Abstract
Bacterial biodiversity occurring in traditional Egyptian soft Domiati cheese was studied by PCR-temporal temperature gel electrophoresis (TTGE) and PCR-denaturing gradient gel electrophoresis (DGGE). Bands were identified using a reference species database (J.-C. Ogier et al., Appl. Environ. Microbiol. 70:5628-5643, 2004); de novo bands having nonidentified migration patterns were identified by DNA sequencing. Results reveal a novel bacterial profile and extensive bacterial biodiversity in Domiati cheeses, as reflected by the numerous bands present in TTGE and DGGE patterns. The dominant lactic acid bacteria (LAB) identified were as follows: Leuconostoc mesenteroides, Lactococcus garvieae, Aerococcus viridans, Lactobacillus versmoldensis, Pediococcus inopinatus, and Lactococcus lactis. Frequent non-LAB species included numerous coagulase-negative staphylococci, Vibrio spp., Kocuria rhizophila, Kocuria kristinae, Kocuria halotolerans, Arthrobacter spp./Brachybacterium tyrofermentans. This is the first time that the majority of these species has been identified in Domiati cheese. Nearly all the dominant and frequent bacterial species are salt tolerant, and several correspond to known marine bacteria. As Domiati cheese contains 5.4 to 9.5% NaCl, we suggest that these bacteria are likely to have an important role in the ripening process. This first systematic study of the microbial composition of Domiati cheeses reveals great biodiversity and evokes a role for marine bacteria in determining cheese type.
Domiati cheese (Gbnah Beeda) is the most popular soft white pickled cheese in Egypt and makes up about 75% of the cheese produced and consumed in that country (52). It differs chiefly from other pickled cheese varieties, such as feta, Brinza, or Telema cheese, in that the milk is salted at the first step of its manufacture. The proportion of salt (5 to 14%) depends on the season of manufacture and on the temperature of cheese ripening (1). The cheese is made from either cow or buffalo whole milk or a mixture of the two. The salted milk can be curdled fresh or sometimes after pasteurization, without the addition of any starter cultures. The cheese can be consumed either fresh or, more often, after pickling in salted whey or a brine solution for up to 2 to 4 months.
During the last decades, several investigators have isolated and identified different lactic acid bacterial species from Domiati cheese, such as Lactococcus lactis subsp. lactis, Lactobacillus delbrueckii subsp. bulgaricus, L. casei (21), L. farciminis, L. alimentarius, Enterococcus faecalis, E. faecium (16), Lactobacillus plantarum, and L. paracasei (15). Other bacterial species were also isolated, including coliforms (2), Micrococcus spp. (20), Arthrobacter spp. (14), Propionibacterium jensenii, Microbacterium lacticum, Brevibacterium linens (16), Staphylococcus aureus (9), and Aeromonas spp. (8). In all the above-mentioned studies, results were obtained using culture methods.
Recently, molecular methods such as temporal temperature gel electrophoresis (TTGE) and/or denaturing gradient gel electrophoresis (DGGE) (5, 17, 38) were successfully used to identify the bacterial biodiversity of different types of cheese such as artisanal Sicilian (43), Stilton (18), mozzarella (6, 19, 34), Beaufort, Saint Nectaire, Morbier, Epoisse (41), Mish (10), Karish (11), and hard Ras cheeses (13). Large-scale analyses of dairy samples in INRA, France, have led to the establishment of a reference database, allowing comparative identification of some 170 bacterial species, including some food pathogens (41). The aim of the present work was to use these molecular methods to characterize the bacterial biodiversity of the popular Egyptian Domiati cheese.
MATERIALS AND METHODS
Sampling.
Eleven samples of Domiati cheese made by traditional methods were collected aseptically from different cheese producers from Alexandria, Behira, and Domiatta governorates. Gross analysis of Domiati cheese samples showed that moisture, salt content, and pH values ranged between 59.83 and 63.15%, 5.46 and 9.50%, and 5.30 and 6.15, respectively.
