Abstract
Certain Campylobacter strains harbor a transcribed intervening sequence (IVS) in their 23S rRNA genes. Following transcription, the IVS is excised, leading to fragmentation of the 23S rRNA. The origin and possible functions of the IVS are unknown. Furthermore, the distribution of IVS-harboring strains within Campylobacter populations is poorly understood. In this study, 104 strains of Campylobacter coli from turkeys, representing 27 different multilocus sequence typing-based sequence types (STs), were characterized in terms of IVS content and erythromycin susceptibility. Sixty-nine strains harbored IVSs in all three 23S rRNA genes, whereas the other 35 strains lacked IVSs from at least one of the genes. The STs of the latter strains belonged to an unusual cluster of C. coli STs (cluster II), earlier found primarily in turkey strains and characterized by the presence of the C. jejuni aspA103 allele. The majority (66/69) of strains harboring IVSs in all three 23S rRNA genes were resistant to erythromycin, whereas none of the 35 strains with at least one IVS-free 23S rRNA gene were resistant. Cluster II strains could be transformed to erythromycin resistance with genomic DNA from C. coli that harbored IVS and the A2075G transition in the 23S rRNA gene, associated with resistance to erythromycin in Campylobacter. Erythromycin-resistant transformants harbored both the A2075 transition and IVS. The findings suggest that the absence of IVS in C. coli from turkeys is characteristic of a unique clonal group of erythromycin-susceptible strains and that IVS can be acquired by these strains via natural transformation to erythromycin resistance.
The thermophilic Campylobacter species Campylobacter jejuni and Campylobacter coli are major bacterial food-borne pathogens. In the United States alone there were 12.9 reported cases of Campylobacter infections per 100,000 persons in 2004 (6). Even though most infections involve C. jejuni, C. coli is also associated with significant disease burden. For instance, recent reports indicated that C. coli accounted for 7% and 18.6% of confirmed human campylobacteriosis cases in the United Kingdom and Germany, respectively (15, 16).
Significant attention has been directed to the emergence of antibiotic resistance in Campylobacter. Fluoroquinolones are no longer recommended for the treatment of Campylobacter infection due to the increasing prevalence of fluoroquinolone resistance among human clinical strains (11, 28). Erythromycin has been shown to be effective in treating patients with Campylobacter enteritis (30) and is the current drug of choice when antibiotic treatment is indicated. However, recent studies indicate that the prevalence of erythromycin resistance (Emr) in C. coli isolated from food animals is significantly higher than in C. jejuni (1, 3, 8, 13, 23, 24). For example, Luangtongkum et al. reported that for samples from conventional poultry farms, 89% of C. coli and 19% of C. jejuni isolates were resistant to erythromycin (23). For Campylobacter, erythromycin resistance has been found to be due to a base substitution in the 23S rRNA gene (A2075G, based on the sequence of the 23S rRNA gene of C. jejuni NCTC 11168) that alters the ribosome target site (14, 20); erythromycin resistance can be disseminated by natural transformation among strains from turkeys and swine (21). Thus, erythromycin-resistant C. coli strains of animal origin may present treatment challenges, should such strains become implicated in human illness. Even though most Campylobacter infections are self-limiting, antibiotic treatment may be indicated for severe cases or for special categories of patients.
Contamination of poultry with Campylobacter has been identified as an important risk factor in human infections obtained through consumption of raw/undercooked poultry or through cross-contamination (19). The human epidemiology of campylobacteriosis, however, remains poorly understood and is complicated especially by the existence of multiple possible hosts for zoonotic transmission, the largely sporadic nature of human illness, and the diversity of strain subtypes colonizing animals and potentially involved in human infection.
Understanding of the population structure of Campylobacter from humans, poultry, and other sources has been compromised by the genotypic diversity of the organisms and the limitations of subtyping schemes, such as pulsed-field gel electrophoresis, that are based on the generation of banding patterns (7). A special challenge has been the difficulty in comparing image-based results among different laboratories. Partly to address this challenge, multilocus sequence typing (MLST) schemes that generate DNA sequence-based (and hence totally portable) data that can be used to subtype strains from diverse sources have been developed (9, 10, 26). MLST appears to be especially promising in the case of C. coli isolated from animal sources. A recent survey of C. coli from turkeys, broilers, cattle, and swine identified several alleles and sequence types (STs) that appeared to be host specific (25).
