Abstract
Bacteria play a major role in marine CO cycling, yet very little is known about the microbes involved. Thirteen CO-oxidizing Stappia isolates obtained from existing cultures, macroalgae, or surf samples representing geographically and ecologically diverse habitats were characterized using biochemical, physiological, and phylogenetic approaches. All isolates were aerobic chemoorganotrophs that oxidized CO at elevated (1,000 ppm) and ambient-to-subambient concentrations (<0.3 ppm). All contained the form I (OMP) coxL gene for aerobic CO dehydrogenase and also the form II (BMS) putative coxL gene. In addition, some strains possessed cbbL, the large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase, suggesting the possibility of lithotrophic or mixotrophic metabolism. All isolates used a wide range of sugars, organic acids, amino acids, and aromatics for growth and grew at salinities from 5 to 45 ppt. All but one isolate denitrified or respired nitrate. Phylogenetic analyses based on 16S rRNA gene sequences indicated that several isolates could not be distinguished from Stappia aggregata and contributed to a widely distributed species complex. Four isolates (of strains GA15, HI, MIO, and M4) were phylogenetically distinct from validly described Stappia species and closely related genera (e.g., Ahrensia, Pannonibacter, Pseudovibrio, and Roseibium). Substrate utilization profiles, enzymatic activity, and membrane lipid composition further distinguished these isolates and supported their designations as new Stappia species. The observed metabolic versatility of Stappia likely accounts for its cosmopolitan distribution and its ability to contribute to CO cycling as well as other important biogeochemical cycles.
The marine genus Stappia encompasses four phylogenetically distinct chemoorganotrophic species in the α-2 subgroup of the Proteobacteria (9, 27, 38). Stappia stellulata and Stappia aggregata were originally isolated from coastal marine water column and sediment samples, assigned to the genus Agrobacterium, and subsequently transferred to the genus Stappia (1, 29, 38). Stappia alba (27), Stappia marina (9), and various Stappia-like isolates have since been obtained from numerous widely distributed sources, including warm temperate surface and permanently cold deep-sea waters, sediments, phytoplankton, macroalgae, invertebrates, and salt marshes (2, 3, 5, 9, 13, 17, 26, 27, 33; Donachie et al., unpublished data). In addition, the presence of Stappia or Stappia-like taxa in a similar range of habitats has also been inferred from cultivation-independent analyses (3, 26; Donachie et al., unpublished).
Neither the original description (29) nor the subsequent work by Uchino et al. (38) addressed the geographic distribution of Stappia or its physiological and ecological attributes. Results of subsequent studies have shown that Stappia-like isolates can account for a significant percentage of cultivable α-Proteobacteria containing dioxygenase genes (3). Stappia or Stappia-like isolates have been reported to produce sodium channel-blocking proteins (5, 34) and a rhizobactin-like siderophore (20). Stappia strains are thus functionally versatile, occupy several ecological niches, and participate in biogeochemical cycles and processes that are important on microscales to global scales (e.g., CO oxidation and denitrification).
To date, all Stappia strains have been obtained from heterotrophic enrichments. Accordingly, the genus has been described as chemoorganotrophic (38). Some strains, however, may function as facultative lithotrophs. All Stappia strains examined to date oxidize carbon monoxide and possess the form I (OMP) coxL gene encoding the large subunit of carbon monoxide dehydrogenase (CODH) (9, 13). Some also contain a gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (cbbL) and may be able to couple CO utilization to CO2 fixation (13).
In spite of their ubiquity, relatively little is known about the phylogenetic and physiological relationships among Stappia isolates or the responses to environmental variables that may affect their distributions and activities. We have enriched and isolated Stappia spp. from geographically diverse habitats and characterized novel strains using biochemical, physiological, and molecular approaches. We have also determined the extent to which isolates oxidize CO at elevated and near ambient concentrations. The results collectively support the designation of four new species: Stappia conradae, Stappia meyerae, Stappia carboxidovorans, and Stappia kahanamokuae.
MATERIALS AND METHODS
Culture sources and isolation.
Stappia aggregata and Stappia sp. strains CV902-700 and CV812-530 were obtained from the Damariscotta River, Maine (2). Stappia sp. strain MIO was obtained from a marine methanotrophic consortium (gift of M. Takeuchi, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan). S. stellulata was obtained from the Deutsche Samlung für Mikroorganizmen und Zellkulturen (Braunschweig, Germany). Stappia sp. strains SE 09 and SE11 were obtained from the Duplin River, Georgia (A. Buchan, University of Tennessee) (3). Stappia strains M4 and M8 were obtained from enrichments based on marine macroalgae as previously described (13).
Additional Stappia strains were obtained from seawater and macroalgae by incubating 20 to 100 ml basal salts medium (PYE) (20) containing 0.01 to 0.05% yeast extract and 25 mM pyruvate with 1 ml of surf water samples from Ka Lae, Hawaii, or 1 to 5 g fresh weight of macroalgae: Gracilaria sp., Ulva sp., Caulerpa sp. or an unidentified, tube-shaped green alga, Ascophyllum nodosum, or Ulva lactuca (the latter was from the Damariscotta River, Walpole, ME). CO was added at 100- to 1,000-ppm concentrations to enrichment flask headspaces, and uptake was monitored using gas chromatography (11). Enrichments positive for CO oxidation were diluted serially and plated onto PYE prepared with artificial seawater (ASW) (MPYE). Colonies were selected arbitrarily and transferred to liquid MPYE and grown to stationary phase. Cultures that oxidized CO were purified by plating serial dilutions on solid MPYE and transferred to liquid medium.
Morphological, physiological, and biochemical characterization.
Growth-supporting substrates were examined using a basal salts medium supplemented with 0.05% yeast extract (21) and various carbon substrates. Growth was assayed spectrophotometrically after 48 h and compared to controls containing 0.05% yeast extract only. Selected biochemical traits were assayed using API 20 NE strips according to the manufacturer's instructions (BioMerieux, Inc., France). Catalase and oxidase tests and Gram staining were performed using standard methods (4). Major membrane lipids of S. aggregata and strains BrT4, GA15, HI, M4, M8, and MIO were obtained from Microbial ID, Inc. (Newark, DE). A Zeiss Axioscope fitted with a Neofluar 100× objective and an AxioCam MR digital camera was used to obtain images of logarithmically growing cells for size estimates and to determine motility.
G+C determination.
Nucleotides were hydrolyzed enzymatically from RNA-free genomic DNA, then dephosphorylated to produce nucleosides. Nucleosides were separated by high-pressure liquid chromatography using a C18 column and an isocratic mobile phase of ammonium phosphate and methanol and detected by UV absorption (254 nm). Reference standards include genomic DNA from Escherichia coli, Burkholderia xenovorans, and Mycobacterium smegmatis for which percent G+C contents are known from genomic sequences.
CO utilization.
To determine maximum CO uptake velocities, Stappia isolates were grown to stationary phase in MPYE, harvested by centrifugation, washed with buffered ASW, and resuspended to an optical density (OD) of 0.5 in buffered ASW containing mineral salts (21) and 0.005% yeast extract. Five-milliliter cell suspensions for each isolate were transferred to triplicate 160-cm3 serum bottles. CO was added to bottle headspaces (1,000 ppm), and cultures were incubated with shaking (200 rpm) at 30°C. CO uptake was monitored at intervals by gas chromatography. Maximum uptake velocities were expressed per milligram of protein after the measurement of culture protein contents using the bicinchoninic acid assay kit (Pierce, Inc.).