Genomic DNA extraction.
Total genomic DNA was extracted from each Domiati cheese sample (7 g) as previously described (41). After undergoing a protein digestion step by the pronase (Boerhinger, Mannheim, Germany), bacterial cells were mechanically lysed. DNA purification was performed as previously described (7). Pellets of DNA were then dissolved in 100 μl Tris-EDTA buffer plus RNase (Sigma, Saint Quentin Fallavier, France) and then analyzed by 0.8% agarose gel electrophoresis.
PCR amplification.
Amplicons for TTGE and DGGE analyses were prepared by performing two successive PCRs using a Gene Amp system model 2400 (PerkinElmer, France) and appropriate primers. First, a 700-bp fragment of the 16S rRNA gene including the V3 region was amplified with primers W01 and W012. Second, the V3 region of 200 bp was amplified using primers HDA1-GC and HDA2. The PCR mixtures and amplification programs were as previously described (40). Sizes and quantities of PCR products were determined by 2% agarose gel electrophoresis (Seakem CTG agarose; TEBU, France) against a standard containing DNA fragments of defined lengths (Smart Ladder, France).
TTGE and DGGE analyses.
TTGE and DGGE analyses of V3 amplicons were applied on 16-cm by 16-cm by 1-mm gels (Bio-Rad DCode universal mutation detection system; Marnes La Coquette, France) and performed as previously described (41). After runs, gels were stained for 15 min with an ethidium bromide solution (0.5 μg/ml of 1× Tris-acetate-EDTA [TAE] buffer), rinsed for 20 min in 1× TAE buffer, and photographed on a UV transillumination table.
Gel analysis and band identification using species database.
TTGE and DGGE gels were analyzed by GelCompar software (Applied-Maths, Belgium) as previously described (41). The software standardizes TTGE and DGGE profiles to minimize migration differences between gels by alignment of the identification ladder with a standard gel (40). Band identifications are performed by comparison to a species database which includes TTGE and DGGE fingerprints of about 170 bacterial species isolated from dairy ecosystems (41). In some cases, specific PCR tests and/or cloning and sequencing were undertaken to confirm species assignments or to distinguish between comigrating species.
Species-specific PCR tests.
Specific PCR tests were carried out using different species-specific primers (Table 1) with DNA obtained from the cheese samples. Primers (MWG Biotech AG, Ebersberg, Germany) were prepared at a final concentration of 60 μM in deionized, autoclaved water. PCR was performed in a GenAmp system model 2400 (PerkinElmer, France), and all reactions were carried out following conditions previously provided by the authors (Table 1). Sizes of PCR products were determined using 1.5% agarose gel electrophoresis (Seakem CTG agarose; TEBU, France).
TABLE 1.
