Abstract
Very little is known regarding the ecology of Nitrosospira sp. strain AF-like bacteria, a unique group of ammonia oxidizers within the Betaproteobacteria. We studied the response of Nitrosospira sp. strain AF-like ammonia oxidizers to changing environmental conditions by applying molecular methods and physiological measurements to Californian grassland soil manipulated in the laboratory. This soil is naturally high in Nitrosospira sp. strain AF-like bacteria relative to the much-better-studied Nitrosospira multiformis-like ammonia-oxidizing bacteria. Increases in temperature, soil moisture, and fertilizer interacted to reduce the relative abundance of Nitrosospira sp. strain AF-like bacteria, although they remained numerically dominant. The overall abundance of ammonia-oxidizing bacteria increased with increasing soil moisture and decreased with increasing temperature. Potential nitrification activity was altered by interactions among temperature, soil moisture, and fertilizer, with activity tending to be higher when soil moisture and temperature were increased. The increase in potential nitrification activity with increased temperature was surprising, given that the overall abundance of ammonia-oxidizing bacteria decreased significantly under these conditions. This observation suggests that (i) Nitrosospira sp. strain AF-like bacteria may respond to increased temperature with an increase in activity, despite a decrease in abundance, or (ii) that potential nitrification activity in these soils may be due to organisms other than bacteria (e.g., archaeal ammonia oxidizers), at least under conditions of increased temperature.
Ammonia oxidizers mediate the transformation of ammonia into nitrite, the first step in the process of nitrification. Nitrification is a “gatekeeper” process in many ecosystems, regulating soil inorganic nitrogen concentrations and linking the decomposition of nitrogenous organic matter to N loss. Studying the ecology of ammonia oxidizers in natural environments is challenging. One approach has been to study the response of pure cultures to various environmental factors in controlled experiments and predict their function and distribution under natural conditions. However, most ammonia oxidizers cannot be cultured. Those that can be cultured are very slow growing, do not always remain active after cultivation, and in cases of successful growth, the yield is often low. This approach has resulted in only a few studies regarding the physiology of pure cultures of ammonia oxidizers (2, 7, 16, 28; reviewed by Koops and Pommerening-Röser [17]) and a relatively poor understanding of their ecology.
Molecular techniques have been used to increase our understanding of the ecology of ammonia oxidizers, especially their distribution in response to environmental conditions. Both ribosomal and “functional” genes (i.e., genes that code for proteins involved directly in ammonia oxidation) have been used as molecular markers for studying the ammonia oxidizers. The gene coding for the α subunit of ammonia monooxygenase (amoA) has been used most frequently as a functional gene marker for studying the phylogeny and distribution of betaproteobacterial ammonia oxidizers (33; reviewed by Avrahami and Conrad [3]). Studies using these two markers have revealed shifts in the community composition of betaproteobacterial ammonia oxidizers in response to changes in the environment, including changes in ammonium concentrations (reviewed by Kowalchuk and Stephen [18]), pH (reviewed by Boer and Kowalchuck [10]), temperature (reviewed by Avrahami and Conrad [3]), and soil moisture (11). However, the interactions among these environmental factors are not well understood.
A number of phylogenetic groups of betaproteobacterial ammonia oxidizers have been defined based on 16S rRNA phylogeny (18, 23). These groups can also be identified based on AmoA phylogeny (4, 6), which is generally concordant with that of 16S rRNA (1, 23). Ecological studies have been performed on members of only a few of these groups. The least understood of these groups is the group “AmoA cluster 10.” This group has a representative pure culture: Nitrosospira sp. strain AF, isolated from red sandy soil (pH 4.0) sampled in Kasama, Zambia (32). Nitrosospira sp. strain AF is distinct from the other studied species (i.e., strains 40KI, L115, and B6) in its extremely slow growth (a generation time of 138 h compared to those of three other strains ranging from 50 to 63 h [16]) and tolerance of low pH (as low as pH 5 [16]). In addition, Nitrosospira sp. strain AF showed a higher temperature optimum (31 to 33°C) than three Nitrosospira strains that had been isolated from Nordic soils (26 to 29°C). Environmental sequences closely related to Nitrosospira sp. strain AF have so far been detected only in soil from Tennessee (9, 30) and in Californian grassland soil (13). However, most of the past studies of betaproteobacrial ammonia oxidizers in nature used PCR primers (24) subsequently shown to be unable to detect amoA sequences related to Nitrosospira sp. strains 24C and AF if mixed with other sequences (6). Those studies that did detect cluster 10 sequences either used a degenerated forward primer modified by Stephen et al. (30) from the amoA-1F primer of Rotthauwe et al. (24) or used seminested-PCR amplification with A189 and amoA-2R primers (12, 13, 24). The failure to detect this cluster in more soils may be due to primer biases (3).
We studied the effect of changes in temperature, soil moisture, and fertilizer concentration on potential nitrification activity and the abundance and community structure of betaproteobacterial ammonia oxidizers by applying molecular methods and physiological measurements to Californian grassland soil manipulated in the laboratory. The high relative abundance of cluster 10 ammonia oxidizers in this soil provided a rare opportunity to study the ecology of this group. Our observations suggest that the response of cluster 10 ammonia-oxidizing bacteria to environmental change is different from that of the much-better-studied Nitrosospira multiformis-like ammonia-oxidizing bacteria.
MATERIALS AND METHODS
Soil sampling and characterization.
