Abstract
The cabbage looper, Trichoplusia ni, is one of only two insect species that have evolved resistance to Bacillus thuringiensis in agricultural situations. The trait of resistance to B. thuringiensis toxin Cry1Ac from a greenhouse-evolved resistant population of T. ni was introgressed into a highly inbred susceptible laboratory strain. The resulting introgression strain, GLEN-Cry1Ac-BCS, and its nearly isogenic susceptible strain were subjected to comparative genetic and biochemical studies to determine the mechanism of resistance. Results showed that midgut proteases, hemolymph melanization activity, and midgut esterase were not altered in the GLEN-Cry1Ac-BCS strain. The pattern of cross-resistance of the GLEN-Cry1Ac-BCS strain to 11 B. thuringiensis Cry toxins showed a correlation of the resistance with the Cry1Ab/Cry1Ac binding site in T. ni. This cross-resistance pattern is different from that found in a previously reported laboratory-selected Cry1Ab-resistant T. ni strain, evidently indicating that the greenhouse-evolved resistance involves a mechanism different from the laboratory-selected resistance. Determination of specific binding of B. thuringiensis toxins Cry1Ab and Cry1Ac to the midgut brush border membranes confirmed the loss of midgut binding to Cry1Ab and Cry1Ac in the resistant larvae. The loss of midgut binding to Cry1Ab/Cry1Ac is inherited as a recessive trait, which is consistent with the recessive inheritance of Cry1Ab/Cry1Ac resistance in this greenhouse-derived T. ni population. Therefore, it is concluded that the mechanism for the greenhouse-evolved Cry1Ac resistance in T. ni is an alteration affecting the binding of Cry1Ab and Cry1Ac to the Cry1Ab/Cry1Ac binding site in the midgut.
Bacillus thuringiensis-based biopesticides have specific spectra of target insect species and are environmentally benign—safe to humans, vertebrate animals, and beneficial arthropods (8, 23). The insecticidal activities of B. thuringiensis are primarily attributed to the proteinaceous Cry toxins. Development of insect resistance to B. thuringiensis toxins threatens the sustained successful application of B. thuringiensis-based biological control tactics (10, 35). Numerous laboratory experiments to select various insect species with B. thuringiensis toxins have shown the high potential for development of B. thuringiensis resistance in insect populations after exposure to B. thuringiensis toxins (10, 37). Currently, populations of two agriculturally important insect species, the diamondback moth (Plutella xylostella) and the cabbage looper (Trichoplusia ni), have evolved B. thuringiensis resistance in open fields or in commercial production greenhouses under selective pressure with application of B. thuringiensis-based biopesticides (20, 32, 34). Effective management of B. thuringiensis resistance development in insect populations requires a thorough understanding of the resistance mechanisms. Understanding B. thuringiensis resistance mechanisms will also provide new insight into the mode of action of B. thuringiensis toxins, which is not yet fully understood.
The pathogenesis of B. thuringiensis toxins in insects involves multiple sequential steps (31). Alteration of any of the steps may affect the toxicity of B. thuringiensis toxins in insects and, consequently, can be potentially involved in B. thuringiensis resistance. Studies of B. thuringiensis resistance in insect strains that were selected with B. thuringiensis toxins in the laboratory have indeed indicated that mechanisms for B. thuringiensis resistance in insects can be complex and diverse (10, 14). Different resistant populations of the same insect species from different origins or selected in different laboratories may have distinctly different mechanisms of resistance to the same B. thuringiensis Cry toxin (1, 15, 25). Alteration of midgut proteases which are critically involved in solubilization and proteolytic processing of Cry proteins in the insect midgut has been found in several B. thuringiensis-resistant insect strains generated by laboratory selections (24, 29). Modification of midgut binding sites for B. thuringiensis toxins, which results in reduced binding of the B. thuringiensis toxins to the midgut targets, has been reported in several B. thuringiensis-resistant insect strains and appears to be a major mechanism for insect resistance to Cry1A toxins (9, 36). In addition, other B. thuringiensis resistance mechanisms have been proposed, including retention of the B. thuringiensis toxin by the midgut peritrophic membrane (16), aggregation of B. thuringiensis toxin proteins by the midgut esterase (15), elevated melanization activity of the hemolymph and midgut cells (25, 30), increased rate of repair or replacement of affected epithelial cells (26), and possibly increased antioxidation activities in B. thuringiensis-resistant insects (5). Therefore, mechanisms for B. thuringiensis resistance in insects can be multifaceted.
Studies of B. thuringiensis resistance by selection of insect populations with B. thuringiensis toxins under laboratory conditions have facilitated studies of mechanisms of B. thuringiensis resistance (11, 29), which has built the main body of the current understanding of the resistance mechanisms. However, B. thuringiensis resistance developed in field insect populations in agricultural practices may involve mechanisms different from those found from laboratory-selected resistance (2). To date, the resistance mechanisms in insects that have developed B. thuringiensis resistance in agricultural practice are not known. The cabbage looper, T. ni, has developed B. thuringiensis resistance in commercial-production greenhouses where B. thuringiensis formulations were frequently applied (20). The trait of resistance to Cry1Ac in the greenhouse T. ni population has been determined to be monogenic, autosomal, and incompletely recessive (22). In this paper, we report the characterization and identification of the biochemical mechanism for the Cry1Ac resistance evolved in a greenhouse T. ni population.
MATERIALS AND METHODS
Insects.
An inbred laboratory colony of T. ni (22) was used as the susceptible strain. This colony has been maintained in the laboratory without exposure to B. thuringiensis for over 20 years. The T. ni Cry1Ac-resistant strain GLEN-Cry1Ac was originally from a B. thuringiensis-resistant population found in commercial greenhouses (20) and has been described by Kain et al. (22). T. ni larvae were reared on a wheat germ-based artificial diet at 27°C with 50% humidity and a photoperiod 16 h of light and 8 h of darkness.
Generation of a Cry1Ac-resistant T. ni introgression strain nearly isogenic to the susceptible laboratory strain.
To introgress the Cry1Ac resistance trait from the greenhouse-derived GLEN-Cry1Ac strain to the susceptible laboratory strain, the GLEN-Cry1Ac strain was crossed with the susceptible laboratory strain and progenies in generations F2, F3, and F4 were selected by feeding on a surface-contaminated artificial diet with 4 μg Cry1Ac/cm2 (2 ml of 100-ppm Cry1Ac spread on the diet surface which is ca. 50 cm2) for F2 larvae and 20 μg Cry1Ac/cm2 (2 ml of 500-ppm Cry1Ac spread on the diet surface) for F3 and F4 larvae. The Cry1Ac-selected hybrid strain was then backcrossed with the susceptible laboratory strain, and their progenies of F2, F3, and F4 were selected again with Cry1Ac as described above. This backcross and the selection procedures were repeated. The backcross line obtained was designated GLEN-Cry1Ac-BCS, a strain nearly isogenic to the susceptible laboratory strain. The T. ni larvae used in this study were from the backcross strain after three or four backcrosses.