An intriguing feature of Campylobacter spp. (primarily C. jejuni, C. coli, C. upsaliensis, C. fetus, and C. sputorum) is the presence, in certain strains, of an intervening sequence (IVS) in at least one copy of the 23S rRNA gene (41). Following transcription, the IVS is excised, leading to fragmentation of the large-subunit (23S) rRNA. The presence of IVSs resulting in such 23S rRNA fragmentation was first described for Salmonella enterica serovars Typhimurium and Arizonae (5), for which it was shown that the IVS was excised posttranscriptionally by endoribonuclease RNase III without religation (5). IVSs have been identified in and characterized for other bacteria, including Yersinia enterocolitica (37), Coxiella burnetii (2), Helicobacter (18), and Haemophilus (39), Brucella (4), Leptospira (35), Proteus, and Providencia (27) spp. Horizontal gene transfer has been postulated to underlie the dissemination of IVSs (34, 36). However, the role or effect of IVSs on the physiology of host cells remains unknown.
C. jejuni and C. coli harbor three rRNA operons on their chromosomes. IVSs are absent from all three 23S rRNA gene copies of C. jejuni NCTC 11168 (32) but were detected in other strains of C. jejuni and C. coli (29, 33, 41) and were also identified in all three copies of the 23S rRNA gene in the recently sequenced genome of C. coli RM2228 (12). When present, the IVS is located only 891 nucleotides (nt) upstream from the site in the 23S rRNA gene where a point mutation (A2075G) has been associated with high-level resistance to erythromycin in Campylobacter (12, 14, 20). Surveys of C. jejuni from broilers showed that only a minority of strains (approximately 10%) contained IVSs (29, 33). The presence of IVSs may be associated with a randomly amplified polymorphic DNA PCR subtype in broilers (33). However, no further data on the prevalence of IVSs in C. jejuni from commercial poultry (broilers and turkeys) are available, and no data have been described for IVSs in C. coli that colonize poultry. In recent studies, we have found that turkeys in eastern North Carolina were frequently colonized by C. coli (22, 38), and a subset of such turkey-derived C. coli was characterized by MLST (25).
In this study, we investigated possible correlations between the MLST-based sequence types, the presence of IVSs, and the erythromycin susceptibility (Ems) of C. coli derived from turkeys. The objectives were to determine whether the presence or absence of IVS was specific to certain MLST-based subtypes and whether the IVS was associated with erythromycin resistance in C. coli from turkeys.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 104 turkey-derived C. coli strains used in this study were part of the Campylobacter culture collection in our laboratory. They had been isolated from cecal or fecal samples from turkeys on various turkey farms in North Carolina during 2001 to 2004 by use of Campylobacter blood-free selective agar with CCDA selective supplement (Oxoid, Basingstoke, Hampshire, England) as described previously (38). Eighty-three of these strains were included in a recent MLST study (25). C. coli strains were grown routinely on Mueller-Hinton agar (MHA; Mueller-Hinton broth [MHB] with 1.2% agar) (Becton Dickinson, Sparks, MD) at 42°C for 40 h microaerobically.
Antibiotic susceptibility and erythromycin MIC determinations.
Antibiotic susceptibility profiles were determined as described previously (22, 38). Strains that grew on media amended with 8 μg/ml of erythromycin were classified as resistant. Resistance to nalidixic acid was determined following growth on media amended with 20 μg/ml of nalidixic acid. C. jejuni ATCC 33560 was used as a negative control for all antibiotic susceptibility determinations, as described previously (22). Erythromycin MIC determinations were done in a range of concentrations (8 μg/ml to 256 μg/ml), as described previously (21).
DNA extractions, IVS PCR, and DNA sequencing.
Genomic DNA was extracted from C. coli by use of a DNeasy kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's protocol. Primers 1162 (5′-TGC GCG GAA AAT ATA ACG GGG CTA-3′) and CG1425 (5′-CTC ACA TTA ATT ATC GCT ACT CAT-3′) were used to amplify a segment of the 23S rRNA gene that has been reported to contain the IVS (41). PCR was done using Taq DNA polymerase (Takara). PCR amplification conditions were as follows: 30 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min. The resulting amplicons were detected by gel electrophoresis on a 1.5% agarose gel buffered with 1× Tris-borate-EDTA. A single amplicon of approximately 160 bp indicated that all three 23S rRNA genes were IVS free, whereas production of a single amplicon of approximately 310 bp indicated that the IVS (approximately 147 bp) was present in all three genes.