The project was initiated with a soil sample taken from the upper 10 cm of a grassland located in the Jasper Ridge Biological Preserve (JRBP). The JRBP is located in the eastern foothills of the Santa Cruz Mountains in northern California. The grassland was dominated by annual grasses (Avena barbata and Bromus hordeaceus) and forbs (Geranium dissectum and Erodium botrys), growing on sandstone-derived soil. The sample was taken adjacent to the Jasper Ridge Global Change Experiment (13, 27) from a location unaffected by the experimental treatments. The main characteristics of JR soil are summarized in Table 1. The soil sample was sieved to an aggregate size of <2 mm and stored at 4°C for 4 to 6 weeks before the laboratory experiment was established. Soil ammonium and nitrate concentrations were determined by extraction with 1 M KCl and analysis using an Alpkem analyzer. The pH was determined after suspension of the soil both in 0.01 M CaCl2 and in water. Soil maximal water-holding capacity (WHC) was determined by standard protocols (25).
TABLE 1.
Main characteristics of grassland soil from Jasper Ridge field
| Parameter | Value |
|---|---|
| Ammonium concn [μg NH4+ (gdw soil)−1] | 13.1 ± 1.2 |
| Nitrate concn [μg NO3− (gdw soil)−1] | 5.7 ± 0.0 |
| pH with water/CaCl2 | 6.0/5.1 |
| 100%WHC [g water · (100 gdw soil)−1] | 25.2 |
| Soil temperature (at 2 cm below surface) | |
| At time of sampling | 27°C |
| 2003-2004 range | 8-34°C |
| 2003-2004 avg | 19°C |
| Parent material | Sandstone |
| Carbon, nitrogen (%) | 1.197 ± 0.298 C, 0.106 ± 0.034 N |
| Location | Santa Cruz Mountains |
| Sampling date | September 2003 |
| References | 13, 27 |
Experimental setup.
The laboratory experiment was set up in a full factorial design with two temperature levels, three soil moisture levels, and three fertilizer concentrations and thus included 18 different treatment combinations. Approximately 100 g of dry soil was placed in 500-ml jars and wetted to 30, 45, or 60% of maximum WHC. The samples were amended with three different concentrations of a commercial fertilizer (Nutralene; Nu-Gro Technologies, Inc., Ontario, Canada) consisting of methylene urea. The fertilizer contains slow-release granules, thus allowing for the gradual release of nitrogen. Low, moderate, and high fertilizer concentrations were designated LF, MF, and HF treatments, respectively (0.05, 0.1, and 0.3% [wt/wt] of fertilizer, respectively). The samples were incubated at two temperatures, 20 and 30°C, which represented typical winter/spring and summer/fall temperatures at Jasper Ridge. These temperatures have been shown to influence the community structure of betaproteobacterial ammonia oxidizers (4, 6). Two replicates of each treatment combination were established. The jars were opened for aeration every 3 days for 10 min to maintain aerobic conditions. The jars were weighed before incubation and every 3 days during incubation. To compensate for evaporation, lost water was added occasionally by spraying. The soils were mixed with a sterile spatula to keep the soil moisture homogeneous. Samples for estimation of ammonium and nitrate concentrations were taken after 1, 3, 5, and 7 weeks of incubation.
Potential nitrification activity.
Potential nitrification activity was measured after 7 weeks of incubation (26, 31; modified by Avrahami et al. [6]). Briefly, sterile Erlenmeyer flasks containing 45 ml phosphate buffer (1 mM; pH 7.4), 0.04 ml (NH4)2SO4 (0.25 M), and 5 g of soil were incubated at 25°C on a shaker for 46 h. Samples were taken at five time points, centrifuged for 5 min, and filtered through Acrodisc-CR 25-mm syringe filters (0.2 μm; Life Sciences). Samples were stored at −20°C until analysis of nitrite and nitrate in a DIONEX ion chromatograph system (Sunnyvale, CA) equipped with an AS11-HC analytical column. The sum of nitrite and nitrate concentrations increased linearly with time. Rates of potential nitrification activity were determined from the slope of a linear regression of nitrite plus nitrate concentration on time.
DNA extraction.
Samples for community analysis were taken at the beginning of the experiment and after 7 weeks of incubation. DNA was extracted from 0.5-g soil samples using the Fast DNA spin kit for soil (BIO 101, Carlsbad, CA) and the FastPrep instrument (twice for 30 s each at speed of 5.5) in accordance with the manufacturer's instructions. DNA was further purified using the Wizard DNA cleanup kit (Promega, Madison, WI).
PCR amplification of amoA.
The forward primer used for PCR amplification both for terminal restriction fragment length polymorphism (T-RFLP) and denaturing gradient gel electrophoresis (DGGE) analyses was amoA-1F*, a degenerated primer modified by Stephen et al. (30) from the amoA-1F primer of Rotthauwe et al. (24). This primer was 5′ end labeled with 5-carboxyfluorescein for T-RFLP analysis, while for DGGE analysis, a GC clamp (21) was added to the 5′ end of the same primer. The degenerated amoA-2R primer of Rotthauwe et al. (24) was used for T-RFLP fingerprinting analysis, while for DGGE, the reverse primer amoA-2R-GG (22) was used. In a previous study, this primer (termed amoAR1) showed the best results for soil samples (6). Amplification was performed by using 1 μM of each primer (QIAGEN, Valencia, CA), 0.48 μg/μl bovine serum albumin (Rosch, Penzberg, Germany), 0.625 units (for T-RFLP) or 1.25 units (for DGGE) AmpliTaq DNA polymerase (AmpliTaq, Applied Biosystems, Foster City, CA), and 12.5 μl (for T-RFLP) or 25 μl (for DGGE) of MasterAmp 2× PCR premix F containing 100 mM Tris-HCl (pH 8.3), 100 mM KCl, 7 mM MgCl2, 400 μM of each deoxynucleoside triphosphate, and PCR enhancer betaine (Epicentre Technologies, Madison, WI). DNA and water (Sigma-Aldrich, Deisenhofen, Germany) were added to a final volume of 25 (for T-RFLP) or 50 μl (for DGGE). All PCRs were performed in triplicate both for T-RFLP and for DGGE. Amplifications were always started by placing PCR tubes into a preheated (94°C) DNA Engine thermal cycler (MJ Research, San Francisco, CA). The thermal profile used for amplification included 5 min at 94°C, followed by 35 cycles of 45 s at 94°C, 60 s at 55°C, and 1 min at 72°C, and 10 min at 72°C for the last cycle.
