Abstract
Here we report associations between secondary metabolite production and phylogenetically distinct but closely related marine actinomycete species belonging to the genus Salinispora. The pattern emerged in a study that included global collection sites, and it indicates that secondary metabolite production can be a species-specific, phenotypic trait associated with broadly distributed bacterial populations. Associations between actinomycete phylotype and chemotype revealed an effective, diversity-based approach to natural product discovery that contradicts the conventional wisdom that secondary metabolite production is strain specific. The structural diversity of the metabolites observed, coupled with gene probing and phylogenetic analyses, implicates lateral gene transfer as a source of the biosynthetic genes responsible for compound production. These results conform to a model of selection-driven pathway fixation occurring subsequent to gene acquisition and provide a rare example in which demonstrable physiological traits have been correlated to the fine-scale phylogenetic architecture of an environmental bacterial community.
Actinomycetes are a remarkably prolific source of structurally diverse secondary metabolites, including many that possess pharmaceutically relevant biological activities (3). The search for new secondary metabolites from microorganisms in general, however, has long been confounded by the observation that different strains belonging to the same species can produce different secondary metabolites (38), while taxonomically diverse strains can produce identical metabolites (22). Contributing to the former are the challenges associated with using traditional taxonomic methods to differentiate among closely related bacteria. These challenges, however, can now be addressed using sequence-based approaches, which provide higher-level taxonomic resolution and opportunities to address the relationships between groups of related strains and the secondary metabolites they produce. These methods can also be used to recreate the evolutionary history of metabolic pathways and thus infer the role of lateral gene transfer (LGT) in the production of similar compounds by unrelated organisms.
The genus Streptomyces is the source of the vast majority of actinomycete secondary metabolites that have been discovered to date. Streptomyces species possess a single linear chromosome (2) consisting of a conserved core flanked by two nonconserved arms (1). The arms of the chromosome contain largely acquired DNA and are the location of most contingency genes, including those that code for nonessential functions, such as secondary metabolite production (1). Evidence that genes involved in secondary metabolism are subject to LGT can be inferred from sequence analysis (9), incongruent phylogenies (21), their occurrence on plasmids (20), and their chromosomal association with mobile genetic elements (33). Given these observations, the apparent lack of species specificity in actinomycete secondary metabolite production suggests that the genes encoding the biosynthesis of these compounds move freely and with little evolutionary effect among species.
LGT is widely recognized to have played an integral role in the evolution of bacterial genomes (8, 29). This mechanism of genetic exchange provides an extremely effective strategy by which bacteria can rapidly exploit new resources (30) and has been proposed as a selective force behind the physical clustering of genes within bacterial genomes (23). While it is increasingly clear that most bacterial genomes include large numbers of acquired genes (21), not all genes are equally subject to this process. As detailed in the complexity hypothesis (14), LGT is largely restricted to contingency genes, such as those involved with metabolism or virulence, and is generally not associated with informational genes, such as RNAs, which are typically members of large, complex systems. While it is clear that the gene clusters responsible for actinomycete secondary metabolite production are among those subject to LGT, the ecological and evolutionary significance of these events remain unknown.
We have been investigating a unique group of marine actinomycetes that are widely distributed in marine sediments (27, 28) and have also been isolated from a marine sponge (18). Phylogenetic analyses of these bacteria reveal three distinct but closely related clades, corresponding to the species Salinispora arenicola, Salinispora tropica (25), and a third species for which the name “S. pacifica” has been proposed (17) but which has yet to be formally described. We recently reported the cultivation of S. arenicola strains from broadly distributed locations, providing new evidence for the cosmopolitan distribution of an individual bacterial species (17). The cooccurrence of various Salinispora species raised the possibility that diversification within this genus is the result of ecological differentiation, not geographical isolation. The ecological basis for this diversification, however, has yet to be determined.
Salinispora strains are a prolific source of structurally diverse secondary metabolites, including salinosporamide A (10), a potent proteasome inhibitor that has entered phase I clinical trials as an anticancer agent. In the present study, detailed chemical analyses of 46 Salinispora strains reveal clear associations between phylotype and secondary metabolite production. This observation has implications for natural product discovery strategies and provides evidence that secondary metabolite production may be linked to ecological differentiation in marine actinomycetes.
MATERIALS AND METHODS
Bacterial strains and genetic analyses.
