Abstract
The health of millions is threatened by the use of groundwater contaminated with sediment-derived arsenic for drinking water and irrigation purposes in Southeast Asia. The microbial reduction of sorbed As(V) to the potentially more mobile As(III) has been implicated in release of arsenic into groundwater, but to date there have been few studies of the microorganisms that can mediate this transformation in aquifers. With the use of stable isotope probing of nucleic acids, we present evidence that the introduction of a proxy for organic matter (13C-labeled acetate) stimulated As(V) reduction in sediments collected from a Cambodian aquifer that hosts arsenic-rich groundwater. This was accompanied by an increase in the proportion of prokaryotes closely related to the dissimilatory As(V)-reducing bacteria Sulfurospirillum strain NP-4 and Desulfotomaculum auripigmentum. As(V) respiratory reductase genes (arrA) closely associated with those found in Sulfurospirillum barnesii and Geobacter uraniumreducens were also detected in active bacterial communities utilizing 13C-labeled acetate in microcosms. This study suggests a direct link between inputs of organic matter and the increased prevalence and activity of organisms which transform As(V) to the potentially more mobile and thus hazardous As(III) via dissimilatory As(V) reduction.
Arsenic poisoning of groundwater used for drinking and irrigation is a global issue, with the risk of harmful human exposure occurring at numerous locations across the Americas, Asia (most notably West Bengal and Bangladesh [7, 33, 34]), and also central Europe (16). Many recent studies have reported arsenic-enriched groundwater within the Ganges-Brahmaputra-Meghna Delta (e.g., see references 3 and 36), with more than 35 million people at risk of arsenic poisoning in Bangladesh alone (34). The weathering of arsenic-rich minerals prevalent in the Himalayas and their gradual transport and deposition in the alluvial deltas below, followed by microbially mediated arsenic solubilization, are thought to be major mechanisms of arsenic mobilization into aquifers within the region. Conditions similarly conducive to the development of arsenic-enriched groundwater are thought to be present within the Red River (4) and Mekong River (28) deltas of Southeast Asia, where elevated concentrations of arsenic have also recently been reported. The present study focuses on the potential causes of changes in arsenic mobility within subsurface sediments taken from the Mekong River Basin, Cambodia, where many tens of thousands of inhabitants could be at risk of exposure to hazardous levels of arsenic (28).
The mechanism of arsenic release from aquifer sediments has been a topic of intense academic debate (2, 22, 26, 33, 43). However, a consensus is developing around the concept of microbially mediated release of arsenic from sediment-bound hydrated ferric oxides as the dominant mechanism of mobilization into groundwater systems of the Ganges Delta (2, 12, 13). These microbial processes may be sustained by predominantly sedimentary organic matter (23) but also influenced by organic matter more recently introduced into surface-derived waters that can percolate into the aquifer (10). Although the precise mechanism of arsenic mobilization remains to be characterized in detail, respiration of sorbed As(V) by dissimilatory As(V)-reducing prokaryotes may play a role, resulting in the formation of potentially more mobile As(III) (25). Dissimilatory As(V)-respiring prokaryotes comprise a diverse phylogenetic group, including Chrysiogenes, Bacillus, Desulfomicrobium, Sulfurospirillum, Shewanella, Citrobacter, and Sulfurihydrogenibium species (25). Although the ability to respire As(V) is spread across several phylogenetic groups, the mechanism of As(V) reduction in these organisms seems to be conserved. The first respiratory As(V) reductase, a periplasmic dimer (87- and 29-kDa subunits) of the dimethyl sulfoxide family of mononuclear molybdenum-containing enzymes, was characterized for Chrysiogenes arsenatis (15) and more recently for Bacillus selenitireducens (1) and Shewanella strain ANA-3 (1). The conserved nature of the characterized respiratory As(V) reductase genes has since been exploited to develop PCR primers (for examples, see Table S1 in the supplemental material). Despite the potential importance of dissimilatory As(V)-reducing prokaryotes in controlling arsenic mobility in the subsurface, there have been no systematic studies of the diversity and activity of these organisms in Southeastern Asian aquifer sediments.
The aim of this study was to use a suite of molecular techniques to identify As(V)-respiring bacteria and their corresponding respiratory As(V) reductase genes in sediments collected from a Cambodian aquifer with elevated aqueous arsenic concentrations. These organisms were stimulated under anaerobic conditions in laboratory microcosms by the addition of acetate as a proxy for organic matter, conditions that have been shown previously to support enhanced rates of arsenic mobilization in analogous sediments from West Bengal (13). The use of stable isotope-labeled [13C]acetate in these experiments and the subsequent isolation of 13C-labeled nucleic acids from the metabolically active fraction of the sediment microbial community allowed the detailed characterization of bacteria coupling acetate oxidization to As(V) reduction. In order to examine the diversity of As(V)-respiring bacteria, a suite of PCR primers were used to amplify 16S rRNA genes and the α-subunit of dissimilatory As(V) reductase genes. This is the first report describing As(V)-respiring bacteria and their corresponding genes in Southeast Asian aquifer sediments and among the first studies to use stable isotope probing (SIP) to dissect a sediment biogeochemical process.
MATERIALS AND METHODS
Experimental outline.
A fine-grained sediment (SY9IIb) was obtained 9 m below the sediment surface in Kean Svay district, Cambodia, 5 mi southeast of Phnom Penh. It has been characterized previously (30, 31), with an arsenic concentration of 13.1 μg g−1 (dry weight).
Three sets of three replicate microcosms were constructed for treatment by combining 20 g of sediment (21% ± 0.4% standard error [SE] water content [wt/vol]) with 40 ml artificial groundwater (MgCl2, 0.34 mM; KH2PO4, 0.01 mM; NaHCO3, 0.51 mM; K2CO3, 0.025 mM; MgSO4, 0.03 mM; KNO3, 0.001 mM; and CaCO3, 1.85 mM; pH 7) in 100-ml serum bottles sealed with butyl rubber stoppers under an N2 atmosphere. Treatments consisted of (i) control treatment, no amendment; (ii) experimental treatment, augmented with 10 mM uniformly labeled [13C]sodium acetate (Sigma-Aldrich, United Kingdom); and (iii) experimental treatment, augmented with 10 mM uniformly labeled [13C]sodium acetate and 10 mM sodium arsenate. The effect of acetate concentration (0, 5, 10, 20, 50, or 100 mM) on nucleic acid yield was also investigated using the same microcosm design. All treatments were incubated in the dark at 20°C.