An internal fragment (1,475 bp) of the 23S rRNA gene was amplified using primers 23SemF (5′-ACC AGG AGG TTG GCT TAG AA-3′) and 23SemR (5′-TCG TCT CTG CTT GAC TTG TAT G-3′). PCR conditions were the same as those described above. PCR amplicons were purified by use of a QIAquick gel extraction kit (QIAGEN Inc., Valencia, CA) and sequenced (Davis Sequencing, Davis, CA). DNA sequences were analyzed by BioEdit (17).
MLST.
The alleles and procedures described before for MLST-based subtyping of 83 of the strains (25) were employed for MLST of 21 additional strains, using total genomic DNA extracted as described above. ST identification was done using the C. jejuni/C. coli MLST database (http://pubmlst.org/campylobacter/); new sequences were entered into the database and were assigned new allele numbers as described previously (25).
Natural transformation.
Broth transformations were done according to the method described by Kim et al. (21) with minor modifications. Genomic DNA from C. coli strain 6818 (ST-1101), an erythromycin-resistant, nalidixic acid-resistant, turkey-derived strain from our laboratory's C. coli collection, was used as the donor DNA. This strain harbored IVSs in all three of its 23S rRNA genes. Seven nalidixic acid-susceptible C. coli strains (6017, 6667, 6979, 7080, 7474, 7755, and 8266) were recipients in natural transformation to nalidixic acid resistance. C. coli 6979 (ST-1150) was the recipient in natural transformation to erythromycin resistance. A colony of the recipient strain was transferred to 5 ml MHB and incubated microaerobically at 42°C for 24 h. Then, 50 μl of this culture was inoculated into 50 ml MHB preconditioned at 42°C and incubated microaerobically at 42°C for 7 h. Donor DNA (3 μg) was mixed with 1 ml of an exponential-phase culture of the recipient strain and incubated microaerobically at 42°C for 5 h. Finally, the DNA-culture mixture was serially diluted and plated on MHA plates and on MHA amended with either nalidixic acid (20 μg/ml) or erythromycin (10 μg/ml) to determine the frequency of transformation to nalidixic acid resistance or erythromycin resistance, respectively. The transformation frequency was defined as the ratio of the CFU/ml on MHA plates to the CFU/ml on MHA amended with the respective antibiotic. Negative controls consisted of the bacterial cultures without donor DNA plated on MHA amended with nalidixic acid (20 μg/ml) or erythromycin (10 μg/ml).
Statistical analysis.
The FREQ procedure of the SAS software (SAS Institute, Inc., Cary, NC) was used to tabulate the data. The uncertainty coefficient (U) was employed to estimate the association between nominal variables of this study (erythromycin susceptibility versus ST, or erythromycin susceptibility versus IVS content) (40). U is one of the indices of proportional reduction in error that are applicable to nominal variables (31). By definition, it is the percent reduction in uncertainty in predicting a variable based on the knowledge of another variable, and its value can range from 0 (no reduction in uncertainty) to 1 (100 percent reduction in uncertainty). In other words, two variables have no relationship when U equals 0, and they have perfect relationship when U equals 1. Since U is an asymmetric statistic, a calculation was done to determine the percent reduction in uncertainty in predicting erythromycin susceptibility based on the knowledge of ST or on the presence/absence of IVSs. Calculation of U was done by the FREQ procedure of the SAS software (SAS Institute, Inc.).
RESULTS
The 104 turkey-derived C. coli strains used in this study were categorized into 27 different STs. The prevalences of the different STs varied noticeably within the strain set. For example, ST-1101, ST-1126, ST-1150, and ST-1161 were detected in 23, 15, 14, and 8 strains, respectively, of the 104 totally characterized. Seventeen STs were unique, i.e., found only once in this strain set.
The majority of C. coli strains from turkeys harbored IVSs in all three 23S rRNA genes.
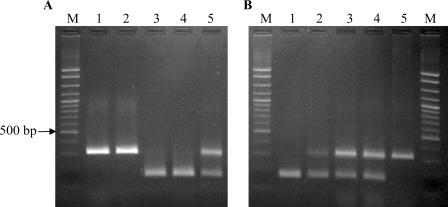
PCR using primers 1162 and CG1425 indicated that the majority (69/104 [66%]) of the strains harbored the IVS in all three 23S rRNA genes, whereas 29 strains (28%) were completely IVS free (Table 1). In the case of six strains (6%), both amplicons were detected, even when the strains were purified from single colonies repeatedly and tested multiple times. Detection of both amplicons in these six strains suggested that the IVS was present in only one or two copies of the 23S rRNA gene and that it was absent from at least one copy (Fig. 1A).