The 46 strains included in this study were obtained in culture as part of previous studies (15-17, 27, 28). They consist of two to nine representatives of each unique 16S rRNA gene variant observed from six broadly distributed collection sites (the Bahamas, the Red Sea, Guam, Palau, the U.S. Virgin Islands, and the Sea of Cortez [17]) with the exception of “S. pacifica” phylotypes “A” and “B,” for which only a single strain was examined. The 16S rRNA gene sequences for all of these strains, including CNS-237, the only strain that is new to this study, were obtained as reported previously (17). Phylogenetic trees were constructed using the neighbor-joining distance algorithm within the PAUP 4.0b10 software package (37).
A 440-b-p segment of the AHBA synthase (rifK) gene was PCR amplified using the forward (MAO1F; 5′-3′, TTCGAGCGGGAGTTTGCSG) and reverse (MAO1R; 5′-3", CSGTCATCAGCTTGCCGTTC) primers designed based on conserved regions of previously reported amino acid sequences (26). PCRs (50 μl) contained 10 to 100 ng DNA template, 0.4 μM (each) MAO1F and MAO1R primers, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 1.25 U AmpliTaq (Applied Biosystems, Foster City, CA), 5 μl MgCl2, 5 μl 10× PCR buffer, and 10% dimethyl sulfoxide. The PCR conditions were as follows: 95°C for 12 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by 72°C for 7 min. PCR products were purified and sequenced (using the MAO1F and MAO1R primers as described above). Sequence contigs were assembled using Sequencher (version 4.5; Gene Codes Corp., Ann Arbor, MI) and phylogenetic analyses performed using PAUP (version 4.0b10).
Fermentation and secondary metabolite screening.
For comparison purposes, all 46 Salinispora strains were cultured under identical fermentation conditions. These consisted of growing each strain in 1 liter of medium 1 [10 g starch, 4 g yeast extract, 2 g peptone, 5 ml Fe2(SO4)3·4H2O at 8 g/liter, 5 ml KBr at 20 g/liter, 1 liter seawater] in 2.8-liter Fernbach flasks with shaking at 230 rpm (gyrotary shaker; 1-in. stroke) at 25 to 27°C. Seed cultures (25 ml in 125-ml Erlenmeyer flasks, medium 1) were grown for 3 to 7 days prior to transfer. All culture flasks were sealed with cheesecloth overlaid with Bio-shield wrap (Allegiance Healthcare, Ontario CA). For each strain, sequential 25-ml aliquots of the fermentation broth were removed from the 1-liter culture either every 2 days from days 3 to 21 (for at least one representative of all phylotypes and collection sites) or from days 7 to 13 (which proved to be the optimal time range to detect all of the compounds). Time point samples were extracted three times with 25 ml EtOAc, the organic layer concentrated to dryness in vaccuo, and the residue resuspended at 10 mg/ml in 50% aqueous methanol.
Chemotype analysis.
Ten microliters of each extract was subjected to chemotype (liquid chromatography-mass spectrometry [LC-MS]) analysis (Agilent 1100) using a linear gradient of 10 to 80% aqueous acetonitrile over 30 min (Hypersil ODS; 4.6- by 100-mm column; flow, 0.7 ml/min; UV detection, 190 to 800 nm). Mass spectra were collected (scanning 100 to 2,000 atomic mass units) in the positive mode (ESI voltage, 6.0 kV; capillary temperature, 200°C; auxiliary and sheath gas pressure, 5 units and 70 lb/in2, respectively). Compounds were identified by comparison of molecular weights, UV spectra, and retention times with authentic standards or, in the case of the rifamycins and staurosporines, by comparison with published UV and MS values. The limits of accurate LC-MS detection were determined by serial dilution of standards to be ca. 5 to 50 μg compound per liter culture, well below the lowest yield at which any of the compounds were recovered (200 μg/liter). Details for compounds 1, 2, and 8, which are new structures isolated from Salinispora strains, are as reported previously (see references 10, 4, and 31, respectively). Details for compounds 5, 6, and 9, which are also new structures, will be published elsewhere. Retention times and pseudomolecular ions for the new Salinispora compounds are as follows: salinosporamide A, 17.0 min, MH+ 314, MNa+ 336; sporolide A, 12.0 min, MH+ 539, MNa+ 561; saliniketal, 15.8 min, MNa+ 418; cyanosporaside A, 10.1 min, MNa+ 440; salinolide A, 19.2 min, MNa+ 827; salinispyrone A, 19.0 min, MNa+ 315.
Nucleotide sequence accession numbers.
Sequence data have been deposited with GenBank and assigned the following accession numbers: AY040617 to AY040623, AY464533, AY464534, DQ224159 to DQ224165, DQ092624, DQ318246, and DQ313184 to DQ313189.
