Abstract
Long-duration consistently Escherichia coli O157:H7 culture-positive cattle were euthanized and necropsied. Tissue and digesta from along the gastrointestinal tract (GIT) were cultured for the bacteria and examined histologically for lymphoid character. E. coli O157:H7 was detected only at the rectoanal junction mucosa and not at any other GIT location.
Escherichia coli O157:H7 causes hemorrhagic colitis and the life-threatening sequela hemolytic-uremic syndrome in humans (6). Healthy cattle are the major reservoir for E. coli O157:H7, and contaminated ground beef, direct animal contact, and bovine manure contamination of other foods or water account for the majority of human infections (2, 3). The predominant colonization site of this microorganism in the bovine gastrointestinal tract (GIT) has been identified as the rectoanal junction (RAJ) mucosa (7). Localization to this site is in contrast with other E. coli serotypes that are present in consistent numbers throughout the large intestine without an increase at the RAJ mucosa. This tissue area comprises a clear junction between the stratified squamous epithelium of the anal canal and the columnar epithelial mucosa of the rectum and is a lymphoid follicle-dense mucosa. Although the mechanisms underlying the tropism of this serotype for this lymphoid follicle-rich site are not known, many studies of experimentally and naturally colonized cattle conclude that the RAJ mucosa plays an important role in E. coli O157:H7 bovine carriage of this human pathogen (5, 7, 8, 10, 11). Also, application of E. coli O157:H7 at the RAJ mucosa results in carriage of the bacteria similar to that by naturally infected animals (10).
Three distinct patterns of E. coli O157:H7 carriage in cattle have been described previously (1, 9, 12). First, animals can be transiently culture positive for short durations of a few days and are considered passive shedders and are likely not colonized at the RAJ mucosa; second, cattle can be colonized and shed the bacteria for an average of 1 month and typically not longer than 2 months; and third, a few rare animals are colonized for a long duration and shed the bacteria from 3 to 12 months or longer. This unique situation in which a few animals develop long-duration colonization (>2 months) with E. coli O157 is likely due to bacterial association at the RAJ mucosa; however, it may be due to unique colonization by the bacteria at a site(s) in addition to the RAJ mucosa. For example, the gall bladder can harbor Salmonella spp. in carrier animals and a recent investigation reports E. coli O157:H7 in the gall bladders of some cattle at slaughter; however, this report does not provide information about the E. coli O157:H7 culture history of the animals tested, so the duration for which each animal was culture positive is unknown (4). For the first time, the current study investigated the colonization site of E. coli O157:H7 along the GIT of animals for which a documented history of long-duration carriage of E. coli O157:H7 was available. To identify animals that fit the criteria for a long-duration-colonized animal, hundreds of cattle were cultured over several years and the rare long-duration, culture-positive animals were used in this case study (data not shown).
Three long-duration E. coli O157:H7 culture-positive Holstein steers, designated no. 711, no. 135, and no. 407, that had been enrolled in different previous trials were naturally or experimentally carrying the bacteria when they were identified for this study. Each animal was 4 to 5 months old when the first samples were cultured. All animal procedures were approved by the Institutional Animal Care and Use Committee. Cattle were penned separately and housed in a large quarantined animal isolation facility at the University of Idaho. Animal no. 135 was carrying E. coli O157 naturally. Dosed animals were given a single challenge of 1010 CFU of E. coli O157:H7 strain ATCC 43894, a known human pathogen isolated from ground beef during an outbreak (10). The E. coli O157 carried by animal no. 135 was unique from ATCC 43894 as determined by pulsed-field gel electrophoresis analysis of digested chromosomal DNA (data not shown). Animal no. 407 was orally dosed, and animal no. 711 was rectally dosed. Both applications lead to bacterial carriage that mimics natural infections (10).
Rectoanal mucosal swab (RAMS) cultures (a reproducible and reliable method) were performed once or twice a week for the first 12 weeks, as previously described (8), to establish the nature of the bacterial colonization. Briefly, RAMS samples were collected, placed in cold Trypticase soy broth, and processed within 2 h. RAMS were cultured directly (quantitative data), and colonies were confirmed to be E. coli O157 colonies by latex agglutination (Pro-Lab Diagnostics, Toronto, Ontario, Canada). Samples negative by direct culture were enriched (qualitative data) at 37°C with aeration for 18 h and similarly analyzed. After the first 3 months, samples were cultured once or twice per month until the animal was euthanized and necropsied.