To confirm that the Cry1Ac resistance gene in GLEN-Cry1Ac-BCS was introgressed from the original B. thuringiensis-resistant greenhouse population, a genetic complementation analysis (38) was performed using the GLEN-Cry1Ac-BCS strain and Cry1Ac-resistant individuals from a new collection of T. ni larvae (collected in 2004) from the same greenhouse where the GLEN strain of T. ni was initially collected in 2001 (20, 22). The newly collected T. ni larvae were maintained on artificial diet without exposure to B. thuringiensis toxins for four generations, and neonate larvae from the fifth generation were placed on Cry1Ac-expressing transgenic broccoli for one generation to eliminate susceptible individuals (22). The T. ni larvae that survived on the transgenic broccoli were collected and crossed with the susceptible laboratory strain and the backcross strain GLEN-Cry1Ac-BCS. The progenies (F1) from these crosses were placed on the transgenic broccoli plants to observe their susceptibilities to the transgenic broccoli.
Bioassays.
A diet overlay assaying method was used to determine the susceptibility of T. ni larvae to Cry1Ac and other Cry toxins as described by Kain et al. (22). For neonate bioassays, an aliquot of 0.2 ml of a Cry toxin was applied to the surface (≈7 cm2) of 5 ml artificial diet in a 30-ml insect-rearing cup. Each bioassay included four to six concentrations of a Cry toxin, which resulted in 10 to 90% larval mortality, and five replications for each concentration. Ten neonate larvae were placed in each assaying cup. Larval growth inhibition (defined as inhibition if larvae did not enter second instar within 4 days) by the Cry toxins was scored after 4 days of feeding on the Cry toxin-treated diet. Doses for inhibition of 50% of the individuals were calculated based on probit analysis, and statistical analyses were performed with the aid of the statistical software packages POLO and SAS as described by Kain et al. (22). The procedures for fifth-instar larva bioassays were similar to the method described for neonate bioassays, except two early-molted fifth-instar larvae were placed in a 30-ml cup with a series of concentrations of Cry1Ac overlaid on diet. Fifteen cups (30 larvae in total) were used for each Cry1Ac concentration, and larval mortalities were observed after 7 days of rearing the larvae on Cry1Ac-treated diet. The sources of B. thuringiensis Cry toxins used for the bioassays were the same as described earlier by Zhao et al. (43), except that Cry1Ac was prepared from B. thuringiensis subsp. kurstaki strain HD-73 (22) and Cry1Ab toxin was purified from inclusion bodies produced in the Escherichia coli PBD-140 strain (Ruud A. de Maagd, Plant Research International B.V., Wageningen, The Netherlands). To prepare Cry1Ab, E. coli PBD-140 was cultured at 37°C in TB medium (17 mM KH2PO4, 72 mM K2HPO4, 10.8 g/liter tryptone, 21.6 g/liter yeast extract, 0.36% glycerol), supplemented with 50 μg/ml of ampicillin. The E. coli was harvested by centrifugation and stored at −20°C after lyophilization. For isolation of Cry1Ab inclusion bodies, a 1-g E. coli pellet was dissolved in a lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 100 mM NaCl, 25 μg/ml lysozyme) at room temperature for 20 min with gentle shaking, followed by addition of 16 mg deoxycholic acid and another incubation for 10 min at 37°C. Then, 0.8 mg DNase I was added, and the incubation was continued for an additional 30 min at 37°C with vigorous shaking. The inclusion bodies of the protoxin were pelleted by centrifugation at 40,000 × g for 10 min, followed by two washes with washing buffer (20 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1 M NaCl) and a final wash with phosphate-buffered saline (PBS; 1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4). The purified inclusion bodies were solubilized by incubation in the solubilization buffer (50 mM sodium carbonate, pH 10, 100 mM NaCl, 10 mM dithiothreitol) for 1 h at 37°C with constant shaking, and solubilized protoxin was recovered in the supernatant after centrifugation at 40,000 × g for 15 min.
Hemolymph melanization assays.
To examine whether hemolymph melanization activity is elevated in the resistant larvae, in vitro hemolymph plasma melanization assays were conducted in accordance with the method described by Rahman et al. (30). Forty microliters of larval hemolymph was collected from a proleg of a fifth-instar larva and immediately diluted in 200 μl ice-cold PBS. Hemocytes were removed by centrifugation at 16,000 × g for 4 min. One hundred microliters of the resulting supernatant was mixed in 100 μl of PBS in a 96-well microtiter plate, and the optical density of the hemolymph plasma suspension was recorded with a microtiter plate reader at 490 nm for 90 min to determine the kinetics of the melanization reaction.
Midgut protease activity assays.
Mid-fifth-instar larvae were immobilized on ice for several minutes and dissected to isolate the complete midgut without loss of its contents. The isolated midgut was weighed and individually stored at −20°C. Midgut homogenates were prepared by thorough homogenization of the midgut in a 1.5-ml microcentrifuge tube and suspended in deionized water to constitute a 10% (wt/vol) suspension.
Total protease activity of the midgut was determined at pH 8 and pH 11 using azocasein (Sigma, St. Louis, MO) as the substrate as described by Broadway (4). Briefly, 50 μl of midgut homogenate was mixed with 150 μl 1% azocasein in 50 mM Tris-HCl buffer (pH 8) or 20 μl of midgut homogenate was mixed with 150 μl 1% azocasein in 50 mM NaHCO3-Na2CO3 buffer (pH 11) and incubated at 28°C for 2 h. The digestion was stopped, and undigested azocasein was precipitated by addition of an equal volume of 10% trichloroacetic acid, followed by incubation at room temperature for 1 h. The precipitated proteins were removed by centrifugation at 16,000 × g for 10 min. The supernatant was collected, and its optical absorbance at 450 nm was measured after mixing with an equal volume of 1 M NaOH.
Midgut elastase activity was assayed with N-succinyl-l-alanyl-l-alanyl-l-alanine-p-nitroanilide (Suc-Ala3-pNA; Sigma) as a chromogenic substrate. For activity assays at pH 8.0, 20 μl of midgut homogenate was mixed with 480 μl of 50 mM Tris-HCl buffer (pH 8.0) and 10 μl of 55 mM Suc-Ala3-pNA. For assays at pH 11, 10 μl of midgut homogenate was mixed with 490 μl of 50 mM NaHCO3-Na2CO3 buffer (pH 11) and 10 μl of 55 mM Suc-Ala3-pNA. The peptidolytic reaction was immediately monitored by recording the increase of optical absorbance at 410 nm at 28°C for 60 min. Trypsin activity in the midgut homogenate was determined by mixing 10 μl of midgut homogenate in 3 ml of 1 mM Na-benzoyl-l-arginine p-nitroanilide (BApNA; Sigma) in 50 mM Tris-HCl (pH 8.0) or by mixing 3 μl of midgut homogenate in 3 ml of 1 mM BApNA in 50 mM NaHCO3-Na2CO3 buffer (pH 11). The enzymatic reaction at 28°C was monitored by recording the optical absorbance at 405 nm for 60 min. Determination of chymotrypsin activity in the midgut homogenates was similar to the method used for the trypsin assays except 1 mM N-succinyl-Ala-Ala-Pro-phenylalanine p-nitroanilide (Sigma) was used as the substrate and 3 μl of 10-fold-diluted midgut homogenate was used for the assays. Aminopeptidase activity in the midgut homogenate was determined using leucine p-nitroanilide (Sigma) as the chromogenic substrate as described by Wang et al. (40). For all the protease assays, samples from five larvae for each strain were individually assayed and the activity for each sample was calculated using the data points in the linear portion of the initial reaction curve, followed by statistical significance analysis.
Zymogram analysis of midgut proteinases.