DGGE fingerprinting analysis.
DGGE was performed as described previously (5) (modified from the method of Muyzer et al. [21]) with slight modifications. We used a DGGE system manufactured by C.B.S. Scientific (Solana Beach, CA). Gels were stained with SYBR green I (Invitrogen-Molecular Probes, Carlsbad, CA) and scanned with a UV scanner and a Kodak camera.
T-RFLP fingerprinting analysis.
T-RFLP fingerprinting analysis was conducted as described previously (13, 15) with some modification. Three PCRs from each sample were pooled and purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA). Purified PCR products (approximately 100 ng) were digested separately with 5 U of the restriction endonuclease Bsh12362I (MBI Fermentas, Hanover, MD), which has the recognition site GCGC. The digestions were carried out in a total volume of 10 μl for 4 h at 37°C, according to the instructions of the manufacturer. Enzyme inactivation was performed by incubation at 65°C for 20 min. The subsequent T-RFLP fingerprinting analysis was performed at the Genomics Technology Support Facility of Michigan State University (East Lansing, MI; http://genomics.msu.edu/), as described previously (14). The relative abundances of individual T-RFs in a given amoA PCR product were calculated based on the peak height of the individual T-RFs in relation to the total peak height of all T-RFs detected in the respective T-RFLP community fingerprint pattern. The peak heights were automatically quantified by the GeneScan software. To verify the assignments of T-RFs to our detected amoA gene types, we also tested individual clones by T-RFLP fingerprinting analysis.
We chose restriction endonuclease Bsh12362I for our T-RFLP analysis based on the analysis of three clone libraries from our soil. One library consisted of 77 clones (accession numbers AY369266 to AY369342) derived from an April 2000 soil sample (described by Horz et al. [13]), one library of 69 clones (accession numbers DQ393198 to DQ393266) derived from an April 2003 soil sample (S. Avrahami and B. Bohannan, unpublished data), and one library of 48 clones (accession numbers DQ396811 to DQ396858) derived from soil samples incubated for 16 to 20 weeks under laboratory conditions (Avrahami and Bohannan, unpublished). All three libraries contained clones from identical phylogenetic groups, although in different relative abundances. The phylogenetic groups which could be recognized via T-RFLP were (i) AmoA cluster 10 (phylogenetic tree in references 4 and 6), with Nitrosospira sp. strain AF as a representative pure culture (indicated by four T-RFs: 60, 133, 329, and 341 bp); (ii) AmoA cluster 3a (denoted previously as clade JR1 by Horz et al. [13]), with Nitrosospira multiformis as a representative pure culture (indicated by one T-RF of 434 bp); and (iii) AmoA cluster 9 (phylogenetic tree in references 4 and 6), with Nitrosospira sp. strain Nsp65 as a representative in pure culture (indicated by T-RFs 427 and 428 bp in length).
Real-time PCR amplification.
Real-time PCR amplification was modified from the method of Horz et al. (13). In contrast to Horz et al. (13), we used the same primer system (amoA-1F* and amoA-2R) for both rounds of PCR amplification. The first round of PCR amplification was performed as described above using 0.3 μl of DNA per 25-μl reaction mixture and the following thermal cycling protocol: 94°C for 5 min; 12 cycles of 94°C for 45s, 55°C for 60s, and 72°C for 180 s; and 72°C for 10 min for the last cycle. Each sample was amplified in triplicate; the resulting samples later were pooled for the second round of PCR amplification. The second round was performed in triplicate under the same conditions for 35 cycles. Each reaction contained 10 μl DyNAmo Hot Start SYBR green quantitative PCR (qPCR) kit (MJ Research, San Francisco, CA), 1 μM of each primer, and 0.3-μl aliquots of the first round in a final volume of 20 μl. The reaction mixtures were placed in hard-shell, thin-wall 96-well microplates (MJ Research, San Francisco, CA) using Ultra Clear caps (MJ Research, San Francisco, CA) and placed in the DNA Engine Opticon 2 system (MJ Research, San Francisco, CA). The primer annealing temperature was determined experimentally using the temperature gradient feature of the DNA Engine Opticon 2 system. In order to avoid any possible primer dimer interference, the fluorescence reading during each cycle was adjusted to 86°C, a temperature above the melting point of the primer dimers. In addition, to confirm nonspecific amplification, we performed melting curve analyses after each PCR amplification, ensuring the detection of a clear single peak at 90°C corresponding to the 491-bp amoA fragment, together with verification of the size of this fragment by agarose gel electrophoresis.
In order to estimate possible PCR interference and/or inhibition from soil substances, we amplified a plasmid with an insert from Nitrosospira multiformis (accession number AY177933) using the same amoA primer system described above. The plasmid was purified using a QIAprep spin miniprep kit (QIAGEN, Valencia, CA) by following the manufacturer's instructions. The dilution of the purified plasmid DNA template was mixed with 0.3 μl of DNA from the experimental sample and subjected to the real-time PCR protocol described above. The quantification analysis was performed as described previously (13).
Statistical analysis.