Analytical techniques.
Sediment slurry (4 ml) was removed prior to the introduction of the Na acetate (0 days) and then after 4, 8, 12, 16, 20, and 30 days of incubation. Fe(II) concentrations were measured spectrophotometrically using the method of Lovley and Phillips (17). Briefly, sediment slurry (100 μl) was added to 4.9 ml HCl (0.5 M) and incubated at room temperature (∼20°C, 1 h). Aliquots of this mixture (50 μl) were added to a ferrozine solution (2 mM ferrozine, 5 mM HEPES; pH 7.0) and incubated at room temperature (∼20°C, 30 s) before the concentration of Fe(II) was measured at 562 nm for comparison to a standard curve prepared with known concentrations of Fe(II) (as ferrous ethylene diammonium sulfate). Acetate was quantified using ion chromatography. An aliquot of sediment suspension (2 ml) was centrifuged (5,600 × g, 3 min) and the supernatant filtered with a 0.45-μm PTFE membrane filter (Whatman, United Kingdom). Concentrations of acetate within the solution were measured using a Dionex DX-120 IC separation center and a Dionex AS-11HC anion-exchange column, with a mobile phase comprising 20 mM NaOH. Samples (200 μl) were injected for analysis at a flow rate of 1.3 ml min−1. Total aqueous arsenic concentrations were measured using inductively coupled plasma optical emission spectroscopy (Horizon; VG-Elemental). The oxidation state of arsenic within the microcosms was assessed using X-ray adsorption near edge structure (XANES) spectroscopy at the K-edge using station 16.5 at the CCLRC Daresbury Laboratory, as previously detailed by Rowland et al. (30).
Isolation of soil RNA and DNA.
Sediment samples (2 g) were removed and stored at −80°C until required for analysis. Sediment DNA was extracted using an Ultraclean soil DNA isolation kit (Mol Bio Labs) according to the manufacturer's instructions. Soil RNA was extracted selectively using the modified method of Griffiths et al. (9), excluding cetyltrimethylammonium bromide (CTAB) from the buffer solution. Extracted nucleic acids were resuspended in sterile, nuclease-free water and stored at −80°C until required for analysis.
PCR regimens.
Various PCR regimens were undertaken in order to characterize bacterial diversity and identify the key organisms present within individual sediment microcosms (see Table S1 in the supplemental material). To characterize the dominant groups of bacteria, a conserved region of the 16S rRNA gene was amplified by PCR using the universal bacterial primers 8Forward and 519Reverse (11). PCR products were purified using a QIAQuick purification kit (QIAGEN Ltd., Crawley, United Kingdom) and cloned into a pCR2.1 vector by using a TA cloning kit (Invitrogen Ltd., Paisley, United Kingdom) and competent Escherichia coli cells (One Shot TOP10; Invitrogen Ltd.) according to the manufacturer's instructions. Recombinant clones were selected by antibiotic (ampicillin) resistance (carried within the vector) and blue/white colony screening before the presence of the 16S rRNA gene fragment was verified by PCR and agarose gel electrophoresis.
Clones were separated into operational taxonomic units (OTUs) based upon the similarity of restriction fragment length polymorphism profiles. PCR products were incubated (16 h, 37°C) with the restriction endonucleases Sau3AI and MspI (0.1 U μl−1; Roche Products Ltd., Welwyn Garden City, United Kingdom) and digested fragments imaged following electrophoresis with agarose gels, stained with ethidium bromide. The nucleotide sequences of each OTU were determined by the dideoxynucleotide method, utilizing an ABI Prism BigDye terminator cycle sequencing kit in combination with an ABI Prism 877 integrated thermal cycler and an ABI Prism 377 DNA sequencer (Perkin Elmer Applied Biosystems, Warrington, United Kingdom).
Changes in the diversity of the bacterial community were assessed using denaturing gradient gel electrophoresis (DGGE). The primers GC338Forward and 530Reverse targeted a smaller region of the same conserved 16S rRNA gene, while the GC-clamped product facilitated community analysis via DGGE, according to the method of van der Gast et al. (42). PCR samples (20 μl) were loaded onto a 10% (wt/vol) polyacrylamide gel with a 40 to 60% denaturing gradient in 0.5%× Tris-acetate-EDTA buffer (pH 8.3). Electrophoresis was undertaken for 16 h at 60°C, and a constant voltage of 100 V was applied using a SciPlas denaturing gradient CDC unit (Wolf Laboratories Ltd., York, United Kingdom). Gels were stained (20 min) in 0.5× Tris-acetate-EDTA buffer containing 2 mg ml−1 SYBR gold (Molecular Probes Inc., OR) and imaged under short-wave UV light. DGGE profiles of sequenced clone fragments were compared to those of mixed microbial communities to facilitate species identification.
Various arsenate-reducing bacteria have been identified previously (25). Primers targeting the functional genes involved in either the bacterial respiration of As(V) (arrA) or resistance to As(V) (arsC) are listed in Table S1 in the supplemental material. PCR was completed by observing published, optimized amplification procedures (20, 38). Nested primers AS1F, AS1R, and AS2F were designed (B. Song et al., unpublished) by comparing conserved regions in the arrA genes from Bacillus selenitireducens, Chrysiogenes arsenatis, Shewanella sp. strain ANA-3, Desulfitobacterium hafniense DCB-2, and Wolinella succinogenes. The first PCR was performed with AS1F and AS1R primers by using a 5-min denaturation step at 94°C, followed by 35 cycles of a 30-s denaturation at 94°C, primer annealing of 30 s at 50°C, and a 1-min extension at 72°C. The second PCR amplification was performed with AS2F and AS1R primers and using the first PCR as templates. The second PCR cycle began with a 2-min denaturation step at 94°C, followed by 30 cycles of a 30-s denaturation at 94°C, primer annealing of 30 s at 55°C, and a 1-min extension at 72°C. The presence of all amplification products was verified by electrophoresis with 1% agarose gels, stained with ethidium bromide.