Primers used in this study for the species-specific PCR assays
| Target | Primers | Sequence (5′-3′) | Annealing temp (°C) | Source or referencea |
|---|---|---|---|---|
| Brevibacterium casei | Brevib, Bcas | Unpublished data | 64 | Furlan |
| Brevibacterium linens | Brevib, Blin | Unpublished data | 64 | Furlan |
| Corynebacterium variabile | Cvar, Corb | Unpublished data | 60 | Furlan |
| Escherichia coli | Eco 223 | ATCAACCGAGATTCCCCCAGT | ||
| Eco 455 | TCACTATCGGTCAGTCAGGAG | 64 | 46 | |
| Enterococcus casseliflavus | EC1, EC2 | Unpublished data | 52 | Firmesse |
| Enterococcus durans | ED1, ED2 | Unpublished data | 52 | Firmesse |
| Enterococcus faecalis | EFS1, EFS2 | Unpublished data | 52 | Firmesse |
| Enterococcus faecium | EFM1, EFM2 | Unpublished data | 52 | Firmesse |
| Enterococcus hirae | EH1, EH2 | Unpublished data | 52 | Firmesse |
| Lactococcus garvieae | 1RL LgR | TTTGAGAGTTTGATCCTGG AAGTAATTTTCCACTCTACTT | 45 | 42 |
| Lactococcus lactis | 1RL LacreR | TTTGAGAGTTTGATCCTGG GGGATCATCTTTGAGTGAT | 45 | 42 |
| Lactococcus lactis subsp. cremoris | CreF LacreR | GTGCTTGCACCGATTTGAA GGGATCATCTTTGAGTGAT | 58 | 42 |
| Lactococcus lactis subsp. lactis | LacF LacreR | GTACTTGTACCGACTGGAT GGGATCATCTTTGAGTGAT | 58 | 42 |
| Lactococcus raffinolactis | 1RL PipLraR | TTTGAGAGTTTGATCCTGG CGTCACTGAGGGCTGGAT | 45 | 42 |
| Lactobacillus acidophilus | Laci01 Laci02 | GACCGCATGATCAGCTTATA AGTCTCTCAACTCGGCTATG | 55 | 25 |
| Lactobacillus brevis | LbBreF LbBreR | CTTGCACTGATTTTAACA GGGCGGTGTGTACAAGGC | 40 | 26 |
| Lactobacillus casei | PrI 16S-23S CasII | CAGACTGAAAGTCTGACGG GCGATGCGAATTTCTTTTTC | 55 | 50 |
| Lactobacillus gasseri | GasI GasII | GAGTGCGAGAGCACTAAAG CTATTTCAAGTTGAGTTTCTCT | 55 | 50 |
| Lactobacillus johnsonii | Joh 16SI 16SII | GAGCTTGCCTAGATGATTTTA ACTACCAGGGTATCTAATCC | 57 | 50 |
| Lactobacillus plantarum | Lfpr16S-23S PlanII | GCCGCCTAAGGTGGGACAGAT TTACCTAACGGTAAATGCGA | 55 | 50 |
| Leuconostoc citreum | Lcit-f Lcit-r | AAAACTTAGTATCGCATGATATC CTTAGACGACTCCCTCCCG | 60 | 30 |
| Leuconostoc mesenteroides | Lnm1 Lnm2 | TGTCGCATGACACAAAGTTA ATCATTTCCTATTCTAGCTG | 58 | 4 |
| Pseudomonas aeruginosa | Paer16SH | AGGGCAGTAAGTTAATACCTTGCTG | 65 | 51 |
| Paer16SIR | CCACCTCTACCGTACTCTAGCTCAG | |||
| Serratia marcescens | Smar16SV | GGGAGCTTGCTCACTGGGTG | ||
| Smar16SWR | GCGAGTAACGTCAGTTGATGAGCGTATTA | 66 | 51 | |
| Staphylococcus aureus | STAA-AuI | TCTTCAGAAGATGCGGAATA | ||
| STAA-AuII | TAAGTCAAACGTTAACATACG | 55 | 24 | |
| Staphylococcus chromogenes | STAC-ChrI | ACGGAATATCGCTTTTAAGC | ||
| STAC-ChrII | CGTTTACATTCGGCTTTCG | 52 | 24 | |
| Staphylococcus epidermidis | STAE-EpI | TCTACGAAGATGAGGGATA | ||
| STAE-EpII | TTTCCACCATATTTTGAATTGT | 52 | 24 | |
| Staphylococcus saprophyticus | fStSap | TCAAAAAGTTTTCTAAAAAATTTAC | ||
| rStSap | ACGGGCGTCCACAAAATCAATAGGA | 55 | 33 | |
| Staphylococcus simulans | STAS-SiI | ATTCGGAACAGTTTCGCAG | ||
| STAS-SiII | ATTGTGAGTAATCGTTTGCC | 55 | 24 | |
| Staphylococcus xylosus | STAX-XyI | TCTTTAGAAGATGACAGAGG | ||
| STAX-XyII | TGACTTTTAACACGACGAAG | 55 | 24 | |
| Streptococcus thermophilus | Sther03 | TTATTTGAAAGGGGCAATTGCT | ||
| Sther08 | GTGAACTTTCCACTCTCACAC | 55 | 25 |
Firmesse, O. Firmesse et al., unpublished data; Furlan, S. Furlan et al., unpublished data.