TABLE 1.
Erythromycin susceptibility, IVS content, and STs of C. coli strains from turkeys
| Erythromycin susceptibility | STs with indicated IVS contenta
|
||
|---|---|---|---|
| IVS present (n = 69) | IVS absent (n = 29) | IVS hybrid (n = 6) | |
| Susceptible | ST-1175 (1) | ST-1150† (11) | ST-1192† (3) |
| ST-1017 (2) | ST-1155† (1) | ST-1150† (3) | |
| ST-1161† (8) | |||
| ST-1188† (2) | |||
| ST-1192† (1) | |||
| ST-1194† (2) | |||
| ST-1199† (1) | |||
| ST-1487† (1) | |||
| ST-1835† (1) | |||
| ST-1242† (1) | |||
| Resistant | ST-889 (8) | ||
| ST-1092 (1) | |||
| ST-1101 (23) | |||
| ST-1110 (1) | |||
| ST-1119 (1) | |||
| ST-1149 (7) | |||
| ST-1126 (15) | |||
| ST-1154 (3) | |||
| ST-1160 (1) | |||
| ST-1163 (1) | |||
| ST-1170 (1) | |||
| ST-1171 (1) | |||
| ST-1184 (1) | |||
| ST-1193 (1) | |||
| ST-1198 (1) | |||
Numbers inside parentheses indicate the number of strains in each category. IVS present, IVS present in all three 23S rRNA genes; IVS absent, IVS absent from all three 23S rRNA genes; IVS hybrid, IVS present in at least one, but not all three, 23S rRNA genes. †, MLST cluster II isolate with the C. jejuni aspA103 allele.
FIG. 1.
Detection of IVS in C. coli strains from turkeys. Primers 1162 and CG1425 were used, as described in Materials and Methods. (A) Agarose gel showing sizes of PCR products from genomic DNA of C. coli strains 6818 (lane 1; ST-1101), 7080 (lane 2; ST-1175), 6667 (lane 3; ST-1161), 7755 (lane 4; ST-1150), and 6017 (lane 5; ST-1150). Lane M is a 100-bp ladder molecular weight marker (exACTGene cloning DNA ladder; Fisher); the location of the 500-bp marker band is indicated by the arrow. Lane 5 shows an example of a strain yielding both 310-bp and 167-bp amplicons. (B) Agarose gel showing PCR products from genomic DNA of C. coli strain 6979 (lane 1; ST-1150) and from genomic DNA of individual colonies of erythromycin-resistant strain 6979 transformants (lanes 2 to 5). Lane M is a 100-bp ladder molecular weight marker, as described above.
Strains lacking IVSs in at least one 23S rRNA gene copy were susceptible to erythromycin.
IVS content varied markedly among strains of different STs. In the case of certain predominant STs, specifically, ST-1101 (n = 23), ST-1126 (n = 15), and ST-889 (n = 8), and for the less common STs ST-1149 (n = 7) and ST-1154 (n = 3), all strains appeared to contain IVS in all three 23S rRNA gene copies, yielding only the 310-bp amplicon with the IVS primers. In contrast, strains with certain other predominant STs, specifically, ST-1150 (n = 14) and ST-1161 (n = 8), were either completely free of IVSs, yielding only the 167-bp amplicon (19 strains), or had one or two IVS-free 23S rRNA genes (three strains) and thus produced both amplicons (167 bp and 310 bp) with the IVS primers (Table 1).
Antibiotic susceptibility determinations showed that the majority (66/69) of strains that harbored IVSs in all three 23S rRNA genes were resistant to erythromycin. In contrast, none of the 35 strains that had at least one IVS-free 23S rRNA gene were resistant to erythromycin (Table 1).
In order to estimate the relationship of erythromycin susceptibility to ST and that of erythromycin susceptibility to IVS content, uncertainty coefficients were calculated. IVS content (presence/absence) was a very good predictor of erythromycin resistance/susceptibility (U = 0.82, or 82% reduction in uncertainty). Uncertainty coefficients also indicated that ST was a strong predictor of resistance/susceptibility to erythromycin (U = 1.00, or 100% reduction in uncertainty).