Stable isotope probing of nucleic acids.
The incorporation of a stable-isotope-labeled substrate (i.e., [13C]sodium acetate) into cellular biomarkers, including nucleic acids, enables substrate-assimilating organisms to be identified via SIP (5, 29). RNA SIP was used in conjunction with DNA SIP, as the greater turnover of RNA within biological cells creates a more responsive biomarker (21) during the early phase of experimental incubation (4 days). DNA SIP was utilized to assess later stages of the incubations (16 days), where a lower rate of turnover is beneficial to prevent the bias of cross-feeding (the secondary use of 13C by organisms utilizing assimilated 13C rather than the labeled acetate).
To confirm that nucleic acids had incorporated 13C, the standard nucleic acid fraction was separated from the higher-density fractions (containing 13C) by equilibrium (isopycnic) density gradient centrifugation, as described by Manefield et al. (21). Briefly, nucleic acids (500 ng) were combined with 5.1 ml cesium trifluoracetate and 166 μl deionized formamide to achieve a solution density of 1.71 or 1.8 g ml−1 for DNA or RNA, respectively, before being loaded into polyallomer bell top quick-seal centrifuge tubes (Beckman Instruments UK, High Wycombe, United Kingdom), which were heat sealed. Samples were then centrifuged (140,000 × g, 36 h, 20°C) (Optima TLX ultracentrifuge; Beckman Instruments UK).
Gradients were fractionated by displacement with water by using a peristaltic pump (205 U; Watson-Marlow Bredel, United Kingdom) at a flow rate of 3.3 μl s−1, collecting fractions from the pierced base of the tube. A density gradient profile was created by weighing known volumes of the solution on a bench-top balance. RNA was precipitated from the solution with 0.6 volumes of isopropanol and centrifuged at 5,600 × g for 10 min. The pellet was washed in ethanol (−20°C) and allowed to air dry before being resuspended in nuclease-free water. Fractions determined to contain predominantly unlabeled or labeled nucleic acids are referred to as light and heavy fractions, respectively.
Reverse transcription.
RNA samples were treated with DNase (RQ1 RNase-free DNase; Promega, WI) to remove genomic DNA contamination. cDNA was synthesized using the treated RNA samples and the necessary reverse primers (see Table S1 in the supplemental material) with an avian myeloblastosis virus reverse transcriptase (Promega) according to the manufacturer's instructions. PCRs were undertaken to amplify the 16S rRNA and arrA genes from the cDNA using the primers listed in Table S1 in the supplemental material. The light and heavy fractions of RNA were successfully separated using isopycnic centrifugation, appearing within two separate gradient zones, as observed by agarose gel electrophoresis of the PCR product. Fragments were cloned and sequenced and the bacterial diversity attained from the RNA SIP methodology compared to that achieved from extracted DNA. All samples included control reactions with avian myeloblastosis virus excluded from the transcription procedure; no products were obtained.
Statistical analysis.
Sequences were analyzed against the NCBI BLAST database to find the most closely matched known gene sequences. Sequences were aligned using the ClustalX v1.8 (40) software package and corrected manually. Phylogenetic distance analysis was performed using the TREECON package (41), employing the Jukes and Cantor correction and bootstrap resampling process, and dendrograms constructed from the matrix via neighbor joining (32). Phylogenetic analysis for translated amino acid sequences of the arrA genes was performed using PAUP 4.0.
XANES data (four scans per sample) were corrected using background subtraction and summed using the Daresbury programs EXCALIB and EXBACK (8). XANES spectra were fitted using the Daresbury laboratory program LINCOM (http://srs.dl.ac.uk/XRS/index.html). t tests were performed as applicable to the data set, using two sets of paired means or nonpaired samples assuming equal variance.
Nucleotide sequence accession numbers.
The 36 sequences obtained in this study have been deposited in the GenBank database under accession no. EF014909 to EF014944.
RESULTS
Acetate utilization and the microbial reduction of Fe(III) and As(V).
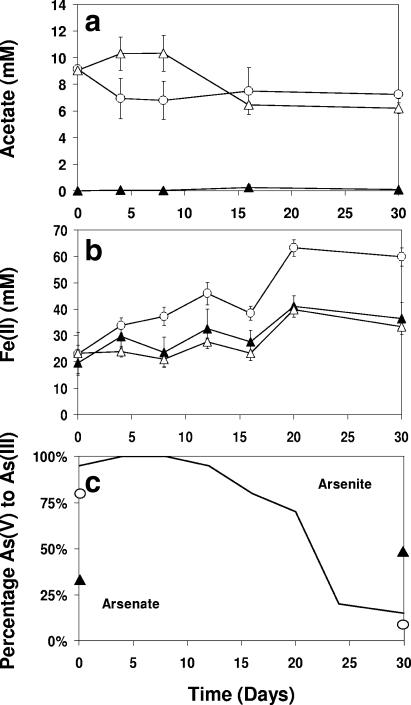
To assess the impact of As(V) and organic carbon (acetate) loadings on microbial communities in sediments from a Cambodian aquifer, microcosms were amended with either acetate (10 mM) or both acetate and arsenate (both 10 mM); nonamended controls were also included. Concentrations of acetate declined more rapidly in non-As(V)-amended microcosms (t = 2.79, df = 10, P < 0.05; 4 days and 8 days, combined) (Fig. 1a), although at the end of the study (30 days), concentrations of acetate were similar with or without added As(V) (t = 2.08, df = 4, P = 0.11). Microcosms augmented with more than 10 mM acetate exhibited no decline in acetate concentration over the duration of the study (P ≥ 0.05) (data not shown).
FIG. 1.