Midgut protease zymogram analysis was performed using the method developed by Garcia-Carreno et al. (12) with modifications. Briefly, 1 μl of 10% midgut homogenate was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis with a 12.5% SDS-PAGE gel to separate the midgut proteins, followed by a washing of the gel with 2% Triton X-100 for 15 min to remove the SDS from the gel. The gel was then incubated with 2% casein (Sigma) in 50 mM Tris-HCl (pH 8.0) for 3 h at room temperature, followed by Coomassie blue staining after a brief rinse of the gel with deionized water to visualize the protease activities in the gel.
Digestion of Cry1Ac protoxin with T. ni larval midgut fluid.
T. ni larval midgut fluid from both susceptible and resistant larvae was collected from fifth-instar larvae by stimulating the larval mouth parts to induce regurgitation. A 10-μl aliquot of Cry1Ac protoxin (5 μg/μl) was mixed with 10 μl of midgut fluid solution in a series of dilutions in 50 mM NaHCO3-Na2CO3 buffer (pH 10.5) and incubated at 26°C for 60 min. The incubation was stopped by addition of 20 μl 2× SDS-PAGE sample buffer followed by heating at 100°C for 5 min. The digestion of Cry1Ac protoxin for conversion to active toxin by midgut fluid was examined by SDS-PAGE analysis, followed by Coomassie blue staining.
Midgut esterase analysis.
Midgut esterases in the resistant and susceptible T. ni larvae were comparatively analyzed by total-activity assays and isozyme profile analysis. Midgut extracts were prepared as described above for midgut protease analysis except that the midgut tissue was homogenized in 0.1% Triton X-100. The midgut homogenates were then clarified by centrifugation at 16,000 × g for 10 min, and the supernatants were collected for esterase analysis. Esterase activities were determined by mixing 5 μl midgut extract in 3 ml reaction solution containing 2.5 mg/ml of Fast Blue RR and 0.1 mg/ml α-naphthyl acetate (Sigma) in 0.1 M sodium phosphate buffer (pH 6.5), followed by monitoring the increase of optical absorbance at a wavelength of 450 nm for 5 min at 26°C (33). The enzymatic activities were calculated as units of optical absorbance increase per minute using the linear part of the recorded reaction curve.
For midgut esterase isozyme profiling, proteins from 10 μl of midgut extract were separated by 7% PAGE, followed by two washings of the gel with 0.1 M sodium phosphate buffer (pH 6.5) for 10 min. The esterase isozyme activities in the gels were visualized by incubation in the reaction solution containing 0.75 mg/ml of Fast Blue RR and 0.2 mg/ml of α-naphthyl acetate in 0.1 M sodium phosphate buffer (pH 6.5). The colorimetric reaction was stopped by fixation of the gel in 5% acetic acid and 10% methanol.
Preparation of midgut BBMVs.
Midgut brush border membrane vesicle (BBMV) preparations were made from the susceptible and resistant strains and the progeny (F1) from the cross of the susceptible (female) and resistant (male) strains. Mid-fifth-instar larvae were immobilized on ice for several min and dissected in cold MET buffer (250 mM mannitol, 17 mM Tris-HCl, 5 mM EGTA, pH 7.5) to isolate the midgut epithelium, free of any attached tissues (fat body and Malpighian tubules), and the peritrophic membrane with food contents. The isolated midgut epithelium was thoroughly rinsed with cold MET buffer and quickly frozen in liquid nitrogen after removal of excess buffer from the dissected tissue. The frozen midgut tissues were shipped on dry ice from Cornell University to the University of Valencia and stored at −80°C before use. Midgut BBMVs were prepared by the differential-magnesium-precipitation method (41) and stored at −80°C after being frozen in liquid nitrogen. BBMV preparations were quantified by the BBMV protein concentration determined with the Bradford method (3).
BBMV-associated aminopeptidase and alkaline phosphatase activities were determined using an aliquot of each BBMV preparation containing 200 ng proteins. Alkaline phosphatase activity was determined using the alkaline phosphatase (DEA) kit from Biosystems (Barcelona, Spain) by following the manufacturer's instructions. Leucine aminopeptidase activity was measured using l-leucil-β-naphthylamide as the substrate as described by Hernández et al. (17).
Preparation, purification, and labeling of Cry1Ac and Cry1Ab for binding analysis.
Cry1Ac toxin was purified from a recombinant B. thuringiensis strain (EG11070) as described by Estela et al. (7). Cry1Ab was prepared as described above. Activated Cry toxins were prepared by digestion with trypsin, followed by dialysis overnight against 20 mM Tris-HCl buffer (pH 8.6) and finally purified by ion-exchange chromatography as described by Estela et al. (7). The purity and quantity of the toxins were examined by SDS-PAGE analysis and consequent densitometric analysis of the toxin protein bands in the gel using bovine serum albumin as a standard. To label the Cry1Ac and Cry1Ab with 125I, 20 μg of each toxin was incubated with 0.25 mCi of Na125I (Nucliber, Madrid, Spain) using chloramine-T (Sigma) as described by Van Rie et al. (39). The specific radioactivities obtained were 110 and 5.1 mCi/mg protein for Cry1Ac and Cry1Ab, respectively.
Binding of 125I-labeled Cry1Ac and Cry1Ab to T. ni midgut BBMVs.
Qualitative binding experiments were performed as previously described by Estada and Ferré (6). Increasing concentrations of BBMV were incubated with 6.3 pM of 125I-Cry1Ac or 130 pM of 125I-Cry1Ab in 0.1 ml binding buffer (PBS-0.1% bovine serum albumin) for 1 h. The BBMVs were then pelleted by centrifugation at 16,000 × g for 10 min and washed twice with binding buffer. The final radioactivity remaining in the BBMV pellets was measured in a 1282 Compugamma CS gamma counter (LKB, Pharmacia). Nonspecific binding was determined in the presence of an excess of unlabeled protein (300 nM). The nonspecific binding of Cry1Ac and Cry1Ab to the BBMVs from the susceptible and F1 larvae accounted for 11.4% and 8.0% and 6.4% and 10.5%, respectively, of the total radioactivity used in the assay. The amount of specific binding was calculated by subtraction of nonspecific binding from the total binding, and the binding experiments were replicated at least twice.
Quantitative binding analysis was performed with BBMVs from susceptible and F1 insects, by means of homologous competition with Cry1Ac. BBMVs (0.05 mg/ml) and a fixed amount of 125I-labeled Cry1Ac were incubated with increasing concentrations of unlabeled Cry1Ac. Data were analyzed with the LIGAND computer program (28).
RESULTS
Introgression of the Cry1Ac resistance trait from the GLEN-Cry1Ac strain to the susceptible laboratory strain.
The Cry1Ac resistance was introgressed into the susceptible laboratory strain by crossing and backcrosses with the susceptible strain. The resulting introgression strain, GLEN-Cry1Ac-BCS, nearly isogenic to the susceptible strain, showed a level of resistance both to Cry1Ac on artificial diet (Table 1) and to Cry1Ac-expressing transgenic broccoli plants (Table 2) similar to that shown by the parental strain, GLEN-Cry1Ac.
TABLE 1.