RESULTS
It was reported previously that strains belonging to the recently described actinomycete genus Salinispora have been cultured from broadly distributed tropical and subtropical locations (Fig. 1) (17). In the present study, these same strains, along with CNS-237 (which is new to this study), were used to address the relationships between actinomycete population structure and secondary metabolite production. Time course LC-MS analyses were performed on 46 strains that originated from 6 global collection sites (the Bahamas, the Red Sea, Guam, Palau, the U.S. Virgin Islands, and the Sea of Cortez) including multiple representatives of all phylotypes cultured from each site and year of collection. The results of >300 LC-MS analyses reveal a high degree of structural diversity (Fig. 2) and an unprecedented association between actinomycete phylotype and secondary metabolite production. To the best of our knowledge, this is the first evidence that fine-scale phylogenetic architecture is correlated to the production of secondary metabolites in bacteria.
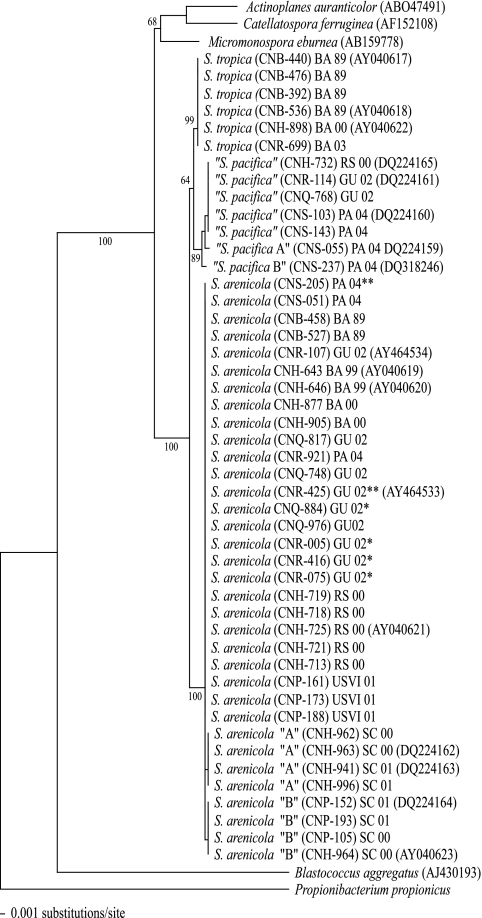
FIG. 1.
Neighbor-joining distance tree created from 46 nearly complete (1,449 nucleotides) Salinispora 16S rRNA gene sequences. The three major Salinispora phylotypes, consisting of the two formally described species S. tropica and S. arenicola and the proposed species “S. pacifica,” are clearly delineated. Species names are followed by strain number, source (BA, Bahamas; RS, Red Sea; GU, Guam; PA, Palau; USVI, U.S. Virgin Islands; SC, Sea of Cortez), year of collection (89 = 1989, etc.), and accession number (for representative sequences). *, produces compound 6; **, produces cyclomarin A (compound 7). P. propionicus and B. aggregatus were used as outgroups.
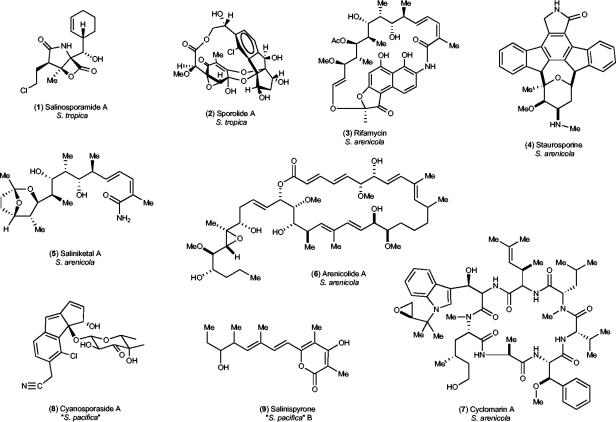
FIG. 2.
Secondary metabolites produced by Salinispora strains. Compound names and the producing species or phylotype are listed under the structures. Compounds 1, 2, 5, 6, 8, and 9 are new secondary metabolites recently discovered from Salinispora strains, while compounds 3, 4, and 7 were previously reported for other actinomycetes (see references 12, 34, and 36, respectively).
Regardless of the geographical origin of the Salinispora strains, secondary metabolite production followed a well-defined pattern. Of the two described species, S. tropica has thus far been cultured only from the Bahamas, and the six strains examined all share 100% 16S rRNA gene sequence identity (17). These six strains, which were obtained in culture during three expeditions spanning 14 years, all produced the same two secondary metabolites (Fig. 2). These molecules are represented by the proteasome inhibitor salinosporamide A (compound 1) (10), which contains a rare bicyclic beta-lactone gamma-lactam ring system, and sporolide A (compound 2), an unprecedented, polyketide-derived macrolide (4) with unknown biological activity. No other secondary metabolites were identified from this species using the fermentation conditions described in this study.