The ability of Stappia isolates to consume CO at ambient concentrations was assessed using triplicate 10-ml cultures of strains M4, GA15, MIO, and HI that were grown to stationary phase and transferred to 160-cm3 serum bottles. The cultures were sealed and incubated at 30°C with rotary shaking (200 rpm) and initial CO concentrations of about 2 ppm. Headspace subsamples were obtained at intervals by needle and syringe for analysis using an RGA-3 reduced gas analyzer (Trace Analytical, Inc.).
Heterotrophic substrate effects on S. aggregata CO uptake.
S. aggregata grown overnight on MPYE was harvested by centrifugation, washed with buffered ASW, and resuspended in basal salts with 0.005% yeast extract. Six sets of sealed triplicate 160-cm3 serum bottles containing 4.5 ml of marine basal salts (21) with glucose concentrations of 0, 0.5, 2.5, 5.0, 10, or 20 mM were inoculated by syringe and needle with 0.5 ml of washed culture. CO was added to the bottle headspaces (1,000 ppm), and absorbance (Abs; optical density at 600 nm [OD600]) and headspace CO concentrations were monitored at suitable intervals. CO uptake rates were determined from analyses of CO concentrations over time using KaleidaGraph software (version 4.0.5; Synergy Software, Inc.) as previously described by Hardy and King (6, 12). Cell biomass was estimated from absorbance using the following empirical expression after correcting for differences in absorbance at 600 and 660 nm (16):
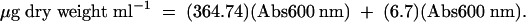 |
Growth response to varied salinities.
Growth responses to salinities ranging from 0 to 45 ppt were determined for strains GA15, HI, M4, and MIO by growing the isolates in 250-ml Erlenmeyer flasks containing 50 ml basal salts medium (20) in artificial seawater supplemented with 0.05% yeast extract; salinities were adjusted to 5, 15, 25, 35, and 45 ppt. Nonmarine media were created using deionized water. A sodium-free medium was created by using nonsodium salts (from 21) and deionized water. Filter-sterilized glucose (25 mM) was added to sterile basal medium. All flasks were inoculated with fresh cultures (<24 h) grown on MPYE. Cultures used for sodium-free medium were harvested by centrifugation (1 min; 13,000 rpm), washed in buffered ASW, and resuspended in sodium-free medium prior to inoculation. Culture absorbance (OD600) was monitored immediately following inoculation and at intervals thereafter. Specific growth rate constants were determined by fitting absorbance data to a modified Gompertz equation (40) using KaleidaGraph software (version 4.0.5; Synergy Software, Inc.) to obtain the fitting parameters. All assays were conducted in triplicate.
DNA extraction, PCR, sequencing, and analysis.
Genomic extracts were obtained using UltraClean microbial DNA isolation kits (MO BIO Laboratories, Inc., Carlsbad, CA) as we have previously described (13). 16S rRNA genes were amplified using primers 27f and 1492r (18, 30). PCRs were carried out using 50-μl volumes (13). A 492- to 495-bp cbbL fragment was amplified and sequenced using primers K2 and V2 as previously described by Nanba et al. (22). OMP coxL genes (form I; about 1,260 bp) and putative coxL genes (BMS, form II; about 1,260 bp) were also amplified from the isolates and sequenced as we have described previously (13). PCR products of the correct size were purified using a MO BIO UltraClean PCR cleanup kit (MO BIO Laboratories, Inc., Carlsbad, CA) and then sequenced bidirectionally with an ABI model 377 sequencer at the University of Maine DNA Sequencing Facility using the amplification primers (Orono, ME).
Phylogenetic analysis.
16S rRNA gene sequences for Stappia isolates and for various phylogenetic neighbors were obtained from the Ribosomal Database Project (www.rdp.cme.msu.edu/). Alignments were imported into ClustalX and further aligned manually as necessary. Aligned sequences were analyzed using maximum parsimony and distance (neighbor joining) algorithms as implemented in PAUP (version 4.0b; Sinauer Associates, Inc., Sunderland, MA). Maximum likelihood analyses were implemented using PHYML (http://atgc.lirmm.fr/phyml/) with an HKY model for base substitution and 100 bootstrap replicates.
BOX PCR.
Genomic extracts were amplified using the BOXAR1 primer (19) in 50-μl PCRs containing recommended concentrations of deoxynucleoside triphosphates, buffers, Mg2+, 1.25 U MasterTaq DNA polymerase (Brinkmann, Inc., Westbury, NY), dimethyl sulfoxide (2.5 μl), and the template. Amplification conditions were as previously described by Louws et al. (19). PCR products were electrophoresed at 70 V on a 1.25% agarose gel for 4 h at 4°C and visualized with GelRed (Biotium, Inc., Hayward, CA). Molecular weights of fragments were determined using Kodak Imaging software. A similarity index was created by dividing two times the number of shared fragments by the total number of fragments of a given pair of strains.
RESULTS
Morphological, physiological, and biochemical characterization.
All strains were gram negative, nonsporing, motile rods with average dimensions of about 2 μm by 0.8 μm (length times width) (Table 1). All strains form irregular to star-shaped aggregates in liquid culture. Strain M4 also produced irregular forms in older cultures, including cells fused into a large spherical aggregate and cells that ballooned centrally or terminally. All strains formed circular, entire, smooth, slightly convex colonies with a light tan color on MPYE agar.
TABLE 1.