Concentrations of total (a) acetate, (b) Fe(II), and (c) arsenic speciation [As(V)/As(III)], derived as percentages of the total As within individual sediment pellets (assessed via XANES analysis) incubated with (○) 10 mM acetate or (▵) 10 mM acetate and 10 mM sodium-arsenate or (▴) not amended. Bars show 1× SE for three replicates.
Sediments incubated with 10 mM acetate only exhibited an increased rate of Fe(III) reduction compared to the nonamended control (60 mM ± 3.5 SE and 36 mM ± 6.0 SE, respectively, at 30 days) (Fig. 1b). These results were in keeping with those of other studies using similar materials (13, 31) and support the hypothesis that the microbial communities in the sediments were limited for electron donor. This increase in the rate and extent of Fe(III) reduction was not noted with the addition of 10 mM As(V), however. Here Fe(III) reduction matched that in the unamended control cultures (t = 0.95, df = 8, P = 0.37), possibly due to toxicity of the As(V) or its use as a competing electron acceptor, nullifying a potential increase in the rate of Fe(III) reduction.
Arsenic speciation was monitored using XANES (Fig. 1c), revealing the initial distribution of arsenic to be 35% As(V) (unamended control) but to progress to 55% over the duration of the study. The proportion of As(V) was expectedly far higher within the As(V)-amended sediment (90%; 0 days). Nevertheless, by the end of the study (30 days), most (80 to 90%) was present as the reduced As(III), as was also noted within experimental treatments amended solely with acetate [10% as As(V)].
Molecular analysis of microcosm bacterial communities.
Concentrations of extractable rRNA appeared low within the sediment (0 days), assessed using agarose gel electrophoresis of total nucleic acids (data not shown). However, with the addition of 10 mM acetate, quantities of rRNA increased within 3 days (data not shown), indicating that the microbial community was rapidly able to assimilate this new source of carbon. The same rapid increase in extractable rRNA was not observed with the further addition of 10 mM sodium arsenate, giving further support to the hypothesis that the metalloid was inhibitory to overall microbial activity.
Nucleic acid stable isotope probing.
Light and heavy fractions of both DNA and RNA were successfully separated using isopycnic centrifugation, being present mainly within fractions exhibiting buoyant densities of 1.790 and 1.805 g ml−1 for RNA and 1.705 and 1.760 g ml−1 for DNA (data not shown), respectively. Although it is not possible to completely separate light and heavy fractions (18), there was no visible evidence of RNA (or rather PCR product, following reverse transcription-PCR) in gradient fractions between these samples. Separation was less distinct for the DNA gradient fractions.
Bacterial community diversity.
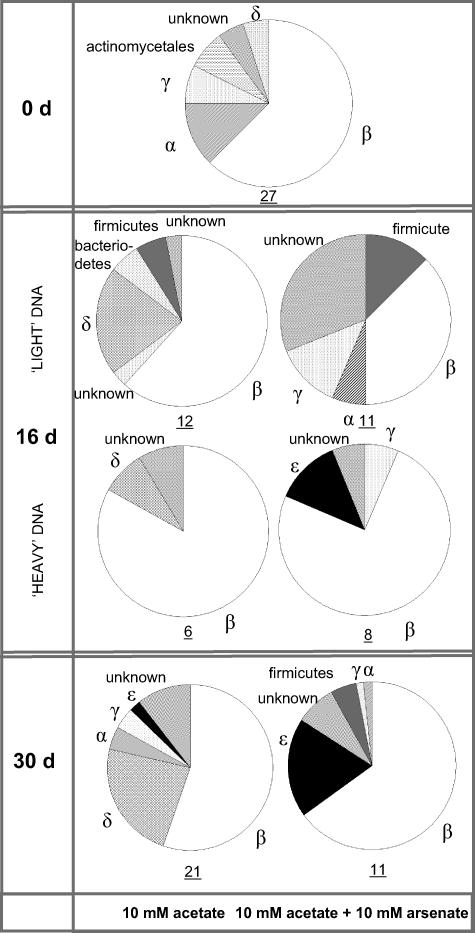
Analysis of 16S rRNA genes present within the initial (0 days) microbial community revealed a varied consortium comprised primarily of the β-proteobacteria (63%) (Fig. 2) (based on OTU abundance), dominated by strains related to the genera Acidovorax (15%) and Comamonadaceae (15%) (clones GL_ARS2 and GL_ARS3, respectively [see Table S2 in the supplemental material]). The examination of 16S rRNA templates revealed a slightly less varied consortium (Fig. 3), similarly comprised mainly of β-proteobacteria, but with α-proteobacteria also prevalent (48 and 38% of the cloned community, respectively). Analysis of sediment rRNA showed the active microbial community to be dominated by strains related to Novosphingobium capsulatum (25%; clone GL_ARS1) and Acidovorax sp. strain JS42 (17%; clone GL_ARS2). Included in the rRNA library were strains linked to the reduction of Fe(III) (clone GL_ARS15, 2%; related to Geobacter sp. [see Table S2 in the supplemental material]; not isolated from sediment DNA) and the oxidation of ammonia (clone GL_ARS3, related to Nitrosospira sp. strain NSp 17, 11%; also comprising 2.5% of the extracted DNA).
FIG. 2.
Compositions of bacterial communities from DNA extracted from original sediments (0 days [0 d]) and after 30 days of incubation with either 10 mM acetate only or 10 mM acetate and 10 mM sodium arsenate, determined by 16S rRNA gene clone libraries. Community compositions of organisms at 16 days have been separated by the buoyant density of DNA from those organisms predominantly utilizing the 13C-labeled acetate (heavy fraction) and the remaining microbial community (light fraction). Values beneath the charts refer to the number of OTUs isolated following the analysis of 30 clones, as a crude measure of diversity.
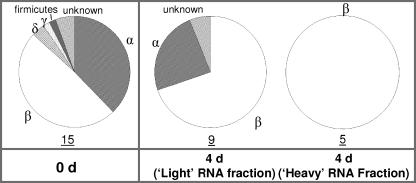
FIG. 3.