Resistance of T. ni GLEN-Cry1Ac-BCS strain to Cry1Ac protoxin
| Straina | Larvae | No. of larvae | Slope (SE) | LC50f (95% CId) (mg/liter) | χ2 (dfe) | RRb | RLc |
|---|---|---|---|---|---|---|---|
| SS | Neonate | 160 | 2.44 (0.38) | 0.067 (0.052-0.083) | 0.94 (2) | 1 | 1 |
| BCS | 200 | 2.06 (0.38) | 58.3 (42.1-75.5) | 0.94 (2) | 870 | 1 | |
| SS | Fifth instar | 120 | 2.40 (0.43) | 1.31 (0.99-1.72) | 0.14 (2) | 1 | 19.6 |
| BCS | 120 | 3.55 (0.52) | 3,070 (2,546-3,731) | 1.60 (2) | 2,344 | 52.7 |
SS, susceptible laboratory strain; BCS, GLEN-Cry1Ac-BCS strain.
RR, resistance ratio (LC50 for BCS/LC50 for SS).
RL, relative LC50 (LC50 for fifth-instar larvae/LC50 for neonate larvae).
CI, confidence interval.
df, degrees of freedom.
LC50, 50% lethal concentration.
TABLE 2.
Survival of T. ni larvae from different strains on Cry1Ac-expressing transgenic broccoli plants
| Strain or crossa | No. of larvae | No. of survivors | % Survival | % Adjusted survivalb |
|---|---|---|---|---|
| SS | 60 | 0 | 0 | 0 |
| GLEN2004 | 40 | 7 | 17.5 | 18.4 |
| GLEN2004-Sel | 60 | 42 | 70.0 | 82.4 |
| BCS | 50 | 46 | 92.0 | 96.0 |
| BCS × SS | 40 | 0 | 0 | 0 |
| BCS × GLEN2004-Sel | 60 | 41 | 68.3 | 80.4 |
SS, susceptible laboratory strain; GLEN2004, greenhouse population collected in 2004; GLEN2004-Sel, F1 larvae from the GLEN2004 individuals that survived on broccoli plants containing B. thuringiensis; BCS, larvae of the GLEN-Cry1Ac-BCS strain; BCS × SS, F1 larvae from cross between GLEN-Cry1Ac-BCS and susceptible strains; BCS × GLEN2004-Sel, F1 larvae from cross between GLEN-Cry1Ac-BCS and GLEN2004-Sel.
Percent survival on broccoli containing B. thuringiensis/percent survival on broccoli not containing B. thuringiensis.
About 18% of the T. ni larvae from the greenhouse population collected in 2004 (GLEN2004) showed resistance to Cry1Ac and survived on Cry1Ac-expressing transgenic broccoli plants (Table 2). Those individuals that survived, referred to as GLEN2004-Sel in Table 2, were used for the genetic complementation test to verify that the Cry1Ac resistance gene in the introgression strain GLEN-Cry1Ac-BCS was the same as that conferring Cry1Ac resistance in the B. thuringiensis-resistant population in the greenhouse (Table 2). Neonate larvae of the strains GLEN-Cry1Ac-BCS and GLEN2004-Sel could feed on Cry1Ac-expressing transgenic broccoli plants with a survival rate of 96% and 82%, respectively. The progenies (F1) from the cross between GLEN-Cry1Ac-BCS and the susceptible laboratory strain exhibited 100% mortality, whereas the progenies (F1) from the cross between GLEN-Cry1Ac-BCS and GLEN2004-Sel showed a survival rate of 80% on the transgenic broccoli plants. These results demonstrated that the strains GLEN-Cry1Ac-BCS and GLEN2004-Sel could not rescue the susceptible phenotype with each other. Therefore, the resistance gene in GLEN-Cry1Ac-BCS is the same as the one conferring Cry1Ac resistance in the resistant greenhouse population. The introgression strain GLEN-Cry1Ac-BCS, nearly isogenic to the susceptible laboratory strain, is an appropriate strain of T. ni for studying Cry1Ac resistance selected in vegetable production greenhouses.
Pattern of cross-resistance of T. ni GLEN-Cry1Ac-BCS larvae to various B. thuringiensis Cry toxins.
Comparative bioassays of neonate larvae of the GLEN-Cry1Ac-BCS strain and its nearly isogenic susceptible strain for their susceptibilities to 11 Cry toxins showed that the GLEN-Cry1Ac-BCS larvae were highly resistant to Cry1Ab and Cry1Ac, with resistance levels of 938- and 870-fold, respectively, and were moderately resistant to Cry1Aa, Cry1E, and Cry1F, with resistance levels ranging from 6.7- to 8.2-fold (Table 3). The GLEN-Cry1Ac-BCS larvae also exhibited reduced susceptibilities to the rest of the tested Cry toxins, but the levels of resistance, ranging from two- to threefold, were minimal or statistically insignificant. This cross-resistance pattern is in agreement with the known pattern of Cry toxin binding to their shared midgut binding sites in T. ni (6, 19).
TABLE 3.
Resistance of GLEN-Cry1Ac-BCS larvae to different B. thuringiensis Cry protoxins
| Protoxin | Straina | n | Slope (SE) | LC50e (95%CIc) (mg/liter) | χ2 (dfd) | RRb |
|---|---|---|---|---|---|---|
| Cry1Aa | SS | 200 | 2.74 (0.32) | 0.174 (0.144-0.209) | 1.58 (3) | 1 |
| BCS | 240 | 1.88 (0.21) | 1.43 (1.14-1.83) | 1.20 (4) | 8.2 | |
| Cry1Ab | SS | 160 | 1.79 (0.24) | 0.664 (0.492-0.910) | 1.72 (2) | 1 |
| BCS | 200 | 2.34 (0.29) | 623 (512-765) | 5.21 (3) | 938 | |
| Cry1Ac | SS | 160 | 2.44 (0.38) | 0.067 (0.052-0.083) | 0.94 (2) | 1 |
| BCS | 200 | 2.06 (0.38) | 58.3 (42.1-75.5) | 0.94 (2) | 870 | |
| Cry1Bb | SS | 200 | 2.10 (0.32) | 2.43 (1.83-3.15) | 0.55 (3) | 1 |
| BCS | 200 | 2.29 (0.31) | 7.92 (5.01-11.7) | 3.20 (3) | 3.3 | |
| Cry1C | SS | 200 | 1.57 (0.24) | 0.345 (0.249-0.455) | 1.86 (3) | 1 |
| BCS | 200 | 1.50 (0.20) | 0.846 (0.617-1.11) | 2.35 (3) | 2.5 | |
| Cry1D | SS | 200 | 1.58 (0.26) | 2.03 (1.02-3.67) | 3.28 (3) | 1 |
| BCS | 200 | 1.49 (0.24) | 4.79 (3.57-6.62) | 1.57 (3) | 2.4 | |
| Cry1E | SS | 240 | 1.78 (0.20) | 13.8 (10.8-17.6) | 1.72 (4) | 1 |
| BCS | 200 | 1.34 (0.17) | 91.8 (32.7-262) | 6.71 (3) | 6.7 | |
| Cry1F | SS | 280 | 1.38 (0.15) | 9.08 (6.81-12.1) | 2.99 (5) | 1 |
| BCS | 200 | 1.24 (0.16) | 70.4 (47.3-102) | 1.87 (3) | 7.8 | |
| Cry1J | SS | 160 | 2.50 (0.48) | 0.083 (0.061-0.107) | 1.14 (2) | 1 |
| BCS | 240 | 2.37 (0.25) | 0.273 (0.224-0.333) | 1.37 (4) | 3.3 | |
| Cry2Ab | SS | 200 | 1.64 (0.25) | 0.062 (0.037-0.138) | 4.08 (3) | 1 |
| BCS | 200 | 2.28 (0.28) | 0.139 (0.112-0.172) | 0.77 (3) | 2.2 | |
| Cry9C | SS | 160 | 2.65 (0.38) | 3.36 (2.77-4.16) | 0.31 (2) | 1 |
| BCS | 160 | 2.75 (0.39) | 6.96 (5.75-8.57) | 0.10 (2) | 2.1 |
SS, susceptible strain; BCS, GLEN-Cry1Ac-BCS strain.