Substrate utilization profiles for S. aggregata, S. stellulata, and strains MIO, HI, BrT4, GA15, M4, and M8a
| Characteristic | S. stellulata | S. aggregata | MIO | HI | BrT4 | GA15 | M4 | M8 |
|---|---|---|---|---|---|---|---|---|
| Average cell size (μm) (length × width) | 1.9 × 0.8 | 1.8 × 0.7 | 2.1 × 0.7 | 2.4 × 0.8 | 2.0 × 0.8 | 1.6 × 0.8 | 2.4 × 0.8 | |
| Sodium requirement | + | + | + | + | + | + | + | + |
| CO Vmax (nmol [mg protein]−1 h−1) | NT | 270 | 350 | 65 | 310 | 400 | 510 | |
| Enzymatic activities: | ||||||||
| Nitrate respiration | − | −* | + | − | − | − | − | − |
| Denitrification | + | +* | − | + | − | + | + | + |
| Indole production | − | NT | − | − | − | − | − | − |
| Fermentation | − | −* | W | − | − | W | W | W |
| Arginine dihydrolase | − | NT | − | − | − | − | − | − |
| Urease | − | NT | + | + | + | + | + | + |
| Εsculine hydrolysis | − | NT | + | + | + | + | + | + |
| Gelatinase | − | NT | − | + | − | − | − | − |
| β-Galactosidase | − | NT | + | + | + | + | + | + |
| Growth substrates: | ||||||||
| Isophthalate | − | W | − | − | − | − | − | W |
| Terephthalate | − | − | + | + | + | + | I | W |
| 4-Hydroxybenzoate | I | W | − | − | − | − | I | + |
| Phthalate | W | − | W | − | − | + | − | − |
| Methanol | − | I | − | − | − | − | − | − |
| Ethanol | − | I | NT | − | − | NT | − | − |
| Isopropanol | − | I | − | − | − | − | − | − |
| Acetone | W | I | − | − | − | − | − | − |
| Formate | − | − | W | − | − | − | − | − |
| Propionate | + | + | + | + | − | + | + | + |
| Glycolate | W | − | NT | + | + | NT | W+ | W+ |
| Malonate | + | + | W | − | W | + | + | + |
| Tartrate | I | − | W | W | + | − | − | − |
| Lactate | W | + | + | + | + | + | + | W+ |
| Gluconate | − | W | + | + | − | + | + | + |
| Glycerol | NT | + | W | + | − | + | + | + |
| Galactose | W | + | + | + | NT | + | + | + |
| Mannose | W | + | + | + | + | + | + | + |
| Ribose | + | + | W | + | + | + | + | + |
| Lactose | + | W | + | + | W | + | + | W |
| Sucrose | + | + | NT | + | + | NT | + | + |
| Glucuronate | W | + | + | + | + | + | + | + |
| Galacturonate | NT | + | NT | + | NT | NT | + | W |
| Mannitol | − | + | + | + | + | + | + | + |
| Glycine | NT | I | − | − | − | − | − | − |
| Alanine | + | + | + | − | − | + | + | + |
| Valine | + | + | W | + | + | + | + | + |
| Glutamate | + | + | + | − | − | + | + | + |
| Aspartate | + | W | W | + | R | + | W+ | W+ |
| Serine | − | − | NT | − | − | NT | I | + |
| Phenylalanine | + | I | − | − | W | W | − | − |
| Benzoate | I | I | − | − | − | − | NT | NT |
| Methylamine | W | I | − | − | − | − | − | I |
| Dimethylamine | W | I | − | − | − | − | − | − |
| Trimethylamine | W | I | I | − | − | − | − | − |
| Betaine | + | W | W | + | + | + | + | + |
| G+C content (%) | 59* | 59* | 58.3 | 60.9 | 56.8 | 57.9 | 57.4 | 57.8 |
+, growth; −, no growth; W, weak growth; I, substrate was inhibitory; NT, substrate was not tested; *, data obtained from Uchino et al. (40); Vmax, maximum rate of metabolism.
All strains grew on acetate, fumarate, citrate, succinate, pyruvate, β-hydroxybutyrate, malate, glucose, fructose, maltose, and proline. Strain M8 also grew on 4-hydroxybenzoate (4-HBA), which supported weak growth by S. aggregata and inhibited the growth of S. stellulata and strain M4 (Table 1). A purple metabolite accumulated transiently during the incubation of S. aggregata and S. stellulata with 4-HBA, indicating at least partial uncoupling of reactions in 4-HBA transformation (not shown). Terephthalate supported growth by some isolates (e.g., those of strains GA15, HI, and MIO), but inhibited the growth of strain M4. Isophthalate supported only weak or no growth, while phthalate supported growth by only strain GA15. None of the isolates grew on methylamines, methanol, isopropanol, acetone, or glycine, which proved inhibitory for S. aggregata (Table 1).
API test results were identical for strains GA15, M4, and M8 (Table 1). Strain MIO differed from the others in its ability to respire nitrate but not denitrify; strain BrT4 neither respired nitrate nor denitrified; strain HI differed in its ability to hydrolyze gelatin (Table 1). All strains were urease, β-glucosidase, and β-galactosidase positive; all strains were negative for indole production and arginine dihydrolase. Only weak evidence of fermentation was observed for strains GA15, M4, and M8.
All isolate membrane lipids were dominated by the fatty acid 18:1ω7c, followed by 18:0 and 11-methyl,18:1ω7c (Table 2). Similarity indices did not provide definitive matches with isolates in the existing Microbial ID database, but did differentiate strains M4 and M8 from S. aggregata and strains GA15, HI, and MIO (not shown). Individual isolates were differentiated from each other by the presence of 17:1ω7c in strains GA15 and BrT4 only, by the presence of 19:0 in all strains except MIO, BrT4, and M8, by the presence of 20:4ω6,9,12,15c in strain HI only, and by the presence of 14:0 and 18:0iso in strains BrT4 and MIO and by 20:0iso and 20:1ω9c in strain MIO only (Table 2).
TABLE 2.
Fatty acid composition (%) for S. aggregata and strains MIO, HI, BrT4, GA15, M4, and M8
| Fatty acid | S. aggregata | MIO | HI | BrT4 | GA15 | M4 | M8 |
|---|---|---|---|---|---|---|---|
| 17:1ω7c | 0.6 | 0.4 | |||||
| 18:1ω5c | 0.2 | 0.2 | 0.3 | 0.2 | 0.2 | ||
| 18:1ω7c | 71.7 | 69.4 | 72.6 | 80.7 | 71.7 | 83.3 | 86.3 |
| 20:1ω9c | 1.0 | ||||||
| 20:4ω6,9,12,15c | 0.4 | ||||||
| 11-methyl 18:1ω7c | 8.6 | 7.7 | 4.5 | 0.6 | 9.6 | 2.2 | 1.0 |
| 12:0 | 0.2 | ||||||
| 14:0 | 0.20 | 0.5 | |||||
| 16:0 | 1.3 | 3.0 | 0.7 | 2.5 | 0.7 | 0.5 | 1.4 |
| 17:0 | 1.3 | 0.1 | 0.4 | 0.3 | 1.8 | 0.2 | 0.5 |
| 18:0 | 8.7 | 10.3 | 10.5 | 5.5 | 9.2 | 7.9 | 7.8 |
| 19:0 | 1.4 | 0.4 | 1.4 | 0.2 | |||
| 20:0 | 1.6 | 3.4 | 7.1 | 0.3 | 1.6 | 1.4 | 1.7 |
| 18:0-3OH | 0.9 | 0.9 | 0.6 | 1.1 | 0.4 | 0.8 | |
| 18:0iso | 0.9 | 0.7 | |||||
| 20:0iso | 0.4 |
G+C contents ranged from 56.8 to 61.2% (Table 1). G+C contents for E. coli strain DH5α, Burkholderia xenovorans, and Mycobacterium smegmatis were 50.8, 65.7, and 65.2%, respectively. Reported values from genomic sequences for several E. coli species and the latter two isolates were 50 to 51, 62.6, and 67.4%, respectively (www.ncbi.nlm.nih.gov/genomes/lproks.cgi).
Growth response to salinity.
Strains GA15, HI, M4, and MIO grew exponentially when incubated in MPYE with salinities ranging from 5 to 45 ppt (Table 3). Analysis of variance revealed significant differences in growth rate constants as a function of strain and salinity (P < 0.0001). For strains GA15, M4, and MIO, maximum growth rate constants decreased at 45 ppt relative to growth rate constants at optimum salinities of 15, 15, and 25 ppt, respectively (Table 3). Strain HI exhibited the highest optimum salinity, 35 ppt. Growth rate constants were greater for strain M4 than for strains GA15 and MIO at all salinities and greater than the values for strain HI from 5 to 15 ppt. In contrast, values for strain HI were greatest at seawater and hypersaline salinities (35 to 45 ppt). All strains grew slowly (linearly) in a nonmarine, basal salts medium, with lower growth yields than those in marine media (not shown). All strains grew negligibly after inoculation into sodium-free media, even though residual sodium may have been present from the washing step (not shown).