Transformation frequency is variable among IVS-free, erythromycin-susceptible strains.
To determine whether the absence of IVSs and erythromycin susceptibility reflects a reduced ability of the bacteria to acquire exogenous DNA (including the IVS) through natural transformation, transformation efficiency to nalidixic acid resistance was determined for a panel of seven C. coli strains. These include three with ST-1150 and two with ST-1161 (Table 2). Certain strains of ST-1150 and ST-1161 (e.g., 7755 and 6667) indeed were among the lowest in transformation frequency (Table 2). On the other hand, C. coli 6017, 6979, and 7474 (also of ST-1150 or ST-1161) had relatively high transformation frequencies, which were similar to those obtained with the positive control, C. coli 8266. C. coli 7080, which had ST-1175 and was susceptible to erythromycin, had relatively low transformation frequency (Table 2). These findings suggested that transformation frequency was a strain-specific attribute, variable among different strains of cluster II STs.
TABLE 2.
Frequency of transformation of C. coli strains to nalidixic acid resistance
| Recipient straina | STb | IVSc | Susceptibility to erythromycin (10 μg/ml) | Mean transformation frequencyd |
|---|---|---|---|---|
| 8266 | 1101 | Present | Resistant | 1.82 × 10−4 |
| 6017 | 1150† | Hybrid | Susceptible | 2.30 × 10−5 |
| 6979 | 1150† | Absent | Susceptible | 4.88 × 10−4 |
| 7755 | 1150† | Absent | Susceptible | 3.02 × 10−6 |
| 6667 | 1161† | Absent | Susceptible | 8.73 × 10−6 |
| 7474 | 1161† | Absent | Susceptible | 1.98 × 10−4 |
| 7080 | 1175 | Present | Susceptible | 7.19 × 10−6 |
Recipient strains were all susceptible to nalidixic acid. The donor strain was 6818 (ST-1101; IVS present, resistant to both erythromycin and nalidixic acid).
†, MLST cluster II isolate with the C. jejuni aspA103 allele.
Present, IVS present in all three 23S rRNA genes; absent, IVS absent from all three 23S rRNA genes; hybrid, IVS present in at least one, but not all three, 23S rRNA genes.
Mean transformation frequencies were calculated from values derived from at least two independent experiments.
IVS and erythromycin resistance determinant can be acquired simultaneously by natural transformation.
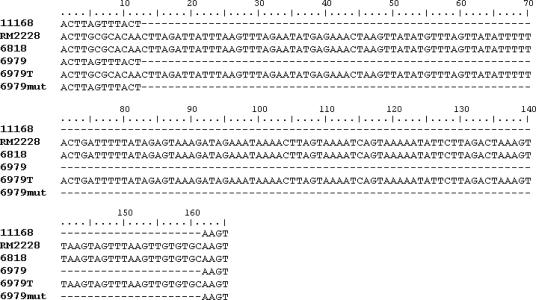
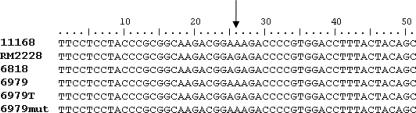
Emr transformants were examined in terms of their simultaneous acquisition of IVS. The mean frequency of the transformation of C. coli 6979, which was Ems and IVS free (IVS−), to erythromycin resistance was 6.84 × 10−6. The erythromycin MIC of the transformants was >256 μg/ml, similar to that of the donor C. coli 6818. PCR with IVS primers, using total genomic DNA extracted from Emr transformants as a template, indicated the presence of IVS in at least one of the 23S rRNA genes of these Emr transformants. Of four randomly chosen Emr transformants, three appeared to harbor both the IVS-free and IVS-plus 23S rRNA genes, and one appeared to harbor IVS in all three copies of the 23S rRNA gene (Fig. 1B). DNA sequencing revealed that IVSs in C. coli 6818 and the Emr transformant of C. coli 6979 were identical (Fig. 2). In contrast, a spontaneous Emr mutant of C. coli 6979, obtained from one of the negative control plates in the transformation experiments, lacked the IVS (Fig. 2). The expected A2075G base substitution responsible for erythromycin resistance was found in DNA sequences of the 23S rRNA gene of C. coli 6818 as well as the Emr transformant of C. coli 6979 but not in the sequence of C. coli 6979 or of the spontaneous Emr mutant of this strain (Fig. 3). The A2075G transition was also identified in the 23S rRNA gene of the recently sequenced C. coli RM2228 (Fig. 3), in agreement with the erythromycin resistance exhibited by this strain. Interestingly, C. coli RM2228 also harbored IVSs identical in sequence and location within the 23S rRNA gene to the IVSs of C. coli 6818 and the Emr transformant of C. coli 6979 (Fig. 2).