Unlike S. tropica, S. arenicola has a cosmopolitan distribution in tropical and subtropical sediments (17). A total of 30 S. arenicola strains from 6 geographically distinct locations were examined, and they all produced compounds in the well-studied rifamycin (compound 3; antibiotic) and staurosporine (compound 4; protein kinase inhibitor) classes, as well as the new, bicyclic compound saliniketal (compound 5), which shares some biosynthetic features with rifamycin (compound 3). These three compounds (3 to 5) were produced by all 30 of the strains examined and represent the core S. arenicola chemotype. Like S. tropica, the S. arenicola strains share 100% 16S rRNA gene sequence identity, with the exception of phylotypes “A” and “B,” both cultured from the Sea of Cortez, each of which differs from all other S. arenicola strains at one of two nucleotide positions (Fig. 1) (17). The secondary metabolites produced by phylotypes “A” and “B” did not differ from the core S. arenicola chemotype.
In addition to compounds 3 to 5, four S. arenicola strains cultured from Guam (CNR-005, CNR-416, CNR-075, and CNQ-884) also produced the unique polyketide-derived macrolide arenicolide A (compound 6), which is accessory to the core chemotype. These four strains, all cultured from different sediment samples, cannot be distinguished phylogenetically based on 16S or gyrB sequence analysis (data not shown). A second deviation from the core S. arenicola secondary metabolite profile was observed in strains CNS-205 (Palau) and CNR-425 (Guam). These strains produced the cyclic peptide cyclomarin A (compound 7), previously reported by our laboratory for a marine-derived Streptomyces species (36), in addition to the core compounds 3 to 5. Like the arenicolide A (compound 6)-producing strains, these two strains are phylogenetically indistinguishable within the S. arenicola clade. Thus, while all S. arenicola strains produced compounds 3 to 5, we identified six cases in which strains produced one additional (accessory) secondary metabolite (compound 6 or 7), and none of these strains could be distinguished phylogenetically based on 16S rRNA gene sequences.
“S. pacifica” is the third major Salinispora phylotype thus far detected, and although it was not as frequently encountered or as widely distributed as S. arenicola, it possesses the greatest intraclade phylogenetic diversity (Fig. 1) (17). The most commonly encountered “S. pacifica” sequence is represented by five strains that share 100% 16S rRNA gene sequence identity. These strains, which were isolated from Guam, Palau, and the Red Sea, all produced cyanosporaside A (compound 8), a novel cyclopenta[a]indene glycoside with a rare cyano functionality and a new 3-keto-pyranohexose sugar (31). Interestingly, two additional “S. pacifica” sequences, each represented by only one strain, have recently been identified. Both of these strains possessed unique secondary metabolite profiles, with phylotype “A” (strain CNS-055) producing a new polyene metabolite, the structure of which is currently under investigation, and phylotype “B” (strain CNS-237) producing a series of unique pyrones characterized by salinispyrone A (compound 9). Neither of these strains produced cyanosporaside A (compound 8). Although the sequences of the “A” and “B” strains differ from that of “S. pacifica” by only two and three nucleotides, respectively, no overlap was detected among the secondary metabolites produced by the three chemotypes. Efforts are currently under way to obtain additional “S. pacifica” strains in culture to better understand the relationships between chemotype and 16S rRNA gene sequence within this relatively diverse phylotype.
Based on the 46 strains examined, we have identified 5 unique Salinispora chemotypes, 1 each from S. tropica and S. arenicola and 3 from “S. pacifica.” All five of these chemotypes produce nonoverlapping suites of secondary metabolites and can be clearly distinguished by 16S rRNA gene sequence analysis. Remarkably, strains within the “S. pacifica” clade that differed by as few as two nucleotides produced unrelated secondary metabolites, indicating that distinct chemotypes can also be resolved at the subspecies level. This level of resolution, however, requires careful corrections for PCR and sequencing errors that include consideration of the secondary structure of the 16S rRNA gene.
The secondary metabolites isolated from Salinispora strains are the single most distinctive phenotypic property that differentiates the three major phylotypes. These compounds have varied biological activities (Table 1); however, they have thus far been tested only in limited, biomedically relevant assays, and therefore their activities in nature remain unknown. Compounds 1, 2, and 7 to 9 were all discovered as part of prior studies and did not display antibacterial activity against methicillin-resistant Staphylococcus aureus or vancomycin-resistant Enterococcus faecium (data not shown). The fact that these compounds do not possess potent, broad-spectrum antibiotic activity suggests that antibiosis may not be a common ecological function of Salinispora secondary metabolites, a concept recently proposed more generally for microbial secondary metabolites (7). Clearly, further tests are required if the ecological functions of these metabolites are to be determined.