TABLE 3.
Growth rate constants versus salinity
| Salinity (ppt) | Growth rate (h−1) of straina:
|
|||
|---|---|---|---|---|
| HI | MIO | M4 | GA15 | |
| 5 | 0.134 (0.004) | 0.136 (0.011) | 0.273 (0.016) | 0.224 (0.005) |
| 15 | 0.176 (0.023) | 0.178 (0.006) | 0.330 (0.006) | 0.231 (0.001) |
| 25 | 0.221 (0.019) | 0.164 (0.006) | 0.336 (0.006) | 0.218 (0.004) |
| 35 | 0.229 (0.006) | 0.148 (0.006) | 0.265 (0.017) | 0.201 (0.003) |
| 45 | 0.256 (0.009) | 0.097 (0.005) | 0.220 (0.008) | 0.159 (0.006) |
Results are averages of triplicate growth rate constants ± 1 standard error for growth rate constants and parts per thousand for salinity. Strains were grown in MPYE.
Carbon monoxide utilization.
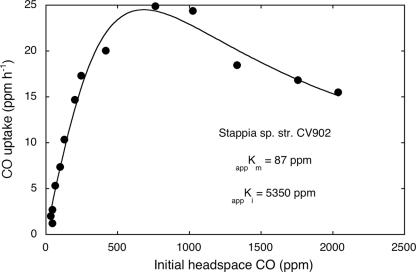
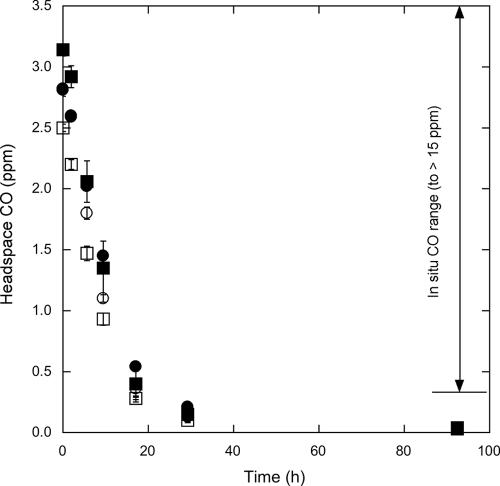
All isolates oxidized carbon monoxide at concentrations of <1,000 ppm (0.1%), but higher concentrations proved inhibitory (Fig. 1). At concentrations of <1,000 ppm, CO uptake was consistent with Michaelis-Menten kinetics (Fig. 1), and apparent maximum uptake rate values ranged from about 270 to 510 nmol CO mg protein−1 h−1 (Table 1). CO was also consumed at ambient concentrations by strains GA15, HI, M4, and MIO (Fig. 2). Uptake rate constants were similar for the four strains, and within 17 h, all were able to reduce CO concentrations to values comparable to those reported for the marine water column. Continued incubation resulted in headspace concentrations of 30 to 40 ppb.
FIG. 1.
CO uptake (ppm h−1) as a function of initial headspace CO concentration for Stappia sp. strain KB902. Data are means of triplicates (±1 standard error).
FIG. 2.
Headspace CO concentrations in cultures of strains GA15 (•), HI (▪), MIO (□), and M4 (○) at near ambient to subambient concentrations versus time (h). Error bars indicate standard errors.
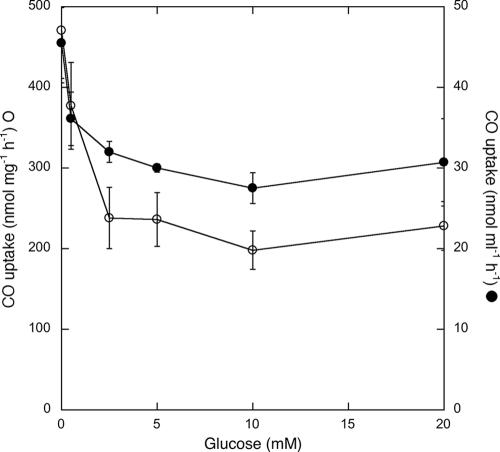
CO consumption by S. aggregata was partially inhibited by incubation with >0.5 mM exogenous glucose (Fig. 3). Consumption rates decreased by up to 40% when expressed per unit volume of culture, even though S. aggregata density increased two- to threefold for glucose concentrations of ≥2 mM. When expressed per unit of cell biomass, increases in cell density were accompanied by decreases in uptake rates of up to 58% (Fig. 3).
FIG. 3.
CO oxidation rates (•, nmol [mgdw biomass]−1 h−1; ○, nmol [ml culture]−1 h−1) for triplicate cultures of Stappia aggregata as a function of the initial glucose concentration of the medium. Error bars indicate standard errors.
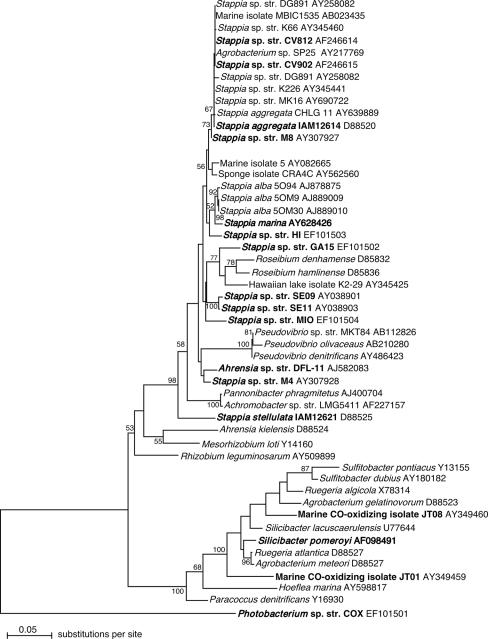
Phylogenetic analyses.
Phylogenetic trees derived from analyses of 16S rRNA gene sequences using maximum likelihood, maximum parsimony, and distance (neighbor-joining) models were topologically similar and characterized by the following consistent patterns (Fig. 4): (i) numerous isolate sequences and clone sequences from uncultured bacteria formed a cluster closely related to S. aggregata; (ii) 16S rRNA gene sequences from S. aggregata were clearly distinct from the sequence for S. stellulata; (iii) 16S rRNA gene sequences from strains GA15, HI, M4, and MIO were distinct from S. aggregata and S. stellulata as well as from S. marina and S. alba and other genera (e.g., Pannonibacter, Pseudovibrio, and Roseibium), which were not distinctly resolved from Stappia phylogenetically (Fig. 4). Other marine bacterial genera that form aggregates in liquid culture, e.g., Ruegeria, also appeared polyphyletic, but distinct from Stappia (Fig. 4). The phylogenetic distinctness of strains GA15, HI, M4, and MIO was consistent with 16S rRNA gene sequence similarities between these taxa and validly described Stappia spp., which ranged from 93.1 to 97.7%.
FIG. 4.