All statistical analyses were performed with SAS version 9.1; analysis of variance (ANOVA) was performed using both the GLM and MIXED procedures in SAS (SAS Institute, Inc., Cary, NC). Means were estimated as least square means, and the degrees of freedom were estimated using the Satterthwaite approximation. The data were transformed to achieve normality. For each data set, the transformation that resulted in the best fit of the data set to a normal distribution was chosen. For most data sets, a log transformation was best, with the exception of the nitrate concentration data (square root transformation), the relative abundance of cluster 10 data (arcsine followed by power of 2 transformation), the relative abundance of cluster 3a data (arcsine followed by power of −9.9 transformation), and the relative abundance of cluster 9 (arcsine followed by an inverse transformation).
RESULTS
Community structure.
The community structure of betaproteobacterial ammonia oxidizers after 7 weeks of incubation was analyzed by both DGGE and T-RFLP analyses of amoA gene fragments. DGGE analysis showed no differences in community structure among the treatments (data not shown). However, differences were evident using T-RFLP analysis; this is consistent with published reports suggesting that T-RFLP is more sensitive than DGGE (20). The numerically dominant population of betaproteobacterial ammonia oxidizers before incubation was AmoA cluster 10 (99.4 ± 0.6% of total abundance). After incubation, this cluster was still dominant (70.6 to 100% of total abundance) in all samples.
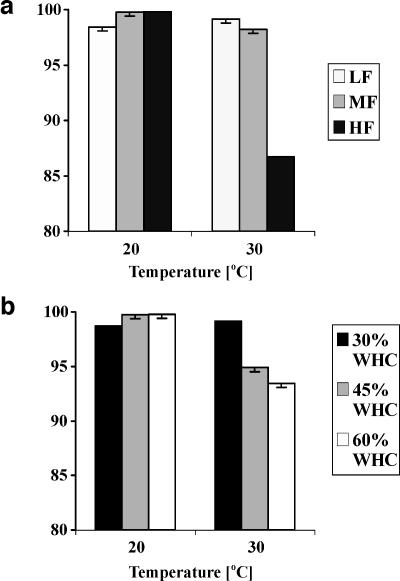
The relative abundance of AmoA cluster 10 was significantly altered by both the temperature and fertilizer treatments (Table 2). There were also significant interactions between these two factors and between temperature and soil moisture (Table 2), such that the effects of either fertilizer or soil moisture were observed at only 30°C (Fig. 1a and b, respectively). At an elevated temperature, either increased soil moisture or increased fertilizer resulted in a decreased relative abundance of cluster 10. When the data from both the 20°C and the 30°C treatments were combined, the relative abundance of cluster 10 did not correlate with any other set of data from the experiment. However, when data from the 30°C treatment were analyzed separately, ammonium concentration was the strongest correlate of the relative abundance of cluster 10 (r = −0.67; P = 0.0023; n = 18), presumably due to the interaction between the fertilizer and temperature treatments. Cluster 3a exhibited the opposite response to temperature, fertilizer, and soil moisture. It increased in its relative abundance at a combination of 30°C and the HF treatment and of 30°C and high soil moisture. Fertilizer treatment was strongly correlated to ammonium concentration (r = 0.92; P < 0.0001). Therefore, the ammonium concentrations were assumed to be due to the fertilizer treatment we imposed. Regression values indicated that ammonium concentration could explain 21% of the variation in the relative abundance of cluster 3a and 58% of the variation when only data from samples incubated at 30°C were analyzed (P = 0.0026 and P = 0.0001, respectively). Cluster 9 also tended to increase in its relative abundance in response to interactions between temperature, soil moisture, and fertilizer (F4,18 = 3.86; P = 0.0195). However, this group could not be detected in the majority of the samples and its highest relative abundance was only 5.6%.
TABLE 2.
ANOVA analysis for the relative abundance of AmoA cluster 10 dataa
| Effect | Num DF | Den DF | F value | Pb |
|---|---|---|---|---|
| T | 1 | 18 | 14.95 | 0.0011 |
| SM | 2 | 18 | 0.73 | 0.4941 |
| F | 2 | 18 | 4.07 | 0.0348 |
| T × SM | 2 | 18 | 5.44 | 0.0142 |
| T × F | 2 | 18 | 10.23 | 0.0011 |
| SM × F | 4 | 18 | 0.52 | 0.7213 |
| T × SM × F | 4 | 18 | 0.99 | 0.4404 |
Num DF, numerator degrees of freedom; Den DF, denominator degrees of freedom; T, temperature; SM, soil moisture; F, fertilizer treatment.
Significant effects (P value < 0.05) are in bold.
FIG. 1.
Effect of (a) temperature and fertilizer concentration and (b) temperature and soil moisture on the relative abundance of AmoA cluster 10. The mean relative abundance of cluster 10 is depicted for all samples, grouped by (a) temperature and soil fertilizer treatments (including all moisture treatments; n = 36) and by (b) temperature and soil moisture treatments (including all fertilizer treatments; n = 36). (a) LF, MF, and HF treatments represent fertilizer concentrations of 0.05, 0.1, and 0.3%, respectively. (b) Soil moisture levels are 30, 45, and 60% of maximum water-holding capacity. Error bars are 95% confidence limits.
Total abundance.
The total abundance of betaproteobacterial ammonia oxidizers was estimated by amplification of the amoA gene using real-time PCR (i.e., qPCR). The abundance of each sample was normalized to a reference sample analyzed in all runs to reduce run-to-run variation, and the ratio of each sample to the reference sample is presented here.