RR, resistance ratio (LC50 for BCS/LC50 for SS).
CI, confidence interval.
df, degrees of freedom.
LC50, 50% lethal concentration.
Melanization activity of hemolymph plasma in vitro.
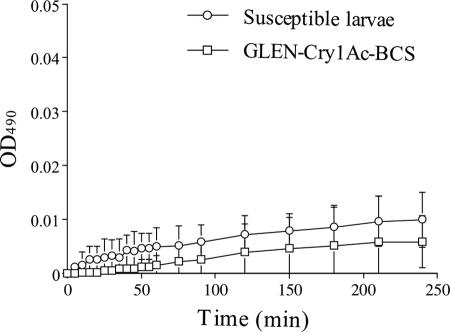
The hemolymph plasma from both the susceptible and the resistant T. ni larvae showed low melanization activity in the in vitro assays (Fig. 1), which is in contrast to the high melanization activities observed in Cry1Ac-resistant Ephestia kuehniella and Helicoverpa armigera larvae (25, 30). There was no significant difference in hemolymph melanization activity between the resistant and the susceptible strains, suggesting that the resistant T. ni larvae did not exhibit an elevated immune response as observed in B. thuringiensis-resistant E. kuehniella and H. armigera (25, 30).
FIG. 1.
In vitro melanization activity of hemolymph plasma from the susceptible and resistant larvae. Error bars indicate 95% confidence intervals of the means from three replications.
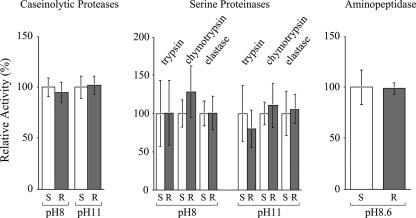
Midgut protease activities.
The caseinolytic protease activities in the midgut extracts from the resistant and susceptible fifth-instar larvae were almost identical (Fig. 2). The midgut serine proteinase activities, including trypsin, chymotrypsin, and elastase activities, from the resistant larvae were also very similar to those from the susceptible larvae. In addition, there was no difference in midgut aminopeptidase activity between the two strains of T. ni larvae.
FIG. 2.
Midgut protease activities from T. ni larvae from the susceptible and GLEN-Cry1Ac-BCS strains. Protease activities are presented as relative activities (the activities from the susceptible strain are presented as 100%). Error bars indicate the 95% confidence intervals for the means from five replications. S and R represent susceptible and resistant larvae, respectively. The pH values indicate the pHs in which the enzymatic activities were assayed.
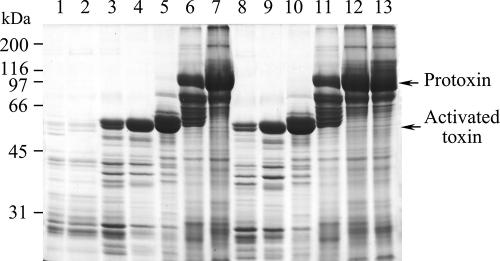
Proteolytic digestion of Cry1Ac protoxin with midgut proteases.
The midgut fluid from larvae of both the resistant and susceptible strains was highly active in proteolytic processing of the Cry1Ac protoxin into the active toxin, which was observed as a reduction of the amount of protoxin and an increase in the amount of activated toxin (Fig. 3). The protoxin was processed into its activated form in 60 min even with 0.1% gut fluid. Meanwhile, overdigestion of the toxin by the midgut fluid led to partial degradation of the activated toxin (Fig. 3, lanes 3 and 8). However, there was no significant difference in activating the protoxins or in degrading the activated toxins between the midgut fluids from the susceptible and resistant strains of T. ni.
FIG. 3.
SDS-PAGE gel showing in vitro digestion of Cry1Ac protoxin by diluted T. ni larval midgut fluid from the susceptible and GLEN-Cry1Ac-BCS strains. Lanes: 1 and 2, 10% larval gut fluid from susceptible and GLEN-Cry1Ac-BCS strains, respectively; 3 to 7, Cry1Ac protoxin incubated with 10%, 1%, 0.1%, 0.01%, and 0.001% gut fluid, respectively, from susceptible larvae; 8 to 12, Cry1Ac protoxin incubated with 10%, 1%, 0.1%, 0.01%, and 0.001% gut fluid, respectively, from GLEN-Cry1Ac-BCS larvae; 13, Cry1Ac protoxin control.
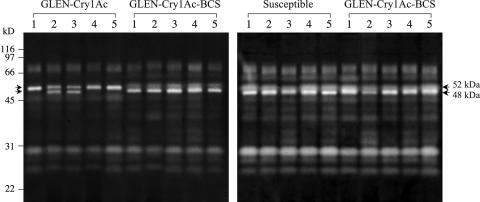
Zymogram analysis of midgut proteases.
T. ni larval midgut proteases could be separated into at least 10 caseinolytic protease bands by SDS-PAGE zymogram analysis (Fig. 4). The zymograms generated from the midgut homogenates of the susceptible larvae exhibited a homogeneous profile among different individuals. However, the midgut proteases from GLEN-Cry1Ac larvae displayed intrastrain heterogeneity among different individuals. Some individuals (GLEN-Cry1Ac larvae 1, 4, and 5 in Fig. 4) showed an intense band at 52 kDa, while others (GLEN-Cry1Ac larvae 2 and 3 in Fig. 4) had an additional major band at 48 kDa and a less intense 52-kDa band. Noticeably, the relative intensity of the 52-kDa protease band from GLEN-Cry1Ac is distinctively higher than the similar one in larvae of the susceptible strain, demonstrating differential protease compositions between the two strains. However, the zymograms from the introgression strain GLEN-Cry1Ac-BCS showed a uniform protease profile identical to the susceptible strain. Therefore, the observed differential protease composition in the original GLEN-Cry1Ac strain is not associated with resistance to Cry1Ac.
FIG. 4.
SDS-PAGE zymograms of larval midgut proteases from the susceptible, GLEN-Cry1Ac, and GLEN-Cry1Ac-BCS strains. Five individual larvae were analyzed in each group.
Midgut esterase activity and isozyme profiles.
The esterase activity in the T. ni larval midgut extract varied considerably among different larvae within the same strain. The average midgut esterase activities from five susceptible larvae and five resistant larvae were 0.060 ± 0.012 and 0.073 ± 0.020 units of optical density at 450 nm/min (average ± 95% confidence interval), respectively, under the experimental conditions. Significance analysis (t test) indicated that the midgut esterase activities for the two strains were not statistically different (P = 0.31).
The midgut esterase isozyme profiles showed four or five major esterase isozymes in the T. ni midgut. Similar to the intrastrain variations in total esterase activities, intraspecific variation of esterase isozyme composition was also observed (Fig. 5). Both the susceptible and resistant larvae showed two types of esterase isozyme gel profiles. Clearly, the observed variation in esterase isozyme composition in the larvae does not correlate with the resistance.
FIG. 5.