Compositions of bacterial communities from RNA extracted from original sediments (0 days [0 d]) and from SIP-labeled gradient fractions of microcosms incubated with 10 mM acetate and 10 mM sodium arsenate, determined by 16S rRNA clone libraries. Values beneath the charts refer to the number of OTUs isolated following the analysis of 30 clones, as a crude measure of diversity.
After only 4 days of incubation with 10 mM As(V), microbial diversity appeared to decrease within the light, unlabeled RNA SIP fraction, being comprised of a greater percentage of the β-proteobacteria (70%) (Fig. 3). Clones related to Acidovorax sp. (GL_ARS2) and Nitrosospira sp. strain Nsp17 (GL_ARS3) remained present within the community (16% and 15%, respectively), while bacteria related to Rhizobium sp. strain OK 55 (18%; clone GL_GLY14) were also now identified. The dominant members of the community were clones related to Comamonadaceae strain PIV-8-1 (36%; GL_ARS4), while the labeled heavy 16S rRNA template was comprised exclusively of clones related to this family (to Comamonadaceae strain PIV-8-1 and Acidovorax sp. [64% and 36%, respectively]).
DNA SIP was undertaken on the samples after 16 days, as As(V) was rapidly transformed to As(III) within the amended microcosms after this time (Fig. 1c). After 16 days, as perhaps would be expected given the dominance of the β-proteobacteria within the sediment 16S rRNA template (4 days), the addition of acetate increased the proportion of clones related to this phylum represented within the labeled DNA to 82% within the acetate only-amended treatments, again dominated by organisms related to the family Comamonadaceae (79%). Absent from the labeled fraction but present in the unlabeled DNA were sequences related to Fe(III)-reducing members of the genus Geobacter (10%; clone GL_ARS15). A smaller contingent of clones associated with the dissimilatory As(V)-reducing bacterium (24) Desulfotomaculum auripigmentum (2%; clone GL_ARS18, GenBank accession no. EF014942, 97% identity, 505 bp) and Desulfosporosinus sp. strain P3 (2%; clone GL_ARS19, GenBank accession no. EF014943, only 93% identity, 291 bp) were also detected. The proportion of clone GL_ARS18 was further increased within the As(V)-enriched sediment, comprising 13% of the unlabeled fraction.
When also incubated with 10 mM As(V), clones related to the As(V)-respiring ɛ-proteobacterium Sulfurospirillum sp. strain NP-4 (clone GL_GLY16) became a significant component of the sediment microbial community, detected solely within the labeled DNA fraction (13% of clones). Sulfurospirillum sp. strain NP-4 is closely related to the bacterium Sulfurospirillum deleyianum, which is similarly known to respire through the dissimilatory reduction of As(V) (25). Clones related to Sulfurospirillum sp. strain NP-4 were observed solely within As(V)-amended microcosms.
Although both communities continued to be dominated by β-proteobacteria after incubation with acetate (30 days), significant differences were apparent in microbial community composition between microcosms with and without amendment of 10 mM As(V). Microbial diversity appeared to be greater in the non-As(V)-amended sediments where the community was dominated by relatives of Aquaspirillum delicatum (30%; clone GL_GLY9) but also included a large percentage of clones related to the family Geobacteraceae (21%; clone GL_ARS15), a member of the group of δ-proteobacteria. No clones closely related to Geobacter species were detected from sediments incubated with 10 mM As(V). Here, the microbial community was once again dominated (29%) by members associated with the family Comamonadaceae (namely, isolate PIV-8-1; clone GL_ARS4) but also the As(V)-respiring ɛ-proteobacterium Sulfurospirillum sp. strain NP-4 (19%; clone GL_GLY16) and members of the genus Acidovorax (16%; clone GL_ARS2).
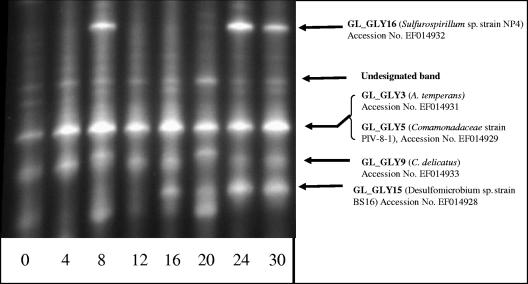
Analysis of the sediment microbial community by DGGE revealed that five main bands dominated microcosms incubated with additional acetate and As(V). The dominant species constituting each band were assigned by comparison of these bands with profiles obtained from cloned and sequenced 16S rRNA gene fragments, previously isolated from the community. Where different clones generated similar banding patterns, it was assumed that the most prevalent clone (based on number of OTUs) contributed most to the intensity of the DGGE band. One new band appeared within the arsenic-enriched sediments after 16 days (Fig. 4), and it appeared to correspond to an increase in the proportion of organisms related to Desulfomicrobium BS16 (clone GL_GLY15) within the microcosms over time. Bands which appeared to represent the increase in number of clone GL_GLY16 (related to Sulfurospirillum strain NP-4) were further observed. The band appearing on day 8 may be caused by heterogeneity in the sample, for example, localized microcolony development, prior to the establishment of bacterium GL_GLY16 as a significant component of the sediment microbial community.
FIG. 4.
Microbial diversity, as assessed by DGGE, of bacterial community within microcosms amended with both 10 mM acetate and 10 mM sodium arsenate. Arrows denote the presence of bands thought to represent named species, as assessed via comparison to the DGGE profiles of cloned and sequenced 16S rRNA gene fragments. The closest relative in GenBank is shown in brackets (see Table S3 in the supplemental material). Numbers at bottom denote days of incubation. C. delicatus, Curvibacter delicatus.