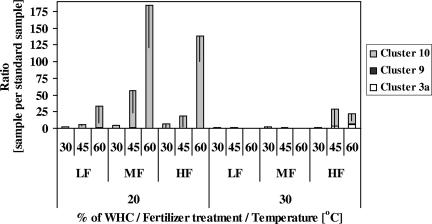
Incubation increased abundance substantially. The value before incubation was on average only 8.3% (±2.1%) of the value of the sample with the lowest abundance after incubation. Both temperature and soil moisture had significant effects on total abundance (Table 3; Fig. 2). Decreasing temperature as well as increasing soil moisture resulted in an increase in abundance (mean ratio values of 128 and 11 at 20 and 30°C, respectively, and of 10, 47, and 109 for 30, 45, and 60% of WHC, respectively). The highest abundances were found in samples incubated at 20°C, 60% of WHC, and with either MF or HF treatment (Fig. 2).
TABLE 3.
ANOVA analysis for total abundance of amoA gene dataa
| Effect | Num DF | Den DF | F value | Pb |
|---|---|---|---|---|
| T | 1 | 18 | 18.29 | 0.0005 |
| SM | 2 | 18 | 5.73 | 0.0118 |
| F | 2 | 18 | 1.08 | 0.3597 |
| T × SM | 2 | 18 | 1.90 | 0.1786 |
| T × F | 2 | 18 | 2.30 | 0.1287 |
| SM × F | 4 | 18 | 1.49 | 0.2475 |
| T × SM × F | 4 | 18 | 0.41 | 0.7981 |
Num DF, numerator degrees of freedom; Den DF, denominator degrees of freedom; T, temperature; SM, soil moisture; F, fertilizer treatment.
Significant effects (P value < 0.05) are in bold.
FIG. 2.
Effect of temperature, soil moisture, and fertilizer concentration on total abundance of betaproteobacterial ammonia oxidizers. The total abundance was normalized to a standard sample (see the text). The stacked bars represent the contribution of each phylogenetic group to the total abundance. Percent WHC represents the percent of the soil's maximum water-holding capacity. LF, MF, and HF treatments represent fertilizer concentrations of 0.05, 0.1, and 0.3%, respectively. Error bars are 95% confidence limits of the total abundance.
In order to estimate the total abundance of cluster 10, we multiplied the total abundance of the amoA gene (estimated by qPCR) by the proportion of AmoA cluster 10 (estimated via T-RFLP); we refer to this product as the “scaled abundance of cluster 10.” The scaled abundance of cluster 10 was significantly reduced by increased temperature and increased by increased soil moisture (mean ratio values of 7.6 and 1.5 at 20 and 30°C, respectively, and of 0.8, 2.9, and 10.2, for 30, 45, and 60% of WHC, respectively) but not by the fertilizer treatments (Table 4).
TABLE 4.
ANOVA analysis for “scaled abundance” of AmoA cluster 10 dataa
| Effect | Num DF | Den DF | F value | Pb |
|---|---|---|---|---|
| T | 1 | 18 | 12.22 | 0.0026 |
| SM | 2 | 18 | 6.85 | 0.0061 |
| F | 2 | 18 | 3.36 | 0.0577 |
| T × SM | 2 | 18 | 3.33 | 0.0590 |
| T × F | 2 | 18 | 3.43 | 0.0547 |
| SM × F | 4 | 18 | 1.21 | 0.3422 |
| T × SM × F | 4 | 18 | 0.68 | 0.6163 |
Scaled abundance of cluster 10 is a multiplication of qPCR data in cluster 10 T-RFLP data, which represents the total abundance of cluster 10. Num DF, numerator degrees of freedom; Den DF, denominator degrees of freedom; T, temperature; SM, soil moisture; F, fertilizer treatment.
Significant effects (P value < 0.05) are in bold.
Potential nitrification activity.
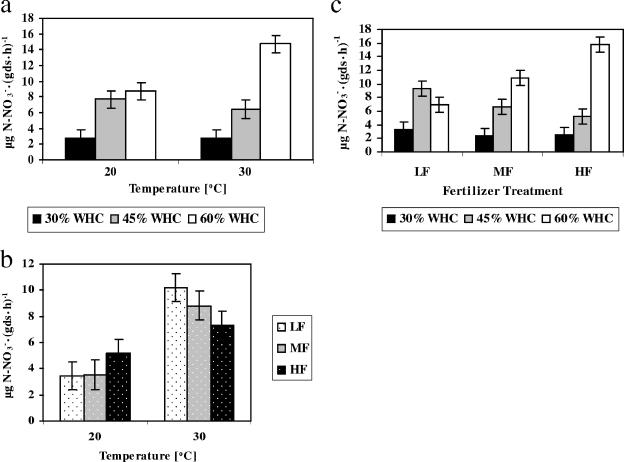
Potential nitrification activity was significantly increased by temperature and soil moisture (Table 5). There were also significant interactions between these two factors and between each of these factors and the fertilizer treatment (Table 5; Fig. 3a, b, and c). Temperature and soil moisture interacted such that the effect of temperature was observed only at 60% WHC (Fig. 3a). Temperature and fertilizer interacted such that the effect of temperature was reduced in the HF treatment (Fig. 3b). Soil moisture and fertilizer interacted such that an increase in soil moisture (i.e., from 30 to 45% WHC) caused an increase in the potential activity in all fertilizer treatments (Fig. 3c). In the higher-concentration fertilizer treatments (MF and HF), a further increase in soil moisture (i.e., from 45 to 60% WHC) caused a further increase in potential activity (Fig. 3c).
TABLE 5.