TABLE 1.
Secondary metabolites isolated from Salinispora spp. and their major biological activities
| Compound no. (name) | Source | Biological activity | Molecular target |
|---|---|---|---|
| 1 (salinosporamide A) | S. tropica | Anticancer | Proteasome |
| 2 (sporolide A) | S. tropica | Unknown | Unknown |
| 3 (rifamycin) | S. arenicola | Antibiotic | RNA polymerase |
| 4 (staurosporine) | S. arenicola | Anticancer | Protein kinase |
| 5 (saliniketal) | S. arenicola | Cancer chemoprevention | Ornathine decarboxylase |
| 6 (arenicolide A) | S. arenicola | Unknown | Unknown |
| 7 (cyclomarin A) | S. arenicola | Anti-inflammatory, antiviral | Unknown, unknown |
| 8 (cyanosporaside A) | “S. pacifica” | Unknown | Unknown |
| 9 (salinispyrone A) | “S. pacifica” B | Unknown | Unknown |
The dramatic structural differences and lack of overlap in the secondary metabolites produced by the five closely related Salinispora phylotypes suggest that the biosynthetic pathways responsible for their production were not inherited vertically from a common Salinispora ancestor. The observation that three of the compounds (rifamycin [compound 3], staurosporine [compound 4], and cyclomarin A [compound 7]) have been reported for strains belonging to other actinomycete families (see references 12, 34, and 36, respectively) further supports this suggestion. To investigate the role of LGT in the acquisition of biosynthetic pathways in Salinispora species and to determine if the secondary metabolites produced in culture by one Salinispora species are predictive of the genetic capacity of that species, we probed all three major phylotypes for the presence of the rifamycin gene cluster, the product of which is consistently observed with S. arenicola. The gene selected to represent this cluster was that encoding 3-amino, 5-hydroxy benzoic acid (AHBA) synthase (rifK), which catalyzes the last step in the biosynthesis of AHBA, the unusual starter unit in the rifamycin (compound 3) mixed polyketide synthase/nonribosomal peptide synthase pathway (12).
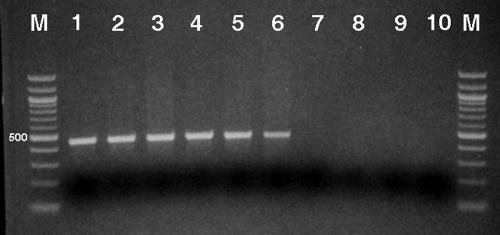
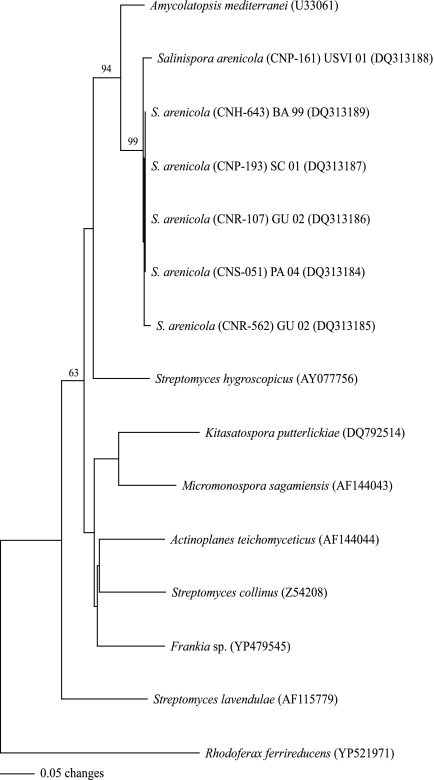
Using rifK-specific primers, six S. arenicola (rifamycin-producing) strains all yielded PCR products of the expected size (440 bp) whose corresponding proteins showed a high level of homology (90% amino acid identity) to the product of the AHBA synthase gene from the rifamycin producer Amycolatopsis mediterranei (Fig. 3). No PCR products were observed from two S. tropica or two “S. pacifica” (non-rifamycin-producing) strains. In addition, no products of the correct size or with rifK sequence homology were obtained from either of these phylotypes when degenerate primers were tested. These results provide evidence that S. arenicola is the only species that possesses the rifK gene and that the LC-MS results for this compound were indicative of genotype as opposed to phenotypic responses to culture conditions. Phylogenetic analysis of the S. arenicola AHBA synthase gene reveals a strongly supported and recently shared evolutionary history with A. mediterranei (Fig. 4), the original source of rifamycin and a member of the actinomycete family Pseudonocardiaceae. The S. arenicola rifK gene sequences did not clade with Micromonospora or Actinoplanes homologues, even though the latter two genera share membership with Salinispora in the family Micromonosporaceae. The high level of sequence homology between the A. mediterranei and S. arenicola rifK genes suggests that the gene cluster responsible for rifamycin biosynthesis has recently moved by LGT between these two species. This result is supported by similar analyses of rifamycin ketosynthase genes from sponge-derived Salinispora strains (19).