Maximum likelihood analysis of partial 16S rRNA gene sequences (100 bootstrap replicates) implemented with phyml using an HYK correction. The phylogenetic tree was visualized with NJplot. Numbers at nodes indicate bootstrap support; values <70% are not shown. Known CO-oxidizing bacteria are indicated in bold. Photobacterium sp. strain (str.) COX was used as an outgroup.
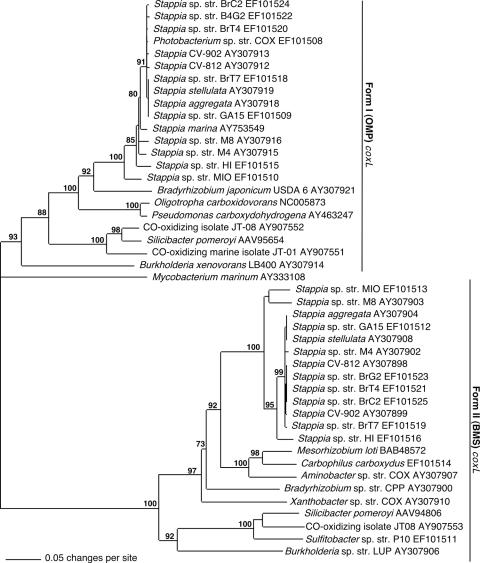
Form I (OMP) coxL sequences were obtained from all isolates examined in this study, while form II (putative, BMS) coxL sequences were obtained from all Stappia isolates but not from a Hawaiian Photobacterium isolate, Mycobacterium marinum, or a Ruegeria isolate (Fig. 5). Phylogenetic analysis revealed distinct OMP and BMS clades, within which sequences from Stappia were differentiated from those of other marine and nonmarine CO oxidizers (Fig. 5). The topology of the form I Stappia coxL cluster was generally similar to that of the form II cluster, with differences primarily in the location of branches for Stappia sp. strains MIO and M8. Within the form I and form II clades, sequences from several Stappia isolates were identical or nearly identical. OMP sequences from S. aggregata, S. marina, and S. stellulata were distinct from sequences for strains HI, MIO, and M4. An OMP coxL sequence from M. marinum, a marine actinobacterium, was distantly related to the proteobacterial sequences. In contrast, an OMP coxL sequence from a Photobacterium (γ-Proteobacteria) was remarkably similar to those of Stappia isolates (Fig. 5).
FIG. 5.
Phylogram from neighbor-joining analysis (1,000 bootstrap replicates) of inferred OMP and BMS putative coxL amino acid sequences implemented with PAUP. Mycobacterium marinum coxL was used as an outgroup. Numbers at nodes indicate bootstrap support; values <70% are not shown.
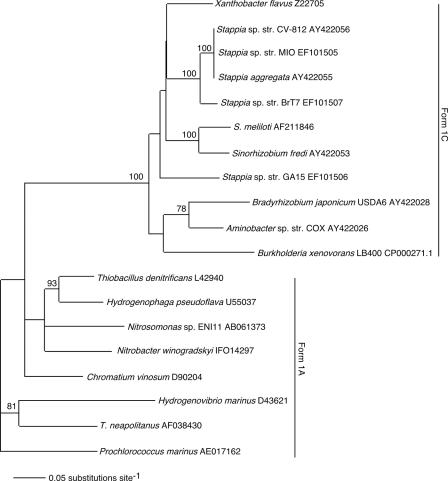
Partial sequences were obtained for the cbbL gene from a subset of Stappia isolates, including S. aggregata and strains BrT7, CV812, GA15, and MIO. PCR products were not obtained from S. stellulata or from strains BrC2, BrG2, BrT4, CV902, HI, M4, and M8. All of the Stappia cbbL sequences clustered with representatives of the form IC RuBisCO clade, based on a phylogenetic analysis (Fig. 6). Sequences from S. aggregata and strains BrT7, CV812, and MIO were nearly identical and clearly distinct phylogenetically from the strain GA15 sequence. Stappia cbbL sequences were not closely related to form IC sequences from other Proteobacteria, including several CO-oxidizing isolates.
FIG. 6.
Phylogram from neighbor-joining analysis (1,000 bootstrap replicates) of aligned nucleotide sequences implemented in PAUP for bacterial form I cbbL with bootstrap support indicated at nodes (values <70% not shown). Prochlorococcus marinus was used as an outgroup to root the tree for visual representation.
BOX-PCR banding patterns were distinctly different for the various Stappia isolates examined and exhibited little similarity (Table 4). Similarity indices (SAB) for pairwise comparisons ranged from 0.0 to 0.4, which indicated substantial differentiation among taxa (Table 3). SAB values derived from comparisons of S. aggregata with all other strains, including S. stellulata, were ≤0.13; values for comparisons of all strains with S. stellulata were ≤0.36. In a number of instances, SAB values were 0 due to the absence of shared bands. The greatest similarities were observed for comparisons of strains HI and M4 (SAB = 0.4) and strains MIO and S. stellulata (SAB = 0.36).
TABLE 4.
SAB derived from BOX genomic DNA fingerprinting analysis for S. aggregata, S. stellulata, and strains M4, M8, CV902, BrT4, GA15, MIO, and HI
| Strain | Index of similarity for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S. aggregata | M4 | M8 | CV902 | BRT4 | S. stellulata | GA15 | MIO | |
| M4 | 0 | |||||||
| M8 | 0 | 0.17 | ||||||
| CV902 | 0.11 | 0.13 | 0.15 | |||||
| BrT4 | 0.13 | 0 | 0 | 0.15 | ||||
| S. stellulata | 0.12 | 0.29 | 0 | 0.13 | 0 | |||
| GA15 | 0.11 | 0 | 0.15 | 0.13 | 0.30 | 0 | ||
| MIO | 0.10 | 0.12 | 0.13 | 0.22 | 0.13 | 0.35 | 0.22 | |
| HI | 0.11 | 0.4 | 0 | 0 | 0 | 0.13 | 0.13 | 0.11 |
DISCUSSION
Our results and those of others indicate that Stappia strains are readily isolated from many marine environments (1, 5, 9, 17, 27, 33). Though Stappia strains have been considered “marine” based on sodium requirements and weak growth in nonmarine media (1, 27, 38), three of four isolates in this study (strains GA15, MIO, and M4) exhibit growth optima in diluted seawater (Table 3). Only strain HI grows optimally in full-strength seawater (35 ppt). While strains GA15, MIO, and M4 occur in coastal environments, isolate growth optima do not reflect in situ salinity regimes since all are derived from systems with relatively constant salinities greater than 30 ppt. Although S. stellulata was originally described as an obligate halophile (1), salinity optima have not been reported for S. aggregata and S. alba. Thus, it is not clear to what extent adaptation to seawater varies within the genus or how Stappia compares in its salinity tolerance with other “marine” genera.
All Stappia isolates examined to date oxidize CO and contain form I coxL (Fig. 5 and Tables 1 and 3) (13). CO uptake capacity has not been reported for S. alba or S. marina, but S. marina possesses a form I CODH gene (9), a strong predictor of its physiological capacity. All Stappia isolates examined to date also contain form II putative coxL (Fig. 5). The form II protein appears to function as a CODH but may have a reduced capacity for CO oxidation and use an alternate substrate preferentially (15). CO utilization by S. aggregata conforms to a simple Michaelis-Menten kinetic model for low to moderate concentrations, but relatively high concentrations result in inhibition (Fig. 1), which may account for the inability of Stappia isolates to grow as typical carboxydotrophs.