Sequencing of bands.
Some bands obtained from TTGE and DGGE analyses of Domiati cheese samples were excised, purified, cloned, and sequenced, as described previously (40). Sequences of the clones of the V3 16S rRNA genes were compared to those present in the Ribosomal Database Project (31) to determine the closest known relative species.
Enumeration of different bacterial groups.
Domiati cheese samples (11 g) were emulsified in 99 ml of sterile 2% (wt/vol) tri-sodium citrate solution (Merck) and homogenized using an Ultra-Turrax mechanical blender (19,000 rpm for 45 s). Serial dilutions were prepared in sterile 1% (wt/vol) peptone water, plated on selective agar medium using a spiral platter (Spiral system; Cincinnati, OH), and incubated at the appropriate temperatures (Table 2). Bacterial enumerations were done on M17 (Difco, Elancourt, France) for lactococci and streptococci, on MRS (Difco) (pH adjusted to 5.2) for mesophilic and thermophilic lactobacilli, on MSE (35) for Leuconostoc, on BEA (Difco) for enterococci, on MSA (Difco) for staphylococci, on BHI (Difco) supplemented with 5% (wt/vol) NaCl for salt-tolerant flora, on YEL (yeast extract-sodium lactate medium) for Propionibacterium spp., on VRBA (Difco) for total coliform, and on cetrimide, fucidin, and cephalosporin (CFC) agar (Oxoid) for Pseudomonas spp.
TABLE 2.
Range of viable counts of different bacterial groups in Domiati cheese samples
| Medium | Group targeted | Incubation temp (°C), time, anaerobic or aerobic conditionsa | Log CFU/gb
|
|
|---|---|---|---|---|
| Lower | Higher | |||
| MSA | Staphylococcus spp. | 37, 48 h, A | 7.09 ± 0.25 | 7.57 ± 0.37 |
| M17 | Mesophilic streptococci | 30, 48 h, A | 5.21 ± 0.23 | 7.40 ± 0.26 |
| BHI + 5% NaCl | Salt-tolerant flora | 25, 48 h, A | 6.36 ± 0.28 | 7.12 ± 0.32 |
| M17 | Thermophilic streptococci | 42, 48 h, A | 5.50 ± 0.19 | 6.86 ± 0.31 |
| MSE | Leuconostoc spp. | 30, 48 h, A | 5.55 ± 0.21 | 6.63 ± 0.22 |
| BEA | Enterococcus spp. | 37, 48 h, An | 6.16 ± 0.18 | 6.62 ± 0.28 |
| YEL | Propionibacterium spp. | 25, 7 days, An | 5.92 ± 0.25 | 6.12 ± 0.21 |
| CFC | Pseudomonas spp. | 30, 48 h, A | 4.02 ± 0.12 | 5.94 ± 0.26 |
| MRS | Mesophilic Lactobacillus | 30, 48 h, An | 4.75 ± 0.16 | 5.57 ± 0.24 |
| MRS | Thermophilic Lactobacillus | 42, 72 h, An | 3.03 ± 0.09 | 5.45 ± 0.14 |
| VRBA | Total coliform | 30, 48 h, A | 2.10 ± 0.15 | 3.78 ± 0.19 |
A, aerobic; An, anaerobic.
The values shown are means ± standard deviations.