FIG. 3.
AHBA synthase gene (rifK) PCR amplification. Presence of the AHBA synthase (rifK) gene in the three major Salinispora phylotypes was assessed using PCR primers specific for a 440-bp region of the gene. M, 100-bp marker; lanes 1 to 6, S. arenicola (strains CNH-643, CNR-562, CNP-161, CNR-107, CNP-193, and CNS-051); lanes 7 and 8, S. tropica (strains CNB-440 and CNB-476); lanes 9 and 10, “S. pacifica” (strains CNH-732 and CNQ-768). Loading dye is seen as a dark band.
FIG. 4.
Neighbor-joining distance tree created from translated AHBA synthase gene sequences (88 amino acids) obtained from six S. arenicola strains and NCBI BLAST homologues. Bootstrap results based on 1,000 resamplings are presented for values greater than 60% at the respective nodes. The tree was rooted using a related aminotransferase gene from R. ferrireducens.
DISCUSSION
A careful analysis of secondary metabolite production within the genus Salinispora reveals clear phylotype specificity, with no overlap among any of the compounds produced by either of the two formally described species (S. tropica and S. arenicola) or by the three sequence variants observed within the third major phylotype, for which the name “S. pacifica” has been proposed. In the case of the “S. pacifica” phylotype, where the greatest intraclade sequence diversity was observed, this metabolite specificity appears to extend to the subspecies level. Species-specific secondary metabolite production within S. arenicola was independent of the geographical site from which the strains were collected, indicating that a suite of secondary metabolites can represent a consistent phenotype associated with a globally distributed population. These observations conform to a model of selection-driven pathway fixation, thus implying that secondary metabolites have important functions in the survival of the producing strains. These results also indicate that the cultivation of phylogenetically distinct strains represents a productive strategy for secondary metabolite discovery, while the cultivation of clonal 16S rRNA gene conspecific strains, regardless of geographical origin, will largely lead to the isolation of redundant chemistry.
The consistency of the core secondary metabolite profiles observed for the two formally described Salinispora species must be carefully interpreted in a broader context, since most currently described bacterial taxa include considerably more intraspecific sequence diversity than the strains analyzed here. If similar associations occur among other actinomycete populations, they would be expected among strains that cluster using a high degree of 16S rRNA gene sequence similarity (e.g., >99%), not in association with most currently characterized species, which generally encompass considerably more genetic diversity. Based on our results for the genus Salinispora, actinomycete secondary metabolite profiles appear to represent useful criteria by which closely related but clearly distinguishable units of diversity can be delineated. The extent to which this concept applies more broadly to other secondary metabolite-producing bacteria, however, requires further sequence-based testing.
The observation that 6 of the 30 S. arenicola strains examined each produced one accessory metabolite (compound 6 or 7) outside of the core chemotype for this species (compounds 3 to 5) warrants discussion. If gene clusters encoding secondary metabolite biosynthesis are periodically acquired by LGT, then it is not surprising that a strain producing a different compound is occasionally encountered. Excluding differential pathway expression among strains, the results observed for these six S. arenicola strains could be an indication of recent gene acquisition or weak selection preventing pathway fixation. Given that compounds 6 and 7 were observed only with strains recovered from Guam or Palau, these pathways appear to have been regionally acquired subsequent to the global fixation of the core chemotype. Although only six strains among the two formally described species were observed to deviate from their respective chemotypes, additional sampling would likely lead to additional examples. It would also not be surprising to find instances where a pathway was inherited vertically from a common ancestor and thus is present in closely related species; however, no such examples were found in the present study. Strains that deviate from their species-specific secondary metabolite profiles may provide a glimpse into the dynamic nature of LGT and clues into the recent evolutionary history of individual biosynthetic gene clusters.