FIG. 2.
23S rRNA gene sequences in the IVS region. The locations and sequences of IVS are shown for C. jejuni NCTC 11168 (11168), C. coli RM2228 (RM2228), C. coli 6818 (6818), C. coli 6979 (6979), C. coli strain 6979 Emr transformant (6979T), and C. coli strain 6979 Emr spontaneous mutant (6979mut). Coordinates correspond to nt 1180 to nt 1195 of the 23S rRNA gene of C. jejuni NCTC 11168.
FIG. 3.
Detection of the A2075G transition in the 23S rRNA gene of Campylobacter. Sequences are from C. jejuni NCTC 11168 (11168), C. coli RM2228 (RM2228), C. coli 6818 (6818), C. coli 6979 (6979), C. coli strain 6979 Emr transformant (6979T), and C. coli strain 6979 Emr spontaneous mutant (6979mut). The A→G transition is indicated by the arrow.
DISCUSSION
In this study, we have shown that the majority of the investigated C. coli strains from turkeys harbored IVSs. Of the 104 strains, 6 harbored IVSs in one or two (but not all three) copies of the 23S rRNA gene. Heterogeneity of 23S rRNA genes is not uncommon and has been shown previously for C. coli (20). It would be interesting to investigate whether homogenization of rrn operons would maintain the IVS or eliminate it from the genome.
The absence of IVSs was detected only in strains of certain STs previously designated as cluster II and found primarily for C. coli from turkeys (25). It is noteworthy that each of the two major cluster II IVS-free STs found in this study (ST-1150 and ST-1161) contained alleles that were mostly turkey/poultry associated (25), suggesting that C. coli strains with these STs were adapted to these hosts. The signature of cluster II is the presence of the aspA103 allele, which is closely related to a C. jejuni aspA allele. Miller et al. suggested that the aspA103 allele was acquired by C. coli from C. jejuni (25). In the region of the country from which the strains originated, turkeys can be colonized frequently with C. coli (22, 38). However, C. coli and C. jejuni can both be present in certain flocks, and both species have been isolated from gastrointestinal tract samples from the same bird (42). Thus, turkeys may represent an avian host that lends itself to the production of C. coli-C. jejuni interspecies hybrids. In contrast, thermophilic campylobacters from swine from the same region have been almost exclusively C. coli strains, and cluster II strains have not been detected among any of these C. coli strains from swine (25).
All strains that harbored at least one IVS-free 23S rRNA gene were susceptible to erythromycin. Conversely, among the 38 strains that were susceptible to erythromycin, all but 3 (a single strain with ST-1175 and 2 strains with ST-1017) belonged to cluster II STs. Moreover, cluster II STs were not found for any of the erythromycin-resistant strains. IVS content and ST were shown to be a good predictor of erythromycin susceptibility of C. coli isolated from turkeys. In other words, if the MLST-based ST or IVS content of a C. coli turkey strain is known, we can predict with confidence whether that particular C. coli strain is susceptible to erythromycin. For instance, cluster II STs could be predicted to lack IVSs (in at least one copy of the 23S rRNA gene) and to be susceptible to erythromycin.
An intriguing finding was that none of the cluster II strains were erythromycin resistant, in contrast to strains of other STs, the majority of which were resistant to this antibiotic. As mentioned above, the MLST data suggested that cluster II C. coli strains acquired DNA in the aspA locus from C. jejuni. It is possible that additional genes in the vicinity of aspA, or in other genomic regions, are of C. jejuni origin and that some of these genes may be related to the extensively documented scarcity of erythromycin resistance in C. jejuni from meat animals (1, 3, 8, 13). C. jejuni can be transformed to erythromycin resistance in the laboratory, using as donor DNA the 23S rRNA fragment from a resistant C. coli strain that harbors the A2075G erythromycin resistance determinant (14). However, it is possible that other C. jejuni genetic determinants negatively impact the competitive fitness of these erythromycin-resistant organisms in natural environments or in the animal host. One may speculate that such putative determinants, if acquired by selected C. coli strains, would also result in a reduced prevalence of erythromycin resistance among these organisms, as observed for cluster II strains. Further characterization of the genomic content of cluster II strains will be necessary to determine the genomic contribution of C. jejuni in these strains.