In addition to using superambient concentrations (e.g., 1,000 ppm), strains GA15, HI, MIO, and M4 oxidize CO at concentrations significantly lower than those reported for seawater (Fig. 2) (37). Similar results have been obtained for other Stappia isolates (not shown). This suggests that CO may serve as a substrate for some Stappia strains under in situ conditions, perhaps supplementing the uptake of heterotrophic substrates, which typically occurs at only nanomolar to low micromolar concentrations.
In vitro CO consumption by Stappia strains and other CO oxidizers depends on incubation conditions (7, 8, 24, 28, 31). In Pseudomonas carboxydoflava, form I CODH is expressed constitutively and CO uptake occurs during batch heterotrophic growth on pyruvate (8). CODH is also expressed constitutively in Mycobacterium sp. strain JC1, and CO uptake occurs in the presence of several heterotrophic substrates, including glucose, but not pyruvate (28). Variability in expression and uptake also occurs for Pseudomonas thermocarboxydovorans (24), Hydrogenophaga pseudoflava (7), and Oligotropha carboxidovorans. Results presented here (Fig. 3) reveal the partial inhibition of activity by the addition of glucose to Stappia aggregata cells that are actively oxidizing CO. This effect appears due to allosteric regulation by glucose or glucose metabolites, repression of CODH synthesis, or both and is consistent with results from O. carboxidovorans CODH expression studies (31).
Decreased CO oxidation due to the addition of 0.5 mM glucose (Fig. 3), which did not stimulate growth of S. aggregata (not shown), supports a role for allosteric inhibition. The repression of synthesis alone would leave cells with CODH levels (and presumably activities) similar to treatments without glucose. The repression of CODH synthesis by glucose concentrations of >0.5 mM is indicated by the fact that CO uptake per unit volume with 20 mM glucose is similar to that with 0.5 mM glucose, but cell density for the former increased by about 2.6-fold relative to the latter. An increase in cell density but not activity is consistent with maintenance of preexisting CODH without new synthesis. Regardless of the mechanism, inhibition is only partial, which suggests that under carbon-limited conditions in situ, CODH may be expressed and active, while diverse heterotrophic substrates are also used.
The metabolic versatility of Stappia strains is illustrated by growth with a wide range of sugars, organic acids, aromatics, and amino acids, among others, and nitrate respiration or denitrification (Table 1) (1, 9, 14, 27, 29, 38). The potential for aromatic utilization reported here is consistent with prior analyses of Stappia-like isolates by Buchan et al. (3), who documented protocatechuic acid degradation. In addition, some Stappia isolates (those of S. aggregata and strains BrT7, CV812, GA15, MIO, and M4) possess cbbL (Fig. 6) and may be able to fix CO2 via the Calvin cycle, possibly in conjunction with CO oxidation. The extent to which these two processes are coupled under in situ conditions remains to be determined.
Variability in substrate utilization and other phenotypic and biochemical traits (Tables 1 and 2), along with phylogenetic analyses, supports the designation of four new species. 16S rRNA gene analyses demonstrate that strains CV812, CV902, M8, BrC2, BrG2, BrT4, and BrT7 cannot be resolved from S. aggregata (Fig. 4) and contribute to a “species complex” that also includes isolates and uncultured bacterial clone sequences obtained from geographically and ecologically diverse sources. Strains GA15, HI, MIO, and M4 are distinct from this complex. Strain GA15 shares <97% 16S rRNA gene sequence similarity with S. aggregata, S. alba, S. marina, and S. stellulata, supporting its designation as a new species, Stappia meyerae sp. nov. It is united with Stappia through growth substrates, lipid profiles, polar flagellation, and enzymatic activities (Tables 1 and 2). Although its closest phylogenetic relatives include two Roseibium isolates, phylogenetic distance from them, growth substrates, gelatinase activity, and the absence of distinct pink pigmentation and peritricious flagellation differentiate Stappia meyerae sp. nov. from Roseibium (35).
Strain MIO 16S rRNA sequence is 96.2 and 97.6% similar to sequences from its closest phylogenetic neighbors, S. aggregata and S. marina, respectively. Phylogenetic distance (Fig. 4), lipid composition (Table 2), and the ability to denitrify and grow on terephthalate (Table 1) differentiate strain MIO from S. aggregata. Lipid composition and growth with glucose, mannose, mannitol, maltose, gluconate, and citrate differentiate strain MIO from S. marina. In addition, previous analyses involving S. alba (27) and S. marina (9) have shown that even 16S rRNA gene sequence similarities as high as 98.9% are associated with DNA-DNA hybridization values of ≪70% for congeneric Stappia species. Thus, these collective observations support the designation of a new species, Stappia conradae sp. nov.
Strain HI and M4 16S rRNA gene sequences share >97% similarity with the S. aggregata 16S rRNA gene sequence (97.6 and 97.7%, respectively), but both strains are clearly distinct from S. aggregata and all other validly described Stappia based on 16S rRNA and coxL phylogenies (Fig. 4 and 5), lipid profiles (Table 2), substrate utilization, and the presence of cbbL (and presumably the Calvin cycle) in S. aggregata but not strains HI or M4. The latter strains also denitrify, while S. marina does not. In addition, BOX-PCR results show substantial divergence between strains HI and MIO and all other Stappia isolates (Table 4). Collectively, these observations support two new species, Stappia kahanamokuae sp. nov. for strain HI and Stappia carboxidovorans sp. nov. for strain M4.
Growth substrate profiles not only provide valuable taxonomic information, but also offer important ecological insights. For instance, aromatic use, though variable among strains, is consistent with the isolation of Stappia spp. from phaeophyte macroalgae, which produce relatively high concentrations of polyphenols (25, 39). The recent identification of putative dehalogenase genes in the draft genome of S. aggregata (G. M. King et al., unpublished data) suggests a possible use of various alkyl or aryl halides, which are produced commonly by phytoplankton, macroalgae, and marine invertebrates (10, 23, 32, 36), in the growth of these organisms. The variable presence of cbbL genes indicates that some Stappia strains can function mixotrophically, supplementing organic carbon and energy sources with CO2 fixation, presumably driven by CO oxidation. Other strains may simply use CO as a supplemental energy source.
In summary, the results presented here indicate that, in addition to its heterotrophic metabolism, the genus Stappia is characterized by its ability to oxidize CO at subambient to superambient levels, with or without mixotrophic CO2 fixation. The ability of Stappia isolates to respire nitrate or denitrify, use numerous organics, and function in low to moderate salinities facilitates participation in both aerobic and anaerobic processes in carbon and nitrogen cycling in a wide range of marine environments. The results also support the establishment of four new species derived from geographically diverse habitats.
Description of Stappia meyerae sp. nov.
Stappia meyerae (meyerae. L. fem. adj. meyerae of Meyer, honoring fundamental contributions by Ortwin Meyer, University of Bayreuth, to the physiology, biochemistry, and molecular biology of CO-oxidizing microbes).