PCR probing for the rifK gene and subsequent sequence analysis provided evidence that the rifamycin biosynthetic gene cluster is present only in S. arenicola and that it was recently the subject of LGT. This observation is further supported by the phylogenetic analysis of ketosynthase gene sequences obtained from S. arenicola strains cultured from a marine sponge (19). The genetic organization of actinomycete secondary metabolite biosynthesis has been studied in detail and reveals a generally ordered and tight packaging of genes into multioperon clusters that include not only structural and regulatory elements but resistance genes that allow the producing strain to avoid autotoxicity (e.g., see reference 35). This genetic arrangement is facilitated by lateral gene transfer and provides a fitting corollary to the selfish operon model (23). Thus, it becomes possible that the lateral transmission of gene clusters involved in actinomycete secondary metabolite production provides an effective strategy for sampling (as opposed to accumulating [29]) genes from a common gene pool (24), the products of which may provide opportunities for immediate access to a new ecological niche (ecotype differentiation) or an effective mechanism to outcompete conspecific strains (selective sweep [6]). The results presented here support previous findings that laterally acquired genes can subsequently be transmitted vertically and thereby serve as stable, species-specific markers (30). While the importance of acquired genes to niche invasion has been discussed in relation to pathogens (13), the data presented here also raise the intriguing possibility that pathway acquisition (i.e., secondary metabolite production) is associated with ecological differentiation and thus represents a previously unrecognized evolutionary force driving diversification within the actinomycetes.
It is now widely recognized that bacterial secondary metabolites have important ecological functions and are not merely artifacts of laboratory culture or metabolic waste products (5, 11). However, outside of quorum sensing (e.g., see reference 32) and iron acquisition (e.g., see reference 39), most of these ecological roles remain uncharacterized. The remarkable diversity of Salinispora secondary metabolites complicates any future interpretation of their ecological significance, since each metabolite may have a distinct ecological function or they may act synergistically, a well-known phenomenon associated with actinomycete secondary metabolism (5). Although the ecological functions of Salinispora secondary metabolites have yet to be determined, metabolite production represents the major phenotypic difference between the two described species (minor differences in sole carbon source utilization were also detected [25]).
Based on recent actinomycete genome sequences (1, 33), it is unlikely that the full biosynthetic potential of the three Salinispora phylotypes was expressed using a single set of fermentation conditions. Nor is it surprising that an individual actinomycete strain produces a diverse suite of secondary metabolites. What is surprising about the results presented here is that careful phylogenetic analyses reveal distinct sequence clusters that are correlated to secondary metabolite production. These results contradict the traditional (pre-molecular systematics) paradigm that actinomycete secondary metabolite profiles represent a strain-specific phenotype (40) and support the recent observation that fungal secondary metabolite profiles represent key taxonomic markers (22).
Further research is required to determine the extent to which secondary metabolite production is correlated to phylogeny in the actinomycetes. Additional data will also be needed to better understand the potential link between the acquisition of secondary metabolite gene clusters and ecological diversification. What is clear, however, from an analysis of the genus Salinispora is that distinct biosynthetic differences can be detected among closely related actinomycete phylotypes and that phylogenetic diversity can be used as an effective tool to guide secondary metabolite discovery. Although it remains unknown how broadly these findings can be applied to other bacteria, it appears that a diversity-based discovery strategy, focused on chemically prolific actinomycetes residing in poorly studied environments, such as marine sediments, represents a productive strategy for the discovery of novel actinomycete products.
Acknowledgments
This work was supported by grants from the University of California Industry University Cooperative Research Program (IUCRP BioSTAR 10354) and the National Institutes of Health (CA44848). DNA sequencing was performed by the Molecular Pathology Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center support grant no. 5P0CA23100-16.
P.R.J. and W.F. are stockholders in and advisors to Nereus Pharmaceuticals, the corporate sponsor of the IUCRP award. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies.