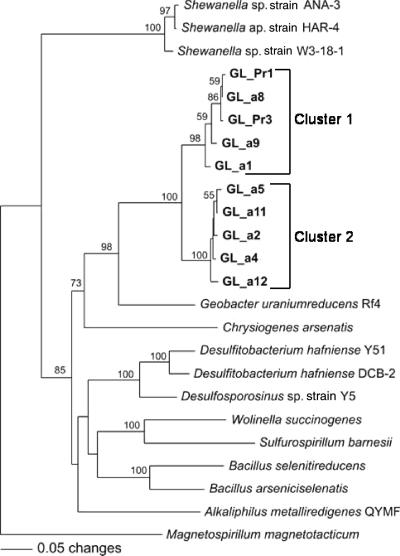
Amplification of sediment nucleic acids (both DNA or mRNA) for the arsC gene fragment, which forms a component of the well-studied ars detoxification system for arsenic (38), yielded no product. Experimental microcosms were further probed using arrA primers as a marker for As(V) respiration in the environment. The method of Malasarn et al. (20) yielded a 165-bp PCR product of arrA genes within As(V)-amended sediments after 30 days of incubation (clone GL_arra1, GenBank accession no. EF014944). The detected gene shared 72.2% amino acid sequence similarity with that in Sulfurospirillum barnesii. Nested-PCR amplification with AS2F and AS1R primers yielded 625-bp products after 30 days of incubation with acetate (no product was detected within the unamended control). PCR product was also obtained from the 13C-labeled DNA of the acetate-enriched microbial community extracted by DNA SIP (16 days). Sequence analysis of the amplified PCR products from 16- and 30-day incubations showed the presence of two distinct groups of arrA genotypes (Fig. 5). The arrA genes belonging to cluster 1 (GL_Pr1, GL_Pr3, GLa1, GLa8, and GLa9) were detected only in the microcosm of the 16-day incubation. Cluster 2 (GLa2, GLa4, GLa5, GLa11, and GLa12) was found from the microcosm of the 30-day incubation. The genes in cluster 1 have 73% and 84.8% amino acid sequence similarities with the arrA genes found in Chrysiogenes arsenatis and Geobacter uraniumreducens, respectively. Cluster 2 genes share 71.2% and 85.7% similarities with those of C. arsenatis and G. uraniumreducens. No product was obtained from any treatment or time point when screening extracted mRNA, possibly the result of a low abundance of the target template within samples.
FIG. 5.
Phylogenetic analysis of translated amino acid sequences from the arrA genes detected within As(V)-amended microcosms (day 16 and day 30). Nearest-neighbor joining of 210 amino acids was used for analysis.
DISCUSSION
The reduction of As(V) to As(III) was observed within microcosms following amendment with acetate. An increase in the proportion of bacteria known to respire As(V) was detected concurrently, with differing microbial community profiles developing within sediments amended with additional As(V) versus within those amended solely with acetate. The initial decreased utilization rate of acetate within microcosms also incubated with 10 mM As(V) is likely a consequence of decreased overall microbial activity and growth in the presence of the metalloid. As(V) is a toxic analogue for inorganic phosphorylation activities (37), entering cells as the toxic oxyanion, where it is capable of uncoupling oxidative phosphorylation and interfering with protein synthesis (39). The decrease in the diversity of 16S rRNA transcript amplified from the As(V)-amended sediment (4 days) indicates a selective pressure on the bacterial community. The growth and activity of less tolerant species, perhaps including those related to Novosphingobium capsulatum, were inhibited, while potentially more resistant organisms, such as those related to Rhizobium sp. (6), increased in dominance. Organisms associated with Acidovorax temperans and Comamonadaceae strain PIV-8-1 also appeared tolerant to the concentrations of arsenic present, increasing in relative proportion over the duration of the study while rapidly utilizing the labeled acetate (within 4 days). It remains possible that bacteria with the ability to respire and reduce As(V) are ubiquitous, but as highlighted in the present study, such bacteria may exist in low numbers until provided with the opportunity to flourish by the addition of new sources of, especially, carbon and possibly the selective pressure of higher concentrations of arsenic.
Similarities in microbial community composition of the acetate-enriched sediment were apparent between the present study and the comparable experiment undertaken by Rowland et al. (31), with the dominance of β-proteobacteria and including a large proportion of δ-proteobacteria affiliated with the family Geobacteraceae. These bacteria are known to respire through the dissimilatory reduction of Fe(III), which is present at significant concentrations in the sediments, but are not thought to mobilize arsenic (14), although at least one Geobacter species (G. uraniumreducens) may harbor the genes for arsenic respiration (C. W. Saltikov, personal communication). Interestingly, As(V) reductase genes affiliated with genes from this sequenced organism were detected in microcosms supplemented with acetate and arsenate. In addition, several bacteria known to respire using arsenic and their As(V) reductase genes were detected in this study. Clone DNA closely matched to Desulfotomaculum auripigmentum (97% match; ∼500 bp) and Sulfurospirillum sp. strain NP-4 (100% match; ∼500 bp) was present after 16 days, suggesting that naturally occurring dissimilatory As(V)-reducing bacteria increased in number/activity within the sediment microcosms. Matches also suggested the presence of members related to the genera Desulfosporosinus (94% match; ∼500 bp) and Desulfomicrobium (98% match; ∼500 bp), similarly known to include dissimilatory As(V)-respiring bacteria, although in common with Desulfotomaculum auripigmentum these organisms can also respire using sulfate as an electron acceptor, which was noted in the groundwater at the sampling site at concentrations of about 10 ppm (0.1 mM). That such strains were not detected within the original sediment (0 or 4 days) suggests they were selectively enriched from a previously low background contingent, which was further supported by DGGE profiling. Genes associated with D. auripigmentum were detected only within sediment enriched solely with acetate, while clones related to Sulfurospirillum sp. strain NP-4 were identified only from sediment also enriched with 10 mM As(V). This indicates that such organisms may be more tolerant to higher concentrations of arsenic within As(V)-amended microcosms or may gain selective advantage from arsenate respiration.