ANOVA analysis for potential nitrification activity dataa
| Effect | Num DF | Den DF | F value | Pb |
|---|---|---|---|---|
| T | 1 | 18 | 99.34 | <.0001 |
| SM | 2 | 18 | 97.40 | <.0001 |
| F | 2 | 18 | 0.42 | 0.6655 |
| T × SM | 2 | 18 | 12.15 | 0.0005 |
| T × F | 2 | 18 | 9.42 | 0.0016 |
| SM × F | 4 | 18 | 8.72 | 0.0004 |
| T × SM × F | 4 | 18 | 0.86 | 0.5092 |
Num DF, numerator degrees of freedom; Den DF, denominator degrees of freedom; T, temperature; SM, soil moisture; F, fertilizer treatment.
Significant effects (P value < 0.05) are in bold.
FIG. 3.
Effect of (a) temperature and soil moisture, (b) temperature and fertilizer concentration, and (c) soil moisture and fertilizer concentration on potential nitrification activity. The mean potential nitrification activity is depicted for all samples grouped by (a) temperature and soil moisture treatments (including all fertilizer treatments; n = 36), (b) temperature and soil fertilizer treatments (including all moisture treatments; n = 36), and (c) soil moisture and soil fertilizer treatments (including all temperature treatments; n = 36). Soil moisture levels are 30, 45, and 60% of the soil's maximum water-holding capacity. LF, MF and HF represent fertilizer concentrations of 0.05, 0.1, and 0.3%, respectively). Rates were calculated per gram of dry soil. Error bars are 95% confidence limits.
Nitrate concentrations.
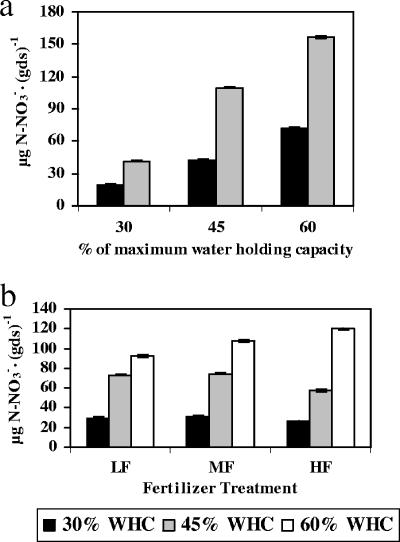
Nitrate concentrations were highly correlated to potential nitrification activity data after 7 weeks of incubation (r = 0.87; P < 0.0001). In addition, nitrate concentrations increased linearly over time in all samples, indicating that the system didn't reach a steady state at 7 weeks of incubation and that there was a constant rate of activity in each sample during the incubation. Therefore, we analyzed nitrate concentrations as another indicator of nitrification activity. Consistent with our observations of potential nitrification activity, the nitrate concentration at week 7 was significantly increased by temperature (F1,18 = 578.99; P < 0.0001) and soil moisture (F1,18 = 494.88; P < 0.0001). These trends were stable during the entire incubation period (i.e., at 1, 3, 5 and 7 weeks of incubation). However, interactions between the factors varied over time. At 7 weeks of incubation, significant interactions were found between soil moisture and both temperature and fertilizer (F4,18 = 3.66, P = 0.0464, and F4,18 = 6.86, P = 0.0015, respectively), such that elevated soil moisture caused an increase in nitrate concentrations, with the effect stronger at 30°C (Fig. 4a) and at a high fertilizer level (Fig. 4b). The scaled abundance of cluster 10 (i.e., qPCR data multiplied by T-RFLP data, as described above) could explain 43 and 38% of the differences in nitrate concentration and potential nitrification activity, respectively, after 7 weeks of incubation at 20°C (P values were 0.002 and 0.004, respectively). However, no such correlations could be found at 30°C.
FIG. 4.
Effect of (a) temperature and soil moisture and (b) soil moisture and fertilizer concentration on nitrate concentrations. The mean nitrate concentration is depicted for all samples grouped by (a) temperature and soil moisture treatments (including all fertilizer treatments; n = 36) and (b) soil moisture and soil fertilizer treatments (including all temperature treatments; n = 36). Temperatures of 20 and 30°C are displayed as black and gray bars, respectively; soil moisture levels are 30, 45, and 60% of the soil's maximum water-holding capacity. Nitrate concentrations were calculated per gram of dry soil. Error bars are 95% confidence limits.
Ammonium concentrations.
Ammonium concentrations were measured after 1, 3, 5, and 7 weeks of incubation. Ammonium concentration values in the LF and MF treatments overlapped and, therefore, were considered as one treatment for statistical analysis. We analyzed each set of time points (1, 3, 5, and 7 weeks) separately. During 7 weeks of incubation, the fertilizer treatment was the only factor that significantly affected ammonium concentration (F1,24 = 125.72, P < 0.001, after 7 weeks of incubation), with higher levels of fertilizer resulting in higher concentrations of ammonium. Mean values after 7 weeks of incubation were 83 and 434 μg N-NH4+·gds−1, where gds indicates gram of dry soil, at the lower fertilizer treatments and the HF treatment, respectively. Note that there was no effect of temperature on ammonium concentration, and therefore any effect of temperature could not be an indirect effect through changes in ammonium concentration (and corresponding changes in pH). This observation is in contrast to those of previous reports (4, 6).