Considering the proximity of the IVS insertion site (position 1184) and the A:G transition (position 2075), one may argue that an IVS− Ems C. coli strain would be readily transformed by donor DNA from an IVS+ Emr C. coli strain to being both IVS+ and Emr if that particular C. coli recipient were naturally competent. Our results showed that IVS− Ems C. coli strains could be indeed transformed to erythromycin resistance and that those transformants also acquired at least one copy of the IVS. Thus, if the majority of Emr strains in a population harbor IVSs, dissemination of erythromycin resistance by transformation is also likely to lead to dissemination of IVSs. Cluster II strains that lack IVS and are susceptible to erythromycin may represent a special subset of C. coli strains that have remained free of the A2075G mutation and the IVSs.
Although the observed limited transformation potential of certain cluster II strains may contribute to their IVS-free, erythromycin-susceptible status, other cluster II strains appeared to have normal competence for transformation, suggesting that reduced competence is not a universal factor limiting the acquisition of IVS and erythromycin resistance by these strains. Instead, the limiting factor may be that the transformants do not compete well with their IVS-free (and erythromycin-sensitive) cluster II counterparts. Although erythromycin-resistant, IVS-harboring transformants of cluster II strains grew normally in the laboratory, it is possible that under other conditions the acquisition of these determinants may negatively impact fitness, possibly in association with C. jejuni components in the genome, as discussed above.
In conclusion, our data have shown that, among C. coli strains from turkeys, only certain strains are free of the IVS in their 23S rRNA genes. These strains constitute a cluster of MLST-based strain types (cluster II STs) characterized by the presence of the C. jejuni aspA103 allele and exhibit susceptibility to erythromycin. Further studies are needed to genetically characterize these C. coli strains and to elucidate the possible impact of IVSs in their antibiotic susceptibility profiles and other adaptations.
Acknowledgments
We appreciate the encouragement and support of all members of our laboratory, especially Robin Siletzky, Carol D'lima, and Sandra Wright for their assistance in the MLST characterization of the strains used in this study.
The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2003-0299.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Aarestrup, F. M., E. M. Nielsen, M. Madsen, and J. Engberg. 1997. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, pigs, cattle, and broilers in Denmark. Antimicrob. Agents Chemother. 41:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afseth, G., Y. Y. Mo, and L. P. Mallavia. 1995. Characterization of the 23S and 5S rRNA genes of Coxiella burnetii and identification of an intervening sequence within the 23S rRNA gene. J. Bacteriol. 177:2946-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., K. N. Kaya, D. D. Hancock, D. R. Call, Y. H. Park, and T. E. Besser. 2005. Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl. Environ. Microbiol. 71:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker, B. J. 2000. Characterization of the three ribosomal RNA operons rrnA, rrnB, and rrnC, from Brucella melitensis. Gene 255:117-126. [DOI] [PubMed] [Google Scholar]
- 5.Burgin, A. B., K. Parodos, D. J. Lane, and N. R. Pace. 1990. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell 60:405-414. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2005. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 sites, United States, 2004. Morb. Mortal. Wkly. Rep. 54:352-356. [PubMed] [Google Scholar]
- 7.de Boer, P., B. Duim, A. Rigter, J. van Der Plas, W. F. Jacobs-Reitsma, and J. A. Wagenaar. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmonts, M. H., F. Dufour-Gesbert, L. Avrain, and I. Kempf. 2004. Antimicrobial resistance in Campylobacter strains isolated from French broilers before and after antimicrobial growth promoter bans. J. Antimicrob. Chemother. 54:1025-1030. [DOI] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, B., D. G. White, P. F. McDermott, W. Girard, S. Zhao, S. Hubert, and J. Meng. 2003. Antimicrobial-resistant Campylobacter species from retail raw meats. Appl. Environ. Microbiol. 69:3005-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibreel, A., V. N. Kos, M. Keelan, C. A. Trieber, S. Levesque, S. Michaud, and D. E. Taylor. 2005. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob. Agents Chemother. 