Cells are aerobic, gram-negative, nonsporing, motile rods with single polar flagellums, 2.0 ± 0.03 μm in length and 0.8 ± 0.03 μm in width. Colonies circular, entire, slightly convex, smooth, cream to tan. Oxidase and catalase positive. Urease, β-galactosidase positive; esculin hydrolysis, gelatinase, indole production from tryptophan and arginine dihydrolase negative. Nitrate reduced to gas. Sodium required; optimum salinity for growth, 5 to 25 ppt. Principle fatty acids are 18:1ω7c and 11-methyl,18:1ω7c; G+C content, 57.8 ± 0.6%. Oxidizes carbon monoxide; contains large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase. Grows with acetate, alanine, aspartate, betaine, citrate, fumarate, fructose, galactose, glucose, gluconate, glucuronate, glutamate, glycerol, β-hydroxybutyrate, lactate, lactose, malate, malonate, maltose, mannitol, mannose, phthalate, propionate, pyruvate, proline, ribose, succinate, terephthalate, and valine. Does not grow with acetone, benzoate, 4-hydroxybenozate, formate, glycine, isophthalate, isopropanol, methanol, tartrate, or mono-, di-, and trimethylamine. Weak growth on phenylalanine.
The type strain GA15 was isolated from Ascophyllum nodosum in the Damariscotta River (Maine).
Description of Stappia conradae sp. nov.
Stappia conradae (conradae. L. fem. adj. conradae of Conrad, honoring important contributions by R. Conrad, Max Planck Institute for Terrestrial Biogeochemistry, Marburg, Germany, to the physiology and ecology of CO-oxidizing microbes in soil and aquatic environments).
Cells are aerobic, gram-negative, non-spore-forming, motile rods with single polar flagellums; 1.8 ± 0.1 μm in length and 0.7 ± 0.1 μm in width. Colonies circular, entire, slightly convex, smooth, cream to tan. Oxidase and catalase positive. Urease, β-galactosidase positive; esculin hydrolysis, gelatinase, indole production from tryptophan and arginine dihydrolase negative. Nitrate respired to nitrite; gas not produced. Sodium required; optimum salinity for growth, 15 to 35 ppt. Principle fatty acids are 18:1ω7c and 11-methyl,18:1ω7c; G+C content 58.2 ± 0.5%. Oxidizes carbon monoxide; contains large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase. Grows with acetate, alanine, citrate, fumarate, fructose, glucose, gluconate, glucuronate, glutamate, β-hydroxybutyrate, lactate, lactose, malate, maltose, mannitol, mannose, propionate, pyruvate, proline, succinate, and terephthalate. Does not grow with acetone, benzoate, 4-hydroxybenozate, glycine, isophthalate, isopropanol, methanol, phenylalanine, or mono- or dimethylamine. Weak growth on aspartate, betaine, formate, galactose, glycerol, malonate, phthalate, ribose, tartrate, and valine. Inhibited by trimethylamine.
The type strain MIO was isolated from a methanotrophic enrichment based on sediment obtained from a 160-m depth in Kagoshima Bay, Japan.
Description of Stappia kahanamokuae sp. nov.
Stappia kahanamokuae (kahanamokuae. L. fem. adj. kahanamokuae of Kahanamokuae, honoring Hawaiian U.S. Olympic gold medalist and pioneering surfer, Duke Pahoe Kahinu Mokoe Hulikohola Kahanamoku).
Cells are aerobic gram-negative, nonsporing, motile rods with single polar flagellums; 2.1 ± 0.1 μm in length and 0.7 ± 0.1 μm in width. Colonies circular, entire, slightly convex, smooth, cream to tan. Oxidase and catalase positive. Urease, β-galactosidase, gelatinase positive; esculin hydrolysis, indole production from tryptophan and arginine dihydrolase negative. Nitrate reduced to gas. Sodium required; optimum salinity for growth, 35 ppt. Principle fatty acids are 18:1ω7c and 11-methyl,18:1ω7c; G+C content, 61.2 ± 0.1%. Oxidizes carbon monoxide; does not contain large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase. Grows with acetate, aspartate, betaine, citrate, fumarate, fructose, galactose, galacturonate, glucose, gluconate, glucuronate, glycerol, glycolate, β-hydroxybutyrate, lactate, lactose, malate, maltose, mannitol, mannose, propionate, pyruvate, proline, ribose, succinate, sucrose, terephthalate and valine. Does not grow with acetone, alanine, benzoate, ethanol, formate, 4-hydroxybenozate, glutamate, glycine, isophthalate, isopropanol, malonate, methanol, phenylalanine, phthalate, serine, or mono-, di-, and trimethylamine. Weak growth on tartrate.
The type strain HI was isolated from a surf water sample at South Point (Ka Lae), Hawaii.
Description of Stappia carboxidovorans sp. nov.
Stappia carboxidovorans (car.box.i.do.vo.rans, L. n. carbo, charcoal, carbon; Gr. adj. oxys, sour, acid; L. v. voro, devour; M. L. part. adj. carboxidovorans, carbon acid devouring).
Cells are aerobic gram-negative, nonsporing, motile rods with single polar flagellums; 1.6 ± 0.1 μm in length and 0.8 ± 0.1 μm in width. Colonies circular, entire, slightly convex, smooth, cream to tan. Oxidase and catalase positive. Urease, β-galactosidase, gelatinase positive; esculin hydrolysis, indole production from tryptophan and arginine dihydrolase negative. Nitrate reduced to gas. Sodium required; optimum salinity for growth, 15 to 25 ppt. Principle fatty acids are 18:1ω7c and 11-methyl,18:1ω7c.; G+C content, 57.4 ± 0.3%. Oxidizes carbon monoxide; does not contain large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase. Grows with acetate, alanine, aspartate, betaine, citrate, fumarate, fructose, galactose, galacturonate, glucose, gluconate, glucuronate, glutamate, glycerol, glycolate, β-hydroxybutyrate, lactate, lactose, malate, malonate, maltose, mannitol, mannose, propionate, pyruvate, proline, ribose, succinate, sucrose, and valine. Does not grow with acetone, ethanol, formate, glycine, isophthalate, isopropanol, methanol, phenylalanine, phthalate, serine, tartrate, or mono-, di-, and trimethylamine. Inhibited by 4-hydroxybenzoate, serine, and terephthalate.
The type strain M4 was isolated from Ascophyllum nodosum from the Damariscotta River, Walpole, ME.
Acknowledgments
We thank K. Boettcher for cultures of S. aggregata and Stappia strains CV-812 and CV-902. We thank M. Takeuchi for the gift of strain MIO. We thank H. Crosby for help with the initial isolation and characterization of strains M4 and M8. We thank K. Johnston and W. Yeung for technical support.
This research was funded by National Science Foundation awards OCE-0425579 and MCB-0348100. C. F. Weber and H. Crosby were partially supported by NSF-REU funds.
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Ahrens, R. 1968. Taxonomische untersuchungen an sternbildenden Agrobacterium-Arten aus der westlichen Ostsee. Kiel. Meeresforsch. 24:147-173. [Google Scholar]
- 2.Boettcher, K. J., B. J. Barber, and J. T. Singer. 2000. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group and colonization of nonaffected animals by Stappia stellulata-like strains. Appl. Environ. Microbiol. 66:3924-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan, A., E. L. Neidel, and M. Moran. 2001. Diversity of the ring-cleaving dioxygenase gene pcaH in a salt marsh bacterial community. Appl. Environ. Microbiol. 67:5801-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Kreig. 1994. Methods for general and molecular bacteriology, p. 791. ASM Press, Washington, DC.