DISCUSSION
In this work, we studied the relatively short-term effects (7 weeks) of temperature, soil moisture and fertilizer concentration on Nitrosospira AmoA cluster 10, represented by Nitrosospira sp. strain AF, which was dominant before and after incubation in a grassland soil from a Mediterranean climate region (coastal California). Consistent with previous studies, betaproteobacterial ammonia oxidizers restricted to cold temperate soils (i.e., Nitrosospira AmoA clusters 1, 2, and 4) (reviewed by Avrahami and Conrad [3]) were not detected before or after incubation. The detection of Nitrosospira sp. strain AF-like sequences in this soil, which annually experiences a long period of warm temperature (up to 45°C), is consistent with observations of Nitrosospira sp. strain AF in the laboratory. Jiang and Bakken (16) observed that the optimum temperature of the isolate Nitrosospira sp. strain AF was higher (31 to 33°C) than that of three other Nitrosospira strains (26 to 29°C) that had been isolated from Nordic soils. However, the close phylogenetic relationship between our soil population to this isolate does not necessarily indicate similar physiological properties. Very little is known regarding the ecology of Nitrosospira AmoA cluster 10, and its prevalence in our soil offers a unique opportunity to study this group.
Nitrosospira AmoA cluster 10 was the most common group in all samples both before and after incubation (representing 70.6 to 100% of the amoA genes detected). Consistent with previous studies of betaproteobacterial ammonia oxidizers, community shifts could not be detected by DGGE after comparable periods of incubation (e.g., 4 to 6.5 weeks [5, 6]). However, we did detect shifts in this study by T-RFLP, which is known to be more sensitive than DGGE (20). The relative abundance of AmoA cluster 10 was reduced through the synergistic effect of increased temperature, fertilizer, and soil moisture, with a parallel increase in the relative abundance of two other phylogenetic groups (AmoA clusters 3a and 9). It is not clear based on our data alone whether the decrease in relative abundance of AmoA cluster 10 is due to direct competition with these two other groups, a decrease in the fitness of AmoA cluster 10 due to suboptimal growth conditions, or both. However, as discussed above, Nitrosospira sp. strain AF has an optimal growth temperature of 31 to 33°C, which was determined at an ammonium concentration over two times higher than our MF treatment and only slightly lower than our HF treatment (16). If this isolate is characteristic of other cluster 10 organisms, these results indicate that high temperature and high ammonium concentrations are not suboptimal for these microorganisms, suggesting that competitive exclusion may underlie the decrease we observed. Further work with pure cultures would be required to test this hypothesis.
The predominance of AmoA cluster 10 in the other treatment combinations essentially provided an “enrichment culture” of cluster 10 organisms and a unique opportunity to study the response of cluster 10 organisms to environmental change.
The total abundance of members of cluster 10 in these treatments was significantly influenced by temperature and soil moisture. The samples at a combination of 20°C, 60% of WHC, and either MF or HF treatment had the highest total abundance, 3 orders of magnitude higher than that of samples from the other treatments. The soil samples before incubation were taken after the summer period, when the soil was exposed to high temperature (as high as 34°C at 2 cm below surface) and extremely low soil moisture (2.3 g/100 g dws, where dws indicates dry weight of soil, immediately after sampling in the field, equivalent to 9.1% of maximum water-holding capacity). The soil at that point had a very low total abundance of betaproteobacterial ammonia oxidizers, 1 order of magnitude lower than the lowest total abundance found in the samples after incubation. A significantly higher total abundance at 20°C than at 30°C must be due to cell growth and demonstrates the potential for proliferation by cluster 10 organisms under these conditions. This observation was consistent with the observation that increased temperature reduced the scaled abundance of AmoA cluster 10 (i.e., total abundance of AmoA cluster 10) in those treatments where this cluster declined over time.
Temperature had a significant effect not only on abundance but also on activity, although in a different direction. Mean potential nitrification activity was significantly higher at 30°C than at 20°C, although there was an antagonistic interaction between temperature and fertilizer level and a synergistic interaction between temperature and soil moisture. The positive effect of increasing soil moisture on nitrification activity is consistent with other studies (e.g., (29). Nitrate concentrations in the soil were highly correlated with potential nitrification activity, suggesting that in situ nitrification activity may respond to environmental change in a manner similar to potential nitrification activity. These similar trends could be explained by the linear increase in nitrate over the course of the experiment, indicating that the system did not reach a steady state after 7 weeks of incubation, presumably as a result of the fertilizer addition preventing the limitation of substrate.
We were surprised to observe that potential nitrification activity increased with temperature, despite decreases in the abundance of betaproteobacterial ammonia oxidizers. What might explain this observation? The diffusion rate of ammonia into the cell might potentially increase with increased temperature. However, high oxidation rates at such high temperatures have not been reported for other ammonia oxidizers. We can hypothesize that the enzymes of cluster 10 organisms are adapted to warm environments and are more active at higher temperatures than at lower temperatures, which is consistent with observations of the representative pure culture. However, based on the low abundance of cluster 10 organisms at 30°C, this high activity does not appear to provide a growth advantage, even after longer incubation (i.e., 16 to 20 weeks [data not shown]). Why might activity and abundance be decoupled? Being active under high temperatures might provide a short-term advantage through increased cell maintenance and survival under high temperatures and/or by decreasing the lag time before growth when conditions improve. This could be a major advantage in highly seasonal environments with hot, dry periods (such as in California's Mediterranean climate). Such a survival strategy has been proposed for Nitrosospira briensis under starvation conditions (8), but it has not previously been suggested for other stressful conditions or for other strains of Nitrosospira.
There is another potential explanation for our observation. It has been recently reported that archaeal ammonia oxidizers may be widespread and abundant in soils (19). Indeed, amoA genes clustering with those of archaeal ammonia oxidizers have recently been discovered in our soil (C. A. Francis et al., unpublished data) and the contribution of such organisms to potential nitrification activity in these soils is currently unknown. If the contribution of such organisms is substantial, especially at high temperatures, this could explain the decoupling of betaproteobacterial ammonia oxidizers' abundance and potential nitrification activity under these conditions. Studies of archaeal ammonia oxidizers in these soils are currently under way.
