wallis. 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect. Immun. 66:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson, P. R., A. V. Gautier, S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 2000. Salmonella enterica serovars Typhimurium and Dublin can lyse macrophages by a mechanism distinct from apoptosis. Infect. Immun. 68:3744-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson, P. R., S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 1995. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect. Immun. 63:2743-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson, P. R., S. M. Paulin, P. W. Jones, and T. S. Wallis. 2000. Interaction of Salmonella serotypes with porcine macrophages in vitro does not correlate with virulence. Microbiology 146:1639-1649. [DOI] [PubMed] [Google Scholar]
- 30.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ygberg, S. E., M. O. Clements, A. Rytkönen, A. Thompson, D. W. Holden, J. C. D. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, S., R. A. Kingsley, R. L. Santos, H. Andrews-Polymenis, M. Raffatellu, J. Figueiredo, J. Nunes, R. M. Tsolis, L. G. Adams, and A. J. Baumler. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]