No clones closely related to Desulfotomaculum (or Desulfosporosinus) species were detected within the labeled DNA fractions. This is perhaps consistent with the findings of Newman et al. (24), who determined that D. auripigmentum was unable to utilize acetate as a sole electron donor, a suggested characteristic of As(V)-respiring bacteria (19), including Sulfurospirillum sp. strain NP-4 (44). However, close relatives Sulfurospirillum arsenophilum strain MIT-13 and S. barnesii strain SES-3 have been observed to utilize acetate as a source of carbon (35) in the presence of other electron donors, including hydrogen. The variant of Sulfurospirillum detected within this study increased proportionally over time to become a dominant member of the microbial community within just 30 days and was detected only within the labeled fraction of the sediment nucleic acids, demonstrating the utilization of acetate-derived 13C.
This study provides a unique insight into the direct influence of exogenous carbon sources on microbial diversity and arsenic speciation within sediments containing naturally elevated levels of arsenic and is among the first to use SIP within the complex environment of natural sediments. A direct link between inputs of carbon and the increased prevalence of organisms which actively convert As(V) to the potentially more mobile As(III) has been identified. An increase in the presence of genes associated with As(V) respiratory reductases suggests that these may provide a reliable marker for dissimilatory As(V) reduction within complex environmental communities. That these reductases have been shown to be comprised of acetate-derived 13C supports the hypothesis that inputs of exogenous organic matter can directly stimulate the growth and activity of bacteria capable of increasing the mobility of arsenic within naturally contaminated aquifer environments, such as those bordering the city of Phnom Penh, Cambodia. Indeed, recent studies of Bangladeshi sediments (10, 27) have suggested biogeochemical processes including the mobilization of arsenic to be driven by inflows of radiocarbon young carbon (e.g., with 14C signatures from bomb testing). Further research is now required to confirm the role of As(V)-respiring prokaryotes, such as those detected in this study, in mobilizing arsenic within aquifers in situ and to understand the factors that control their metabolism in aquifers, including critical biogeochemical controls on the delivery of electron donors and other limiting nutrients.
Supplementary Material
Acknowledgments
Financial support from EPSRC (grant GR/S30207/01) and NERC (grant NE/D013291/1) is acknowledged.
XANES analysis was carried out under CCLRC Daresbury beamtime awards 42/217, 43/322, and 44/258 to D.A.P. and A.G.G. We thank Bob Bilsborrow and John Charnock (CCLRC Daresbury) for assistance with XANES data acquisition and analysis, Paul Lythgoe and Alastair Bewsher for chemical analysis, and Vibol Long, Richard Pattrick, Helen Rowland, Roy Wogelius, and Som Yen for assistance with the drilling for and the collection of samples in the field.
Footnotes
Published ahead of print on 17 November 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Afkar, E., J. Lisak, C. Saltikov, P. Basu, R. S. Oremland, and J. F. Stolz. 2003. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Akai, J., K. Izumi, H. Fukuhara, H. Masuda, S. Nakano, T. Yoshimura, H. Ohfuji, H. M. Anawar, and K. Akai. 2004. Mineralogical and geomicrobiological investigations on groundwater arsenic enrichment in Bangladesh. Appl. Geochem. 19:215-230. [Google Scholar]
- 3.Anawar, H. M., J. Akai, K. Komaki, H. Terao, T. Yoshioka, T. Ishizuka, S. Safiullah, and K. Kato. 2003. Geochemical occurrence of arsenic in groundwater of Bangladesh: sources and mobilization sourced. J. Geochem. Explor. 77:109-131. [Google Scholar]
- 4.Berg, M., H. C. Tran, T. C. Nguyen, H. V. Pham, R. Schertenleib, and W. Giger. 2001. Arsenic contamination of groundwater and drinking water in Vietnam: a human health threat. Environ. Sci. Technol. 35:2621-2626. [DOI] [PubMed] [Google Scholar]
- 5.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graff, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 6.Carrasco, J. A., P. Armaric, E. Pajuelo, A. Burgos, B. A. Caviedes, R. Lopez, M. A. Chamber, and A. J. Palomares. 2005. Isolation and characterisation of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcollar pyrite mine. Soil Biol. Biochem. 37:1131-1140. [Google Scholar]
- 7.Chakraborti, D., M. Rahman, K. Paul, U. K. Chowdhury, M. K. Sengupta, D. Lodh, C. R. Chanda, K. C. Saha, and S. C. Mukherjee. 2002. Arsenic calamity in the Indian subcontinent. What lessons have been learned? Talanta 59:3-22. [DOI] [PubMed] [Google Scholar]
- 8.Dent, A., and J. F. W. Mosselmans. 1992. A guide to EXBACK, EXCALIB and EXCURV92. Daresbury Laboratory, Warrington, United Kingdom.