We thank C. Kauffman (UCSD) for general technical assistance and T. Mincer (MIT) for contributions to strain isolation and sequencing.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárranga, G. L. Challis, N. R. Thompson, K. D. James, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., and J. Parkhill. 2004. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 38:771-791. [DOI] [PubMed] [Google Scholar]
- 3.Bérdy, J. 2005. Bioactive microbial metabolites. J. Antibiot. 58:1-26. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan, G. O., P. G. Williams, R. H. Feling, C. A. Kauffman, P. R. Jensen, and W. Fenical. 2005. Sporolides A and B: structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org. Lett. 7:2731-2734. [DOI] [PubMed] [Google Scholar]
- 5.Chalis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100:14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 7.Davies, J. 2006. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33:496-499. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree of life. Science 284:2124-2128. [DOI] [PubMed] [Google Scholar]
- 9.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 2001. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Leeuwenhoek 79:127-133. [DOI] [PubMed] [Google Scholar]
- 10.Feling, R. H., G. O. Buchanan, T. J. Mincer, C. A. Kauffman, P. R. Jensen, and W. Fenical. 2003. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 42:355-357. [DOI] [PubMed] [Google Scholar]
- 11.Firn, R. D., and C. G. Jones. 2000. The evolution of secondary metabolism—a unifying model. Mol. Microbiol. 37:989-994. [DOI] [PubMed] [Google Scholar]
- 12.Floss, H. G., and T. W. Yu. 2005. Rifamycin—mode of action, resistance, and biosynthesis. Chem. Rev. 105:621-632. [DOI] [PubMed] [Google Scholar]
- 13.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 14.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, P. R., R. Dwight, and W. Fenical. 1991. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 57:1102-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, P. R., E. Gontang, C. Mafnas, T. J. Mincer, and W. Fenical. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 7:1039-1048. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, P. R., and C. Mafnas. 2006. Biogeography of the actinomycete genus Salinispora. Environ. Microbiol. 8:1881-1888. [DOI] [PubMed] [Google Scholar]
- 18.Kim, T. K., Garson, M. J., and J. A. Fuerst. 2005. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 7:509-518. [DOI] [PubMed] [Google Scholar]
- 19.Kim, T. K., A. K. Hewavitharana, P. N. Shaw, and J. A. Fuerst. 2006. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 72:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinashi, H., M. Shimaji, and A. Sakai. 1987. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis. Nature 328:454-456. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, T. O., J. Smedsgaard, K. F. Nielsen, M. E. Hansen, and J. C. Frisvad. 2005. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 22:672-695. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5:355-359. [DOI] [PubMed] [Google Scholar]
- 24.Maiden, M. C. J., B. Malorny, and M. Achtman. 1996. A global gene pool in the neisseriae. Mol. Microbiol. 21:1297-1298. [DOI] [PubMed] [Google Scholar]
- 25.Maldonado, L., W. Fenical, M. Goodfellow, P. R. Jensen, C. K. Kauffman, and A. C. Ward. 2005. Salinispora gen nov., sp. nov., Salinispora arenicola sp. nov., and S. tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Appl. Microbiol. 55:1759-1766. [DOI] [PubMed] [Google Scholar]
- 26.Mao, Y., M. Varogula, and D. H. Sherman. 1999. Genetic localization and molecular characterization of two key genes (mitAB) required for biosynthesis of the antitumor antibiotic mitomycin C. J. Bacteriol. 181:2199-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mincer, T. J., W. Fenical, and P. R. Jensen. 2005. Cultured and culture-independent diversity within the obligate marine actinomycete genus Salinispora. Appl. Environ. Microbiol. 71:7019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mincer, T. J., P. R. Jensen, C. A. Kauffman, and W. Fenical. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial evolution. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Ochman, H., E. Lerat, and V. Daubin. 2005. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 102:6595-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh, D.-C., P. G. Williams, C. A. Kauffman, P. R. Jensen, and W. Fenical. 2006. Cyanosporasides A and B, cyano- and chloro-cyclopenta [a]indene glycosides from the marine actinomycete “Salinispora pacifica.” Org. Lett. 8:1021-1024. [DOI] [PubMed] [Google Scholar]
- 32.Okada, M., I. Sata, S. J. Cho, H. Iwata, T. Nishio, D. Dubmau, and Y. Sakagami. 2005. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1:23-24. [DOI] [PubMed] [Google Scholar]
- 33.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, et al. 2001. Genome sequencing of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omura, S., Y. Sasaki, Y. Iwai, and H. Takeshima. 1995. Staurosporine, a potentially important gift from a microorganism. J. Antibiot. 48:535-548. [DOI] [PubMed] [Google Scholar]
- 35.Pojet, F., L. Shu-Ming, and L. Heide. 2002. Molecular cloning and sequence analysis of the chlorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 36.Renner, M. K., Y.-C. Shen, X.-C. Cheng, P. R. Jensen, W. Frankmoelle, C. A. Kauffman, W. Fenical, E. Lobkovsky, and J. Clardy. 1999. Cyclomarins A-C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Am. Chem. Soc. 121:11273-11276. [Google Scholar]
- 37.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA.
- 38.Waksman, S. A., and E. Bugie. 1943. Strain specificity and production of antibiotic substances. Proc. Natl. Acad. Sci. USA 29:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkelmann, G. 2002. Microbial siderophore-mediated transport. Biochem. Soc. Trans. 30:691-696. [DOI] [PubMed] [Google Scholar]
- 40.Zahner, H. 1979. What are secondary metabolites? Folia Microbiol. 24:435-443. [DOI] [PubMed] [Google Scholar]