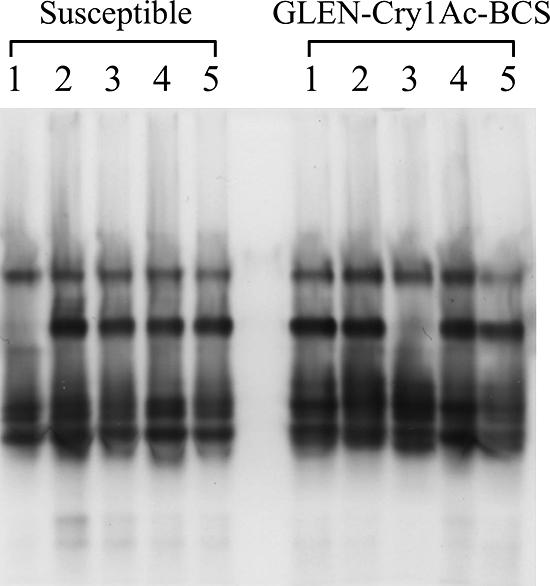
Midgut esterase isozyme profiles of T. ni larvae from the susceptible and GLEN-Cry1Ac-BCS strains. Five individual larvae were analyzed for each strain.
Binding of Cry1Ab and Cry1Ac to the midgut brush border membranes.
The midgut BBMVs prepared from the resistant and susceptible strains and their F1 larvae showed similar levels of enzymatic activities of alkaline phosphatase and aminopeptidase, which are BBMV marker enzymes and are also putative B. thuringiensis toxin receptors (14). Alkaline phosphatase activities in the BBMVs from the susceptible, resistant, and F1 larvae were 6.5, 6.9, and 7.6 mmol substrate/min/mg protein, respectively, without significant differences (P > 0.05). Similarly, no significant differences were found in leucine aminopeptidase activity in the susceptible, F1, and resistant BBMVs (120, 125, and 120 μmol substrate/min/mg, respectively).
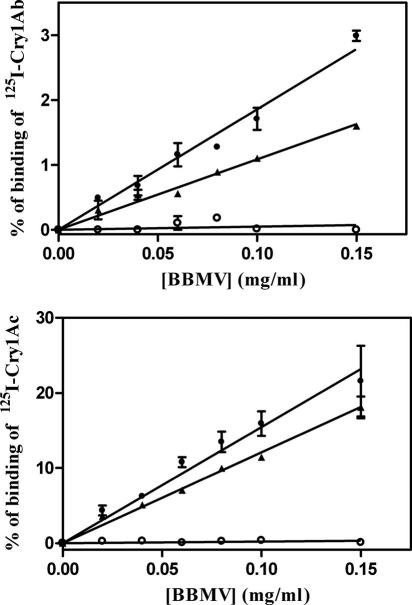
Determination of the binding of 125I-labeled Cry1Ab and Cry1Ac to BBMVs from the susceptible, resistant, and F1 T. ni larvae showed that the binding of 125I-Cry1Ac to the BBMVs from susceptible and F1 larvae increased linearly with increasing concentrations of BBMVs within the range of concentrations tested, reaching maximum specific binding levels of 22% and 18.1%, respectively, at the highest concentration of BBMVs tested (Fig. 6). In contrast, the same binding assay using BBMVs from resistant insects did not show any specific binding of 125I-Cry1Ac. Similarly, no specific binding of 125I-Cry1Ab to the BBMVs from resistant larvae was detected, although 125I-Cry1Ab bound to the BBMVs from the susceptible and F1 larvae specifically with maxima of 3% and 1.6%, respectively (Fig. 6). Therefore, the midgut BBMVs from the resistant larvae lack specific affinity for binding to Cry1Ab and Cry1Ac.
FIG. 6.
Specific binding of 125I-Cry1Ac and 125I-Cry1Ab as a function of midgut BBMV concentration from susceptible (•), resistant (○), and F1 (▴) larvae. Each data point represents the mean of two replicates. Error bars indicate standard deviations of the data points.
Assays of homologous competition binding (Fig. 7) of Cry1Ac to the BBMVs from the susceptible and F1 larvae allowed us to estimate the dissociation constants (Kd) and concentration of binding sites (Rt). The values obtained for the susceptible insects were as follows: Kd = 5 ± 1 nM (mean ± standard deviation [SD]) and Rt = 11.7 ± 2.3 pmol/mg (mean ± SD); for the F1 insects the values were as follows: Kd = 7.5 ± 1.5 nM (mean ± SD) and Rt = 10.8 ± 6.7 pmol/mg (mean ± SD). There was no significant difference between the susceptible and the F1 larvae in both Kd and Rt values. The overall affinity (Rt/Kd), a parameter that combines Kd and Rt, was slightly higher for susceptible insects (2.2 ± 0.8 pmol/mg/nM) than for F1 insects (1.4 ± 0.6 pmol/mg/nM), although this difference was not significant.
FIG. 7.
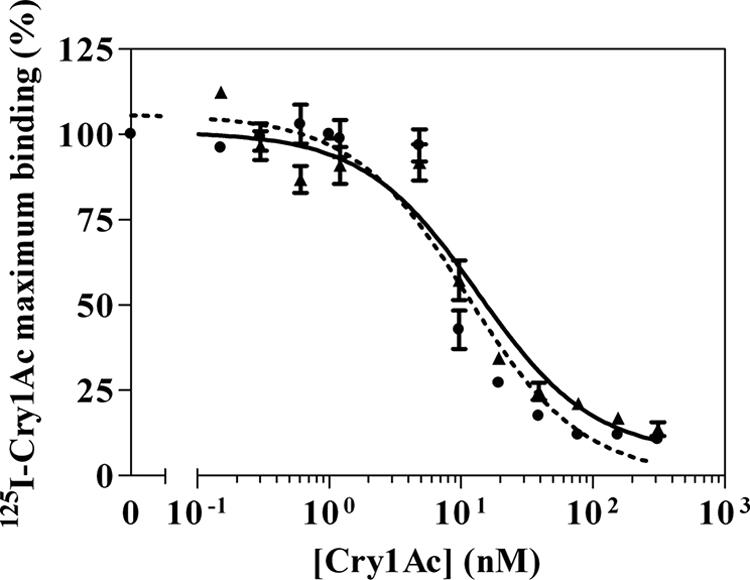
Competitive binding of 125I-Cry1Ac to midgut BBMVs from susceptible (•) and F1 (▴) larvae in the presence of nonlabeled Cry1Ac. Each data point represents the mean of two replicates. Error bars indicate standard deviations of the data points.
DISCUSSION
Comparative analysis of B. thuringiensis-resistant populations with a susceptible reference population is a logical approach to identify resistance mechanisms. However, global genetic variations between the populations in their genetic backgrounds can complicate the analysis and may even lead to data misinterpretation. In this study, the Cry1Ac resistance in the greenhouse-evolved B. thuringiensis-resistant T. ni population was introgressed into an inbred laboratory T. ni strain, and the introgression proved to be effective to limit genetic variations unrelated to Cry1Ac resistance (Fig. 4). Therefore, GLEN-Cry1Ac-BCS and its nearly isogenic susceptible strain provided us with a more reliable biological system to investigate the mechanism of Cry1Ac resistance evolved in T. ni populations under agricultural conditions.