- 5.Groben, R., G. J. Doucette, M. Kopp, M. Kodama, R. Amann, and L. K. Medlin. 2000. 16S rRNA targeted probes for the identification of bacterial strains isolated from cultures of the toxic dinoflagellate Alexandrium tamarense. Microb. Ecol. 39:186-196. [DOI] [PubMed] [Google Scholar]
- 6.Hardy, K., and G. M. King. 2001. Enrichment of a high-affinity CO oxidizer in Maine forest soil. Appl. Environ. Microbiol. 67:3671-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang, B. S., and Y. M. Kim. 1999. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 181:5581-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiessling, M., and O. Meyer. 1982. Profitable oxidation of carbon monoxide and hydrogen during heterotrophic growth of Pseudomonas carboxydoflava. FEMS Microbiol. Lett. 13:333. [Google Scholar]
- 9.Kim, B., J. Park, J. Bae, S. Rhee, K. Kim, J. Oh, and Y. Park. 2006. Stappia marina sp. nov., a marine bacterium isolated from the Yellow Sea. Int. J. Syst. Evol. Microbiol. 56:75-79. [DOI] [PubMed] [Google Scholar]
- 10.King, G. M. 1986. Inhibition of microbial activity in marine sediments by a bromophenol from a hemichordate. Nature (London) 323:257-259. [Google Scholar]
- 11.King, G. M. 1999. Attributes of atmospheric carbon monoxide oxidation by Maine forest soils. Appl. Environ. Microbiol. 65:5257-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, G. M. 2000. Impacts of land use on atmospheric carbon monoxide consumption by soils. Global Biogeochem. Cycles 14:1161-1172. [Google Scholar]
- 13.King, G. M. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, G. M. Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 15.King, G. M., and C. F. Weber. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol., in press. [DOI] [PubMed]
- 16.Koch, A. L. 1994. Growth measurements, p. 248-276. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Kreig (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, DC.
- 17.Kurahashi, M., and A. Yokota. 2002. A preliminary report of phylogenetic diversity of bacterial strains isolated from marine creatures. J. Gen. Appl. Microbiol. 48:251-259. [DOI] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 19.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. de Bruijn. 1994. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez, J. S., J. N. Carter-Franklin, E. L. Mann, J. D. Martin, M. G. Haygood, and A. Butler. 2003. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc. Natl. Acad. Sci. USA 100:3754-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, O., and H. G. Schlegel. 1978. Reisolation of the carbon monoxide utilizing hydrogen bacterium Pseudomonas carboxydovorans (Kistner) comb. nov. Arch. Microbiol. 118:35-43. [DOI] [PubMed] [Google Scholar]
- 22.Nanba, K., G. M. King, and K. Dunfield. 2004. Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microbiol. 70:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neidleman, S. L., and J. Geigert. 1986. Biohalogenation: principles, basic roles and applications, p. 203. Ellis Horwood Ltd., Chichester, United Kingdom.
- 24.Pearson, D. M., C. O'Reilly, J. Colby, and G. W. Black. 1994. DNA sequence of the cut A, B, and C genes, encoding the molybdenum containing hydroxylase carbon monoxide dehydrogenase, from Pseudomonas thermocarboxydovorans strain C2. Biochim. Biophys. Acta 1188:432-438. [DOI] [PubMed] [Google Scholar]
- 25.Pereira, R. C., and Y. Yoneshigue-Valentin. 1999. The role of polyphenols from the tropical brown alga Sargassum furcatum on the feeding by amphipod herbivores. Bot. Mar. 42:441-448. [Google Scholar]
- 26.Pichon, D., V. Gaia, M. D. Norman, and R. Boucher Rodoni. 2005. Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (Cephalopoda: Lolignidae and Idiosepiidae). Mar. Biol. 147:1323-1332. [Google Scholar]
- 27.Pujalte, M. J., M. C. Macian, D. R. Arahal, and E. Garay. 2005. Stappia alba sp. nov., isolated from Mediterranean oysters. Syst. Appl. Microbiol. 28:672-678. [DOI] [PubMed] [Google Scholar]
- 28.Ro, Y. T., and Y. M. Kim. 1993. Constitutive expression of carbon monoxide dehydrogenase in Acinetobacter sp. strain JC1 DSM 3803. Korean J. Microbiol. 31:214-217. [Google Scholar]
- 29.Rueger, H.-J., and M. G. Hoefle. 1992. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov., nom. rev.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int. J. Syst. Bacteriol. 42:133-143. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Santiago, B., U. Schübel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 32.Scarratt, M. G., and R. M. Moore. 1996. Production of methyl chloride and methyl bromide in laboratory cultures of marine phytoplankton. Mar. Chem. 54:263-272. [Google Scholar]
- 33.Sfanos, K., D. Harmody, P. Dang, A. Ledger, S. Pomponi, P. McCarthy, and J. Lopez. 2005. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst. Appl. Microbiol. 28:242-264. [DOI] [PubMed] [Google Scholar]
- 34.Smith, E. A., F. H. Mackintosh, F. Grant, and S. Gallacher. 2002. Sodium channel blocking (SCB) activity and transformation of paralytic shellfish toxins (PST) by dinoflagellate-associated bacteria. Aquat. Microb. Ecol. 29:1-9. [Google Scholar]
- 35.Suzuki, T., Y. Muroga, M. Takahama, and Y. Nishimura. 2000. Roseibium denhamense gen. nov., sp. nov. and Roseibium hamlinense sp. nov., aerobic bacteriochlorophyll-containing bacteria isolated from the east and west coasts of Australia. Int. J. Syst. Evol. Microbiol. 50:2151-2156. [DOI] [PubMed] [Google Scholar]
- 36.Tait, V. K., and R. M. Moore. 1995. Methyl chloride (CH3Cl) production in phytoplankton cultures. Limnol. Oceanogr. 40:189-195. [Google Scholar]
- 37.Tolli, J. D., and C. D. Taylor. 2005. Biological CO oxidation in the Sargasso Sea and Vineyard Sound, Massachusetts. Limnol. Oceanogr. 50:1205-1212. [Google Scholar]
- 38.Uchino, Y., A. Hirata, A. Yokota, and J. Sugiyama. 1998. Reclassification of marine Agrobacterium species: proposals of Stappia stellulata gen. nov., comb. nov., Stappia aggregata sp. nov., nom. rev., Ruegeria atlantica comb. nov., and Ahrensia kieliense gen. nov., sp. nov., nom. rev. J. Gen. Appl. Microbiol. 44:201-210. [DOI] [PubMed] [Google Scholar]
- 39.Van Alstyne, K. L., and V. J. Paul. 1990. The biogeography of polyphenolic compounds in marine macroalgae: temperate brown algal defenses deter feeding by tropical herbivorous fishes. Oecologia 84:158-163. [DOI] [PubMed] [Google Scholar]
- 40.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. Van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]