- 9.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey, C. F., C. H. Swartz, A. B. M. Badruzzaman, N. Keon-Blute, W. Yu, M. Ashraf Ali, J. Jay, R. Beckie, V. Niedan, D. Brabander, P. P. Oates, K. N. Ashfaque, S. Islam, H. F. Hemond, and A. M. Feroze. 2002. Arsenic mobility and groundwater extraction in Bangladesh. Science 298:1602-1606. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, D. E., K. T. Finneran, and D. R. Lovley. 2002. Enrichment of Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horneman, A., A. van Geen, D. V. Kent, P. E. Mathe, Y. Zheng, R. K. Dhar, S. O'Connell, M. A. Hoque, Z. Aziz, M. Shamsudduha, A. A. Seddique, and K. M. Ahmed. 2004. Decoupling of As and Fe release to Bangladesh groundwater under reducing conditions. Part 1. Evidence from sediment profiles. Geochim. Cosmochim. Acta 68:3459-3473. [Google Scholar]
- 13.Islam, F. S., A. G. Gault, C. Boothman, D. A. Polya, J. M. Charnock, D. Chatterjee, and J. R. Lloyd. 2004. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68-71. [DOI] [PubMed] [Google Scholar]
- 14.Islam, F. S., R. L. Pederick, A. G. Gault, L. K. Adams, D. A. Polya, J. M. Charnock, and J. R. Lloyd. 2005. Interactions between the Fe(III)-reducing bacterium Geobacter sulfurreducens and arsenate, and capture of the biogenic metalloid by biogenic Fe(II). Appl. Environ. Microbiol. 71:8642-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krafft, T., and J. M. Macy. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255:647-653. [DOI] [PubMed] [Google Scholar]
- 16.Lindberg, A.-L., W. Goessler, E. Gurzau, K. Koppova, P. Rudnai, R. Kumar, T. Fletcher, G. Leonardi, K. Slotova, E. Gheorghiu, and M. Vahter. 2006. Arsenic exposure in Hungary, Romania and Slovakia. J. Environ. Monit. 8:203-208. [DOI] [PubMed] [Google Scholar]
- 17.Lovley, D. R., and E. R. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lueders, T., M. Manefield, and M. W. Freidrich. 2004. Enhanced sensitivity of DNA- and RNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 19.Macy, J. M., J. M. Santini, B. V. Pauling, A. H. O'Neill, and L. I. Sly. 2000. Two new arsenate/sulfate-reducing bacteria: mechanisms of arsenate reduction. Arch. Microbiol. 173:49-57. [DOI] [PubMed] [Google Scholar]
- 20.Malasarn, D., C. W. Saltikov, K. M. Campbell, J. M. Santini, J. G. Hering, and D. K. Newman. 2004. arrA is a reliable marker for As(V) respiration. Science 306:455. [DOI] [PubMed] [Google Scholar]
- 21.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McArthur, J. M., D. M. Banerjee, K. A. Hudson-Edwards, R. Mishra, R. Purohit, P. Ravenscroft, A. Cronin, R. Howarth, A. Chatterjee, T. Talukder, D. Lowry, S. Houghton, and D. K. Chadha. 2004. Natural organic matter in sedimentary basins and its relation to arsenic in anoxic groundwater: the example of West Bengal and its worldwide implications. Appl. Geochem. 19:1255-1293. [Google Scholar]
- 23.McArthur, J. M., P. Ravenscroft, S. Safiullah, and M. Thirlwall. 2001. Arsenic in groundwater: testing pollution mechanisms for sedimentary aquifers in Bangladesh. Water Resour. Res. 37:109-117. [Google Scholar]
- 24.Newman, D. K., E. K. Kennedy, J. D. Coates, D. Ahmann, D. J. Ellis, D. R. Lovley, and F. M. M. Morrel. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168:380-388. [DOI] [PubMed] [Google Scholar]
- 25.Oremland, R. S., and J. F. Stolz. 2005. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 13:45-49. [DOI] [PubMed] [Google Scholar]
- 26.Oremland, R. S., and J. F. Stolz. 2003. The ecology of arsenic. Science 300:939-944. [DOI] [PubMed] [Google Scholar]
- 27.Polizzotto, M. L., C. F. Harvey, S. R. Sutton, and S. Fendorf. 2005. Processes conducive to the release and transport of arsenic into aquifers of Bangladesh. Proc. Natl. Acad. Sci. USA 102:18819-18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polya, D. A., A. G. Gault, N. J. Bourne, P. R. Lythgoe, and D. A. Cooke. 2003. Coupled HPLC-ICP-MS analysis indicates highly hazardous concentrations of dissolved arsenic species are present in Cambodian wellwaters. R. Soc. Chem. Spec. Publ. 288:127-140. [Google Scholar]
- 29.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 30.Rowland, H., A. G. Gault, J. M. Charnock, and D. A. Polya. 2005. Preservation and XANES determination of the oxidation state of solid-phase arsenic in shallow sedimentary aquifers in Bengal and Cambodia. Mineral. Mag. 69:825-839. [Google Scholar]
- 31.Rowland, H. A. L., D. A. Polya, A. G. Gault, J. M. Charnock, R. L. Pederick, and J. R. Lloyd. 2004. Microcosm studies of microbially mediated arsenic release from contrasting Cambodian sediments. Geochim. Cosmochim. Acta 68:A390. [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Smedley, P. L., and D. G. Kinniburgh. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17:517-568. [Google Scholar]
- 34.Smith, A. H., E. O. Lingas, and M. Rahman. 2000. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. W. H. O. 78:1093-1103. [PMC free article] [PubMed] [Google Scholar]
- 35.Stolz, J. F., D. J. Ellis, J. S. Blum, D. Ahmann, D. R. Lovley, and R. S. Oremland. 1999. Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the epsilon proteobacteria. Int. J. Syst. Bacteriol. 49:1177-1180. [DOI] [PubMed] [Google Scholar]
- 36.Stuben, D., Z. Berner, D. Chandrasekharam, and J. Karmakar. 2003. Arsenic enrichment in groundwater of West Bengal, India: geochemical evidence for mobilisation of As under reducing conditions. Appl. Geochem. 18:1417-1434. [Google Scholar]
- 37.Summers, A. O., and S. Silver. 1978. Microbial transformations of metals. Annu. Rev. Microbiol. 32:637-672. [DOI] [PubMed] [Google Scholar]
- 38.Sun, Y., E. A. Polishchuk, U. Radoja, and W. R. Cullen. 2004. Identification and quantification of arsC genes in environmental samples by using real-time PCR. J. Microbiol. Methods 58:335-349. [DOI] [PubMed] [Google Scholar]
- 39.Tamaki, S., and W. T. Frakenberger, Jr. 1992. Environmental biochemistry of arsenic. Rev. Environ. Contam. Toxicol. 124:79-110. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 42.van der Gast, C. J., C. J. Knowles, M. A. Wright, and I. P. Thompson. 2001. Identification and characterisation of bacterial populations of an in-use metal-working fluid by phenotypic and genotypic methodology. Int. Biodeterior. Biodegrad. 47:113-123. [Google Scholar]
- 43.Van Geen, A., Y. Zheng, M. Stute, and K. M. Ahmed. 2003. Comment on “Arsenic mobility and groundwater extraction in Bangladesh” (II). Science 300:584. [DOI] [PubMed] [Google Scholar]
- 44.Weldon, J., and J. MacRae. 2006. Correlations between arsenic in Maine groundwater and microbial populations as determined by fluorescence in situ hybridisation. Chemosphere 63:440-448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.