With the nearly isogenic resistant and susceptible T. ni strains, two approaches were taken to examine the involvement of the midgut binding to the Cry toxins: characterization of the pattern of cross-resistance of Cry1Ac-resistant T. ni to various Cry toxins and direct determination of specific binding of the toxins to the midgut BBMVs. The results from both approaches indicated that Cry1Ac resistance in T. ni is conferred by loss of affinity for midgut binding to the Cry1Ac toxin. It is known that Cry1Ab and Cry1Ac share the same high-affinity binding site in the T. ni midgut and that Cry1Aa and Cry1F also bind to the Cry1Ab/Cry1Ac shared binding site with a low binding affinity (6, 19). The pattern of cross-resistance of GLEN-Cry1Ac-BCS larvae to 11 tested Cry toxins (Table 3) showed that the levels of resistance to the Cry toxins in the resistant T. ni larvae correlated with the lack of the Cry1Ab/Cry1Ac shared binding site in the larvae. Consistently, the direct binding analysis demonstrated that the midgut BBMV preparation from the resistant T. ni larvae lacked affinity for binding to Cry1Ab/Cry1Ac. Moreover, the F1 larvae from the cross of the susceptible and resistant parents, which were susceptible to Cry1Ac, exhibited affinity for midgut binding to the toxins, further confirming that loss of the Cry1Ab/Cry1Ac binding affinity in the larval midgut is the mechanism conferring the resistance.
Current understanding of B. thuringiensis resistance in insects has indicated that mechanisms for B. thuringiensis resistance can be diverse and complex and that a resistant strain may have multiple resistance mechanisms (10, 13, 14, 18, 21). Therefore, it is necessary to examine all possible mechanisms for B. thuringiensis resistance in the greenhouse-evolved T. ni populations, in addition to binding of B. thuringiensis toxins to the midgut. Results in this study showed that heightened insect immune response, as determined by measuring hemolymph melanization activity, and increased midgut esterase production are not the mechanism for Cry1Ac resistance in the T. ni greenhouse population (Fig. 1 and 5). Differential midgut protease compositions, profiled by zymogram analysis, between the greenhouse-originated T. ni larvae (GLEN-Cry1Ac) and the laboratory susceptible larvae were observed. However, this difference arose from the different genetic backgrounds of the two strains but was not associated with Cry1Ac resistance (Fig. 4). In agreement with the midgut protease zymogram analysis, no significant difference was observed in larval midgut protease activities between the resistant and susceptible strains (Fig. 2). Furthermore, the results from in vitro activation of Cry1Ac protoxin and degradation of activated Cry1Ac toxin by the larval midgut digestive fluid also indicated that alteration of midgut proteases is not the mechanism of Cry1Ac resistance in the greenhouse-evolved B. thuringiensis-resistant T. ni population (Fig. 3). Therefore, it is clear that the mechanism for greenhouse-evolved Cry1Ab/Cry1Ac resistance resulted from the loss of affinity for the binding of the midgut epithelium to the toxins Cry1Ab and Cry1Ac. Notably, a previously reported Cry1Ab-resistant T. ni strain obtained by laboratory selection did not exhibit cross-resistance to Cry1Ac regardless of the fact that the Cry1Ab and Cry1Ac share the same binding sites (6). This observation demonstrates that field-evolved resistance may involve a mechanism different from laboratory-selected resistance.
Results from this study showed that the loss of affinity for the binding of the midgut to Cry1Ab/Cry1Ac is inherited as a recessive trait, which is in agreement with the recessive inheritance that we have previously observed for the resistance trait (22) and consistent with a loss-of-function mechanism. Reduced binding of Cry toxins to the midgut brush border membranes has been observed in a number of insect species with high levels of resistance to Cry1A toxins (9, 18). Suggested midgut receptors for Cry1A include the midgut cell membrane proteins cadherin, aminopeptidase N, alkaline phosphatase, and a 252-kDa high-molecular-weight protein and midgut glycolipids. The loss of binding sites in the insect larval midgut could be due to the absence of the midgut cadherin as a result of premature truncation of the cadherin gene transcript or mutations in the Cry toxin binding sites in the cadherin sequences (11, 27, 42). However, in Cry1Ac-resistant diamondback moth strains originated from field-developed resistant populations from two geographical regions, the reduced binding of Cry1A toxins is not conferred by cadherin gene mutations (2). In this study, it became clear that the Cry1Ac resistance in the greenhouse T. ni population is conferred by alteration of the larval midgut binding to the Cry toxins. What serves as the receptor for the Cry1Ab and Cry1Ac in T. ni larvae and what alteration has occurred to the receptor in the resistant population require further investigation in order to understand the molecular genetic basis for the greenhouse-evolved resistance in T. ni populations.
Acknowledgments
This work was supported in part by the Cornell University Agricultural Experiment Station federal formula funds, project no. NYG-621510, received from Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture (to P.W.), and by the Council of Culture, Education and Sports from the Valencian Autonomic Government (reference GRUPOS2004-21) and the Spanish Ministry of Education and Science for International Cooperation (reference AGL2004-0367-E) (to J.F.).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Akhurst, R. J., W. James, L. J. Bird, and C. Beard. 2003. Resistance to the Cry1Ac δ-endotoxin of Bacillus thuringiensis in the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 96:1290-1299. [DOI] [PubMed] [Google Scholar]
- 2.Baxter, S. W., J.-Z. Zhao, L. J. Gahan, A. M. Shelton, B. E. Tabashnik, and D. G. Heckel. 2005. Novel genetic basis of field-evolved resistance to Bt toxins in Plutella xylostella. Insect Mol. Biol. 14:327-334. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Broadway, R. M. 1997. Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J. Insect Physiol. 43:855-874. [DOI] [PubMed] [Google Scholar]
- 5.Candas, M., O. Loseva, B. Oppert, P. Kosaraju, and L. A. Bulla, Jr. 2003. Insect resistance to Bacillus thuringiensis, alterations in the Indianmeal moth larval gut proteome. Mol. Cell. Proteomics 2:19-28. [DOI] [PubMed] [Google Scholar]
- 6.Estada, U., and J. Ferré. 1994. Binding of insecticidal crystal proteins of Bacillus thuringiensis to the midgut brush border of the cabbage looper, Trichoplusia ni (Hübner) (Lepidoptera: Noctuidae), and selection for resistance to one of the crystal proteins. Appl. Environ. Microbiol. 60:3840-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estela, A., B. Escriche, and J. Ferré. 2004. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 70:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federici, B. 2003. Effects of Bt on non-target organisms, p. 11-30. In M. Metz (ed.), Bacillus thuringiensis: a cornerstone of modern agriculture. Food Products Press, Binghamton, NY.