RESULTS
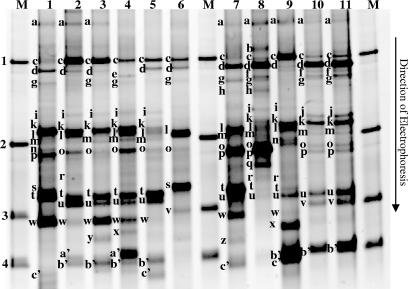
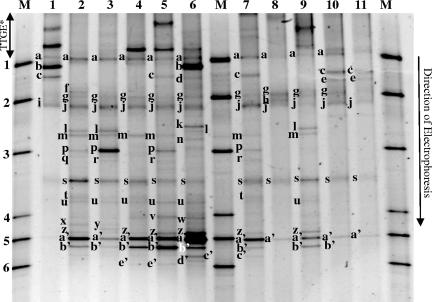
Considerable bacterial biodiversity existed within the 11 samples of Domiati cheese that were analyzed by TTGE and DGGE (Fig. 1 and 2). In TTGE profiles, all cheese samples showed complex profiles ranging from 7 bands (Fig. 1, sample 6) to 18 bands (Fig. 1, sample 8). In DGGE profiles, some Domiati cheese samples showed relatively simple profiles with 5 bands (Fig. 2, sample 1), whereas other samples showed 17 bands (Fig. 2, sample 6).
FIG. 1.
TTGE electrophoresis of V3 16S rDNA fragments from different samples of Domiati cheese. Samples 1 to 11 correspond to V3 16S rDNA regions PCR amplified from genomic DNA extracted from cheese samples (see Materials and Methods). After standardization of the gel by GelCompar software, bands are identified by comparison with the species database. Band a, PCR artifact; b, unidentified band; c, Lactococcus garvieae; d, Aerococcus viridans; e, unidentified band; f, Lactobacillus johnsonii, Lactobacillus gasseri; g, Lactobacillus plantarum, Lactobacillus pentosus; h, Acinetobacter lwoffii; i, Leuconostoc citreum; j, Enterococcus casseliflavus, Staphylococcus lentus; k, Leuconostoc mesenteroides; l, Staphylococcus epidermidis, Staphylococcus aureus, Staphylococcus simulans; m, Lactococcus raffinolactis, Staphylococcus equorum, Staphylococcus capitis; n, unidentified band; o, Enterococcus faecium group, Pseudomonas fluorescens, Leuconostoc pseudomesenteroides; p, Leuconostoc lactis, Staphylococcus xylosus, Lactobacillus brevis; q, Pseudomonas putida; r, Lactobacillus acidophilus group; s, Staphylococcus saprophyticus, Pseudomonas aeruginosa; t, Pediococcus pentosaceus, Macrococcus caseolyticus; u, Streptococcus uberis, Moraxella bovis; v, Enterococcus faecalis, Staphylococcus warneri; w, unidentified band; x, unidentified band; y, Staphylococcus chromogenes, Hafnia alvei, Pseudomonas alcaligenes; z, unidentified band; a′, Acinetobacter spp.; b′, Lactococcus lactis; c′, Streptococcus thermophilus. Markers: 1, Lactococcus garvieae CNRZ1323; 2, Lactococcus raffinolactis CNRZ1214; 3, Enterococcus faecalis CE17; 4, Lactococcus lactis subsp. lactis bv. diacetilactis CNRZ260. Lane M, TTGE standardization ladder; lane 1 to lane 11, cheese samples from 1 to 11.
FIG. 2.