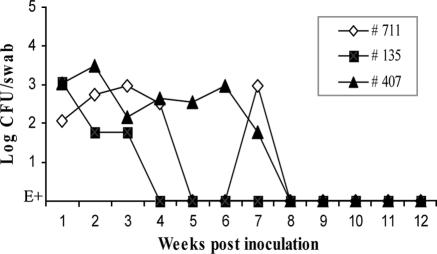
The results of RAMS culture showed that animals no. 135, no. 407, and no. 711 were consistently E. coli O157:H7 culture positive until they entered this study at 12 months, 8 months, and 3 months, respectively. Figure 1 shows the culture results for the first 3 months. During this period, the animals were not shedding unusual levels of E. coli O157:H7 and had CFU/swab typical of the concentration of E. coli O157:H7 carried by naturally and experimentally infected cattle in general, ranging from 103 CFU/swab to levels detectable only by enrichment culture (10). Table 1 shows monthly culture results until the animals were euthanized and necropsied. Interestingly, animal no. 135 carried E. coli O157:H7 naturally and remained culture positive for 12 months although the bacteria were detectable only by enrichment culture of RAMS samples after the first 4 weeks, indicating the samples had ≤30 CFU/swab (the limit of the direct plate technique) (10). It is not known if or for how long the animal was culture positive before day zero. This is one of the few descriptions of a naturally infected animal consistently shedding low numbers of E. coli O157 for 12 months. Digested chromosomal DNA obtained from select E. coli O157:H7 isolates each month was analyzed by pulsed-field gel electrophoresis, which confirmed that each animal was carrying the original strain of E. coli O157:H7 and no other strain throughout the study (data not shown).
FIG. 1.
Detection of E. coli O157:H7 by RAMS cultures from cattle. Three 5-month-old Holstein steers were penned separately. RAMS samples were cultured for E. coli O157 as indicated. Animal no. 135 was carrying a natural bovine E. coli O157 and was not challenged with a bacterial dose. Animal no. 711 received a single oral dose and animal no. 407 received a single rectal dose of 1010 CFU E. coli O157:H7 ATCC 43894 at day zero. E+, culture positive by enrichment culture only.
TABLE 1.
RAMS culture of long-duration E. coli O157:H7 culture-positive cattle
| Montha | Resultb for animal no.
|
||
|---|---|---|---|
| 711 | 135 | 407 | |
| 1 | D+ | D+ | D+ |
| 2 | D+ | E+ | D+ |
| 3 | E+ | E+ | E+ |
| 4 | E+ | E+ | |
| 5 | E+ | E+ | |
| 6 | E+ | E+ | |
| 7 | E+ | E+ | |
| 8 | E+ | ||
| 9 | E+ | ||
| 10 | E+ | ||
| 11 | E+ | ||
Weekly or biweekly RAMS cultures were analyzed each month.
D+, positive by direct RAMS culture; E+, positive by enrichment RAMS culture only.
Animals were euthanized and the necropsy performed. The contents of the digesta or swab or tissue samples were collected from various sections throughout the GIT from the rumen to the rectum. Three grams of digesta/tissue and a tissue swab sample were collected from each site and added to 27 ml or 3 ml Trypticase soy broth, respectively. Samples were held on ice and cultured within 2 h of collection. Tissue samples for histology were fixed in 10% neutral-buffered formalin, and representative sections from each level of GIT were trimmed into cassettes, dehydrated through a series of graded alcohols, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin. Sections were considered positive for gut-associated lymphoid tissue (GALT) if the mucosa contained either solitary lymphatic nodules or aggregated lymphatic nodules (Peyer's patches). The results are shown in Table 2. The total samples for cultures were 71, 83, and 83 throughout the GIT of animals no. 711, no. 135, and no. 407, respectively. Among them, E. coli O157:H7 was found only at RAJ sites even though many other GIT locations possessed lymphoid tissue, indicating this criterion alone did not suffice for E. coli O157:H7 colonization (Table 2). Interestingly, two of three animals were culture negative in fecal samples, although they were culture positive by RAMS culture, indicating the superior sensitivity of this technique. Studies by Naylor et al. found the RAJ mucosa to be the principal site of colonization of E. coli O157:H7 by necropsy after 14 to 28 days postinoculation (7). However, they also cultured E. coli O157:H7 in the contents and tissue samples from rumen, ileum, cecum, and colon, albeit at low levels (7). Also, Cray et al. showed the recovery of E. coli O157:H7 all along the GIT in the rumen, reticulum, abomasum, small intestine, ileum, cecum, and colon from cattle inoculated with this organism 2 to 18 days previously (1). These previous reports used cattle that had been culture positive for E. coli O157:H7 for less than 1 month.