- 9.Ferré, J., M. D. Real, J. Van Rie, S. Jansens, and M. Peferoen. 1991. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc. Natl. Acad. Sci. USA 88:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferré, J., and J. Van Rie. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501-533. [DOI] [PubMed] [Google Scholar]
- 11.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Carreno, F. L., L. E. Dimes, and N. F. Haard. 1993. Substrate-gel electrophoresis for composition and molecular weight of proteinases or proteinaceous proteinase inhibitors. Anal. Biochem. 214:65-69. [DOI] [PubMed] [Google Scholar]
- 13.González-Cabrera, J., S. Herrero, and J. Ferré. 2001. High genetic variability for resistance to Bacillus thuringiensis toxins in a single population of diamondback moth. Appl. Environmen. Microbiol. 67:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffitts, J. S., and R. V. Aroian. 2005. Many roads to resistance: how invertebrates adapt to Bt toxins. Bioessays 27:614-624. [DOI] [PubMed] [Google Scholar]
- 15.Gunning, R. V., H. T. Dang, F. C. Kemp, I. C. Nicholson, and G. D. Moores. 2005. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl. Environ. Microbiol. 71:2558-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa, T., Y. Shitomi, K. Miyamoto, and H. Hori. 2004. GalNAc pretreatment inhibits trapping of Bacillus thuringiensis Cry1Ac on the peritrophic membrane of Bombyx mori. FEBS Lett. 576:331-335. [DOI] [PubMed] [Google Scholar]
- 17.Hernández, C. S., A. Rodrigo, and J. Ferré. 2004. Lyophilization of lepidopteran midguts: a preserving method for Bacillus thuringiensis toxin binding studies. J. Invertebr. Pathol. 85:182-187. [DOI] [PubMed] [Google Scholar]
- 18.Herrero, S., B. Oppert, and J. Ferré. 2001. Different mechanisms of resistance to Bacillus thuringiensis toxins in the Indianmeal moth. Appl. Environ. Microbiol. 67:1085-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iracheta, M. M., B. Pereyra-Alferez, L. Galan-Wong, and J. Ferre. 2000. Screening for Bacillus thuringiensis crystal proteins active against the cabbage looper, Trichoplusia ni. J. Invertebr. Pathol. 76:70-75. [DOI] [PubMed] [Google Scholar]
- 20.Janmaat, A. F., and J. Myers. 2003. Rapid evolution and the cost of resistance to Bacillus thuringiensis in greenhouse populations of cabbage loopers, Trichoplusia ni. Proc. R. Soc. Lond. B Biol. Sci. 270:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurat-Fuentes, J. L., and M. J. Adang. 2006. Cry toxin mode of action in susceptible and resistant Heliothis virescens larvae. J. Invertebr. Pathol. 92:166-171. [DOI] [PubMed] [Google Scholar]
- 22.Kain, W. C., J.-Z. Zhao, A. F. Janmaat, J. Myers, A. M. Shelton, and P. Wang. 2004. Inheritance of resistance to Bacillus thuringiensis Cry1Ac toxin in a greenhouse-derived strain of cabbage looper (Lepidoptera: Noctuidae). J. Econ. Entomol. 97:2073-2078. [DOI] [PubMed] [Google Scholar]
- 23.Kough, J. 2003. The safety of Bacillus thuringiensis for human consumption, p. 1-10. In M. Metz (ed.), Bacillus thuringiensis: a cornerstone of modern agriculture. Food Products Press, Binghamton, NY.
- 24.Li, H., B. Oppert, R. A. Higgins, F. Huang, K. Y. Zhu, and L. L. Buschman. 2004. Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 34:753-762. [DOI] [PubMed] [Google Scholar]
- 25.Ma, G., H. Roberts, M. Sarjan, N. Featherstone, J. Lahnstein, R. Akhurst, and O. Schmidt. 2005. Is the mature endotoxin Cry1Ac from Bacillus thuringiensis inactivated by a coagulation reaction in the gut lumen of resistant Helicoverpa armigera larvae? Insect Biochem. Mol. Biol. 35:729-739. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Ramírez, A. C., F. Gould, and J. Ferré. 1999. Histopathological effects and growth reduction in a susceptible and a resistant strain of Heliothis virescens (Lepidoptera: Noctuidae) caused by sublethal doses of pure Cry1A crystal proteins from Bacillus thuringiensis. Biocontrol Sci. Technol. 9:239-246. [Google Scholar]
- 27.Morin, S., R. W. Biggs, M. S. Sisterson, L. Shriver, C. Ellers-Kirk, D. Higginson, D. Holley, L. J. Gahan, D. G. Heckel, Y. Carriere, T. J. Dennehy, J. K. Brown, and B. E. Tabashnik. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 100:5004-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munson, P. J., and D. Rodbard. 1980. LIGAND: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107:220-239. [DOI] [PubMed] [Google Scholar]
- 29.Oppert, B., K. J. Kramer, R. W. Beeman, D. Johnston, and W. H. McGaughey. 1997. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J. Biol. Chem. 272:23473-23476. [DOI] [PubMed] [Google Scholar]
- 30.Rahman, M. M., H. L. S. Roberts, M. Sarjan, S. Asgari, and O. Schmidt. 2004. Induction and transmission of Bacillus thuringiensis tolerance in the flour moth Ephestia kuehniella. Proc. Natl. Acad. Sci. USA 101:2696-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelton, A. M., J. L. Roberson, J. D. Tang, C. Perez, S. D. Eigenbrode, H. K. Preisler, W. K. Wilsey, and R. J. Cooley. 1993. Resistance of diamondback moth (Lepidoptera: Plutellidae) to Bacillus thuringiensis subspecies in the field. J. Econ. Entomol. 86:697-705. [Google Scholar]
- 33.Stauffer, C., T. Shiotsuki, W. Chan, and B. D. Hammock. 1997. Characterization of the esterase isozymes of Ips typographus (Coleoptera, Scolytidae). Arch. Insect Biochem. Physiol. 34:203-221. [Google Scholar]
- 34.Tabashnik, B. E., N. L. Cushing, N. Finson, and M. W. Johnson. 1990. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 83:1671-1676. [Google Scholar]
- 35.Tabashnik, B. E. 1994. Evolution of resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 39:47-79. [Google Scholar]
- 36.Tabashnik, B. E., N. Finson, F. R. Groeters, W. J. Moar, M. W. Johnson, K. Luo, and M. J. Adang. 1994. Reversal of resistance to Bacillus thuringiensis in Plutella xylostella. Proc. Natl. Acad. Sci. USA 91:4120-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabashnik, B. E., Y. Carriere, T. J. Dennehy, S. Morin, M. S. Sisterson, R. T. Roush, A. M. Shelton, and J.-Z. Zhao. 2003. Insect resistance to transgenic Bt crops: lessons from the laboratory and field. J. Econ. Entomol. 96:1031-1038. [DOI] [PubMed] [Google Scholar]
- 38.Tabashnik, B. E., Y.-B. Liu, D. C. Unnithan, Y. Carriere, T. J. Dennehy, and S. Morin. 2004. Shared genetic basis to Bt toxin Cry1Ac in independent strains of pink bollworm. J. Econ. Entomol. 97:721-726. [DOI] [PubMed] [Google Scholar]
- 39.Van Rie, J., S. Jansens, H. Hofte, D. Degheele, and H. Van Mellaert. 1989. Specificity of Bacillus thuringiensis δ-endotoxins. Importance of specific receptors on the brush border membrane of the midgut of target insects. Eur. J. Biochem. 186:239-247. [DOI] [PubMed] [Google Scholar]
- 40.Wang, P., X. Zhang, and J. Zhang. 2005. Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 35:611-620. [DOI] [PubMed] [Google Scholar]
- 41.Wolfersberger, M. G., P. Luethy, P. Maurer, P. Parenti, V. F. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301-308. [Google Scholar]
- 42.Xu, X., L. Yu, and Y. Wu. 2005. Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 71:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, J.-Z., Y. X. Li, H. L. Collins, J. Cao, E. D. Earle, and A. M. Shelton. 2001. Different cross-resistance patterns in the diamondback moth (Lepidoptera: Plutellidae) resistant to Bacillus thuringiensis toxin Cry1C. J. Econ. Entomol. 94:1547-1552. [DOI] [PubMed] [Google Scholar]