DGGE electrophoresis of V3 16S rRNA gene fragments from different samples of Domiati cheese. Samples 1 to 11 correspond to V3 16S rRNA gene regions PCR-amplified from genomic DNA extracted from cheese samples (see Materials and Methods). After standardization of the gel by GelCompar software, bands are identified by comparison with the species database. Band a, Klebsiella pneumoniae; b, Enterobacter amnigenus, E. coli; c, Citrobacter freundii; d, unidentified band; e, unidentified band; f, Serratia marcescens, Raoultella planticola; g, Corynebacterium variabile; h, Enterobacter cloacae, Klebsiella oxytoca; i, Serratia liquefaciens; j, unidentified band; k, Corynebacterium flavescens; l, Microbacterium gubbeenense; m, Lactobacillus casei; n, Corynebacterium vitaeruminis, Kluyvera ascorbata; o, Clostridium sporogenes; p, Micrococcus spp., Microbacterium spp.; q, unidentified band; r, Corynebacterium ammoniagenes; s, Arthrobacter spp., Brachybacterium tyrofermentans; t, unidentified band; u, Brevibacterium spp.; v, unidentified band; w, Brevibacterium linens (50%); x, unidentified band; y, Brevibacterium casei, Corynebacterium bovis; z, Propionibacterium freudenreichii; a′, Kocuria kristinae, Brevibacterium linens (50%); b′, Propionibacterium acidipropionici, Kocuria spp.; c′, unidentified band; d′, unidentified band, e′, unidentified band. Markers, 1, Bacillus pumilus ATCC 7725; 2, Klebsiella oxytoca ATCC 103434T; 3, Kytococcus sedentarius CNRZ880; 4, Arthrobacter citreus CNRZ928T; 5, Kocuria kristinae CNRZ872; 6, Propionibacterium jensenii Z87. Lane M, DGGE standardization ladder; lane 1 to lane 11, cheese samples from 1 to 11. TTGE*, bands separated by TTGE method.
Biodiversity among low-G+C-percentage species present in Domiati cheeses.
TTGE profiles of the Domiati cheese samples revealed 29 different bands (Fig. 1). Among them, 22 could be potentially identified using the species database, while 7 others (Fig. 1, bands a, b, e, n, w, x, and z) were novel. Results showed different major bands corresponding to Leuconostoc mesenteroides (band k), Lactococcus garvieae (band c), and Aerococcus viridans (band d) (Fig. 1). High band intensities and frequencies for these species may reflect their strong dominance in Domiati cheese samples. The presence of these three species was further confirmed using species-specific PCR tests (Table 3) and/or a cloning and sequencing strategy (Table 4). Other major bands (Fig. 1, bands p, t, u, and w) gave ambiguous assignments using the reference database and were then sequenced for some Domiati samples. They were identified as close relatives of Staphylococcus sciuri (band p), Pediococcus inopinatus and/or Macrococcus caseolyticus (band t), Lactobacillus versmoldensis (band u), and Vibrio spp. (band w) (Table 4). We note that some bands may correspond to different bacterial species (Table 4).
TABLE 3.
Presence of bacterial species in individual Domiati cheese samples using specific primer tests
| Primer-specific species | Presence of bacteria in indicated Domiati cheese samplea
|
Frequency (samples/total)b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| Brevibacterium casei | − | + | + | − | − | − | + | − | − | − | − | 3/11 |
| Brevibacterium linens | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Corynebacterium variabile | + | + | + | + | + | + | + | − | + | + | + | 10/11 |
| Escherichia coli | + | + | + | + | + | + | + | + | − | + | 9/11 | |
| Enterococcus casseliflavus | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Enterococcus durans | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Enterococcus faecalis | + | + | + | − | − | − | − | + | − | − | + | 5/11 |
| Enterococcus faecium | + | − | − | − | − | − | − | + | − | − | − | 2/11 |