49:2753-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurtler, M., T. Alter, S. Kasimir, and K. Fehlhaber. 2005. The importance of Campylobacter coli in human campylobacteriosis: prevalence and genetic characterization. Epidemiol. Infect. 133:1081-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Hurtado, A., J. P. Clewley, D. Linton, R. J. Owen, and J. Stanley. 1997. Sequence similarities between large subunit ribosomal RNA gene intervening sequences from different Helicobacter species. Gene 194:69-75. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs-Reitsma, W. 2000. Campylobacter in the food supply. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 20.Jensen, L. B., and F. M. Aarestrup. 2001. Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. S., D. K. Carver, and S. Kathariou. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, B. C., N. Reimers, H. J. Barnes, C. D'Lima, D. Carver, and S. Kathariou. 2005. Strain persistence and fluctuation of multiple-antibiotic resistant Campylobacter coli colonizing turkeys over successive production cycles. Foodborne Pathog. Dis. 2:103-110. [DOI] [PubMed] [Google Scholar]
- 23.Luangtongkum, T., T. Y. Morishita, A. J. Ison, S. Huang, P. F. McDermott, and Q. Zhang. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luber, P., J. Wagner, H. Hahn, and E. Bartelt. 2003. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001-2002 from poultry and humans in Berlin, Germany. Antimicrob. Agents Chemother. 47:3825-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, W. L., K. Pabbaraju, and K. E. Sanderson. 2000. Fragmentation of 23S rRNA in strains of Proteus and Providencia results from intervening sequences in the rrn (rRNA) genes. J. Bacteriol. 182:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nachamkin, I., H. Ung, and M. Li. 2002. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982-2001. Emerg. Infect. Dis. 8:1501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesbit, E. G., P. Gibbs, D. W. Dreesen, and M. D. Lee. 2001. Epidemiologic features of Campylobacter jejuni isolated from poultry broiler houses and surrounding environments as determined by use of molecular strain typing. Am. J. Vet. Res. 62:190-194. [DOI] [PubMed] [Google Scholar]
- 30.Nolan, C. M., K. E. Johnson, M. B. Coyle, and K. Faler. 1983. Campylobacter jejuni enteritis: efficacy of antimicrobial and antimotility drugs. Am. J. Gastroenterol. 78:621-626. [PubMed] [Google Scholar]
- 31.Olszak, M., and G. Ritschard. 1995. The behaviour of nominal and ordinal partial association measures. Statistician 44:195-212. [Google Scholar]
- 32.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 33.Payne, R. E., M. D. Lee, D. W. Dreesen, and H. M. Barnhart. 1999. Molecular epidemiology of Campylobacter jejuni in broiler flocks using randomly amplified polymorphic DNA-PCR and 23S rRNA-PCR and role of litter in its transmission. Appl. Environ. Microbiol. 65:260-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pronk, L. M., and K. E. Sanderson. 2001. Intervening sequences in rrl genes and fragmentation of 23S rRNA in genera of the family Enterobacteriaceae. J. Bacteriol. 183:5782-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ralph, D., and M. McClelland. 1993. Intervening sequence with conserved open reading frame in eubacterial 23S rRNA genes. Proc. Natl. Acad. Sci. USA 90:6864-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralph, D., and M. McClelland. 1994. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J. Bacteriol. 176:5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skurnik, M., and P. Toivanen. 1991. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol. Microbiol. 5:585-593. [DOI] [PubMed] [Google Scholar]
- 38.Smith, K., N. Reimers, H. J. Barnes, B. C. Lee, R. Siletzky, and S. Kathariou. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J. Food Prot. 67:1463-1468. [DOI] [PubMed] [Google Scholar]
- 39.Song, X. M., A. Forsgren, and H. Janson. 1999. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene 230:287-293. [DOI] [PubMed] [Google Scholar]
- 40.Theil, H. 1970. On the estimation of relationships involving qualitative variables. Am. J. Sociol. 76:103-154. [Google Scholar]
- 41.Trust, T. J., S. M. Logan, C. E. Gustafson, P. J. Romaniuk, N. W. Kim, V. L. Chan, M. A. Ragan, P. Guerry, and R. R. Gutell. 1994. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J. Bacteriol. 176:4597-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesley, I. V., W. T. Muraoka, D. W. Trampel, and H. S. Hurd. 2005. Effect of preslaughter events on prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys. Appl. Environ. Microbiol. 71:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]