TABLE 2.
Culture and lymphoid characteristics of gastrointestinal tract samples from long-duration E. coli O157:H7 culture-positive cattle
| Sample source | Animal no. 711
|
Animal no. 135
|
Animal no. 407
|
||||
|---|---|---|---|---|---|---|---|
| Sample typesa | No. of positive samplesb/total no. of samples | Sample types | No. of positive samples/total no. of samples | No. of GALT samples/total no. of samples | Sample types | No. of positive samples/total no. of samples | |
| Rumen | C, S, T | 0/18 | C, T | 0/4 | 0/2 | C, T | 0/4 |
| Reticulum | C, T | 0/2 | 0/1 | C, T | 0/2 | ||
| Omasum | C, T | 0/2 | 0/1 | C, T | 0/2 | ||
| Abomasum | C, T | 0/2 | 0/1 | C, T | 0/2 | ||
| Duodenum | C, T | 0/4 | 0/2 | C, T | 0/4 | ||
| Proximal jejunum | C, T | 0/6 | 0/3 | C, T | 0/6 | ||
| Middle jejunum | C, T | 0/6 | 1/3 | C, T | 0/6 | ||
| Distal jejunum | C, T | 0/6 | 1/3 | C, T | 0/6 | ||
| Ileum | C, T | 0/4 | 2/2 | C, T | 0/4 | ||
| Cecum | C, S, T | 0/9 | C, T | 0/4 | 2/2 | C, T | 0/4 |
| Gall bladder | C, S, T | 0/9 | C, T | 0/4 | C, T | 0/4 | |
| Proximal colon | C, S, T | 0/9 | C, T | 0/10 | 5/5 | C, T | 0/10 |
| Middle colon | S, T | 0/6 | C, T | 0/10 | 4/5 | C, T | 0/10 |
| Distal colon | S, T | 0/12 | C, T | 0/10 | 5/5 | C, T | 0/10 |
| RAJ | S, T | 3/5 | S, C, T | 3/5 | 3/5 | S, C, T | 4/6 |
| Feces | 0/3 | 0/4 | 1/3 | ||||
| Total | 3/71 | 3/83 | 23/40 | 5/83 | |||
C, samples from contents; S, samples from swab; T, samples from tissues.
Positive direct or enrichment culture.
This study showed that among animals with documented histories of long-duration E. coli O157:H7 colonization, the bacteria were cultured only from the RAJ mucosa and not from the gall bladder or other GIT locations even though these tissues were rich in GALT.
Acknowledgments
We thank Lonie Austin for technical assistance and animal handling.
This work was supported, in part, by the Idaho Agriculture Experiment Station, the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (grants 99-35201-8539 and 04-04562), Public Health Service grants U54-AI-57141, P20-RR16454, NO1-HD-0-3309, and P20-RR15587 from the National Institutes of Health, and grants from the United Dairymen of Idaho and the Idaho Beef Council.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gansheroff, L. J., and A. D. O'Brien. 2000. Escherichia coli O157:H7 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated. Proc. Natl. Acad. Sci. USA 97:2959-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong, K. C., M. Y. Kang, C. Heimke, J. A. Shere, I. Erol, and C. W. Kasper. 26 August 2006. Isolation of Escherichia coli O157:H7 from the gall bladder of inoculated and naturally-infected cattle. Vet. Microbiol. 119:339-345. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 5.Low, J. C., I. J. McKendrick, C. McKechnie, D. Fenlon, S. W. Naylor, C. Currie, D. G. Smith, L. Allison, and D. L. Gally. 2005. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 71:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice, D. H., H. Q. Sheng, S. A. Wynia, and C. J. Hovde. 2003. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 41:4924-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanderson, M. W., T. E. Besser, J. M. Gay, C. C. Gay, and D. D. Hancock. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet. Microbiol. 69:199-205. [DOI] [PubMed] [Google Scholar]
- 10.Sheng, H., M. A. Davis, H. J. Knecht, and C. J. Hovde. 2004. Rectal administration of Escherichia coli O157:H7: novel model for colonization of ruminants. Appl. Environ. Microbiol. 70:4588-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng, H., J. Y. Lim, H. J. Knecht, J. Li, and C. J. Hovde. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Baale, M. J., J. M. Sargeant, D. P. Gnad, B. M. DeBey, K. F. Lechtenberg, and T. G. Nagaraja. 2004. Effect of forage or grain diets with or without monensin on ruminal persistence and fecal Escherichia coli O157:H7 in cattle. Appl. Environ. Microbiol. 70:5336-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]