Acknowledgments
We are grateful to three anonymous reviewers for comments on previous drafts of this article. We thank Doug Turner and Margaret Gentile for technical assistance and Nutralene, Nu-Gro Technologies, Inc., Ontario, Canada, for supplying us with a fertilizer sample and for helpful advice. MJ Research, Inc., generously provided the equipment used for the quantitative PCR assays, and we are grateful to Richard Kurtz for advice on its operation.
This work was supported by awards from the Mellon Foundation, the David and Lucile Packard Foundation, and the National Science Foundation (DEB-0221838 and DEB-0108556).
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Aakra, A., J. B. Utaker, and I. F. Nes. 2001. Comparative phylogeny of the ammonia monooxygenase subunit A and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237-242. [DOI] [PubMed] [Google Scholar]
- 2.Aakra, A., J. B. Utaker, I. F. Nes, and L. R. Bakken. 1999. An evaluated improvement of the extinction dilution method for isolation of ammonia-oxidizing bacteria. J. Microbiol. Methods 39:23-31. [DOI] [PubMed] [Google Scholar]
- 3.Avrahami, S., and R. Conrad. 2005. Cold-temperate climate: a factor for selection of ammonia oxidizers in upland soil? Can. J. Microbiol. 51:709-714. [DOI] [PubMed] [Google Scholar]
- 4.Avrahami, S., and R. Conrad. 2003. Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl. Environ. Microbiol. 69:6152-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. [DOI] [PubMed] [Google Scholar]
- 7.Bollmann, A., and H. J. Laanbroek. 2001. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations. FEMS Microbiol. Ecol. 37:211-221. [Google Scholar]
- 8.Bollmann, A., I. Schmidt, A. M. Saunders, and M. H. Nicolaisen. 2005. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. J., A. Hussain, J. R. Stephen, M. D. Mullen, D. C. White, and A. Peacock. 2001. Impact of herbicides on the abundance and structure of indigenous β-subgroup ammonia-oxidizer communities in soil microcosms. Environ. Toxicol. Chem. 20:2462-2468. [DOI] [PubMed] [Google Scholar]
- 10.De Boer, W., and G. A. Kowalchuk. 2001. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 33:853-866. [Google Scholar]
- 11.Hastings, R. C., C. Butler, I. Singleton, J. R. Saunders, and A. J. McCarthy. 2000. Analysis of ammonia-oxidizing bacteria populations in acid forest soil during conditions of moisture limitation. Lett. Appl. Microbiol. 30:14-18. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, A. J., Costello, A., Lidstrom, M. E., and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 13.Horz, H. P., A. Barbrook, C. B. Field, and B. J. M. Bohannan. 2004. Ammonia-oxidizing bacteria respond to multifactorial global change. Proc. Natl. Acad. Sci. USA 101:15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horz, H. P., V. Rich, S. Avrahami, and B. J. M. Bohannan. 2005. Methane-oxidizing bacteria in a California upland grassland soil: diversity and response to simulated global change. Appl. Environ. Microbiol. 71:2642-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Q. Q., and L. R. Bakken. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171-186. [DOI] [PubMed] [Google Scholar]
- 17.Koops, H. P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 18.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 19.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 20.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer, G., T. Brinkhoff, U. Nuebel, C. Santegoeds, H. Schafer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. DeBruijn (ed.), Molecular microbial ecology manual. Kluwer, Dordrecht, The Netherlands.
- 22.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 23.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlichting, E., and H. P. Blume. 1966. Bodenkundliches Praktikum. Paul Parey, Hamburg, Germany.
- 26.Schmidt, E. L., and L. W. Belser. 1994. Autotrophic nitrifying bacteria, p. 160-177. In J. M. Mickelson, M. Weaver, S. Angle, P. Bottomley, D. Bezdicek, S. Smith, A. Tabatabai, and A. Wollum (ed.), Methods in soil analysis. American Society of Agronomy, Soil Science Society of America, Madison, WI.
- 27.Shaw, M. R., E. S. Zavaleta, N. R. Chiariello, E. E. Cleland, H. A. Mooney, and C. B. Field. 2002. Grassland responses to global environmental changes suppressed by elevated CO2. Science 298:1987-1990. [DOI] [PubMed] [Google Scholar]
- 28.Sorokin, D., T. Tourova, M. C. Schmid, M. Wagner, H. P. Koops, J. G. Kuenen, and M. Jetten. 2001. Isolation and properties of obligately chemolithoautotrophic and extremely alkali-tolerant ammonia-oxidizing bacteria from Mongolian soda lakes. Arch. Microbiol. 176:170-177. [DOI] [PubMed] [Google Scholar]
- 29.Stark, J. M., and M. K. Firestone. 1995. Mechanisms for soil-moisture effects on activity of nitrifying bacteria. Appl. Environ. Microbiol. 61:218-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephen, J. R., Y. J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stienstra, A. W., P. K. Gunnewiek, and H. J. Laanbroek. 1994. Repression of nitrification in soils under a climax grassland vegetation. FEMS Microbiol. Ecol. 14:45-52. [Google Scholar]
- 32.Utaker, J. B., L. Bakken, Q. Q. Jiang, and I. F. Nes. 1996. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA gene sequences. Syst. Appl. Microbiol. 18:549-559. [Google Scholar]
- 33.Yeager, C. M., D. E. Northup, C. C. Grow, S. M. Barns, and C. R. Kuske. 2005. Changes in nitrogen-fixing and ammonia-oxidizing bacterial communities in soil of a mixed conifer forest after wildfire. Appl. Environ. Microbiol. 71:2713-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]