| Enterococcus hirae | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Lactococcus garvieae | + | + | + | + | + | + | + | + | + | + | + | 11/11 |
| Lactococcus lactis | + | + | + | + | + | + | + | + | + | + | + | 11/11 |
| Lactococcus lactis subsp. cremoris | + | + | − | + | − | − | − | − | − | − | − | 3/11 |
| Lactococcus lactis subsp. lactis | + | + | + | + | + | + | + | + | + | + | + | 11/11 |
| Lactococcus raffinolactis | − | − | − | − | − | + | − | − | − | − | − | 1/11 |
| Lactobacillus acidophilus | − | + | + | + | − | − | − | + | + | − | − | 5/11 |
| Lactobacillus brevis | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Lactobacillus casei | + | + | + | − | + | + | + | + | + | + | + | 10/11 |
| Lactobacillus gasseri | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Lactobacillus johnsonii | + | + | + | − | + | − | + | − | + | + | + | 8/11 |
| Lactobacillus plantarum | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Leuconostoc citreum | + | + | + | + | + | + | + | − | + | + | + | 10/11 |
| Leuconostoc mesenteroides | + | + | + | + | + | + | + | + | + | + | + | 11/11 |
| Pseudomonas aeruginosa | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Serratia marcescens | + | + | − | − | − | + | − | − | − | − | − | 3/11 |
| Staphylococcus aureus | − | + | − | − | − | − | − | − | − | − | − | 1/11 |
| Staphylococcus chromogenes | − | + | − | − | − | − | − | − | − | − | − | 1/11 |
| Staphylococcus epidermidis | − | + | + | − | − | − | − | − | − | − | − | 2/11 |
| Staphylococcus saprophyticus | + | + | − | − | + | + | + | + | − | + | + | 8/11 |
| Staphylococcus simulans | − | − | + | + | − | − | − | + | − | − | − | 3/11 |
| Staphylococcus xylosus | − | − | − | − | − | − | − | − | − | − | − | 0/11 |
| Streptococcus thermophilus | + | − | + | + | − | − | + | − | + | − | − | 5/11 |
+, positive; −, negative.
Frequency, ratio of numbers of positive samples to numbers of tested cheese samples.
TABLE 4.
Bacterial species identified in Domiati cheese samples using cloning and sequencing strategy
| Band | TTGE or DGGE | Bacterial species or groups assigned by species database | Domiati sample no. | Closest sequence relative species | Identity (%) | GenBank accession no. |
|---|---|---|---|---|---|---|
| c | TTGE | Lactococcus garvieae | 2, 3, 6, | Lactococcus garvieae | 100 | AY699289.1 |
| d | TTGE | Aerococcus viridans | 3, 7 | Aerococcus viridans | 100 | DQ402378.1 |
| g | TTGE | Lactobacillus plantarum, Lactobacillus pentosus | 5 | Lactobacillus plantarum, Lactobacillus pentosus | 100, 100 | DQ860149, AY362458 |
| k | TTGE | Leuconostoc mesenteroides | 1, 3 | Leuconostoc mesenteroides | 99 | AB120036.1 |
| n | TTGE | Unidentified | 9 | Marine sediment bacterium | 99 | AY669172.1 |
| p | TTGE | Staphylococcus xylosus, Latobacillus brevis, Lactobacillus lactis | 7, 8 | Staphylococcus sciuri | 98 | AB233332 |
| s | TTGE | Staphylococcus saprophyticus, Pseudomonas aeruginosa | 6 | Staphylococcus saprophyticus | 99 | AY375294.1 |
| t | TTGE | Pediococcus pentosaceus, Macrococcus caseolyticus | 4 | Pediococcus inopinatus | 100 | AF404741.1 |
| 7 | Macrococcus caseolyticus | 98 | AY15711 | |||
| u | TTGE | Streptococcus uberis, Moraxella bovis | 4, 10 | Lactobacillus versmoldensis | 100 | AJ496791.1 |
| v | TTGE | Enterococcus faecalis, Staphylococcus warneri | 11 | Enterococcus faecalis | 99 | AY850358.1 |
| w | TTGE | Unidentified | 7 | Vibrio spp. | 98 | AY836815.1 |
| 1, 3 | Staphylococcus spp., Staphylococcus arlettae | 100, 100 | DQ888572, DQ872460.1 | |||
| x | TTGE | Unidentified | 9 | Streptococcus parauberis | 100 | AY584477.1 |
| b′ | TTGE | Lactococcus lactis | 10 | Lactococcus lactis subsp. lactis | 100 | AF515224.1 |
| c′ | TTGE | Streptococcus thermophilus | 7 | Streptococcus thermophilus | 100 | CP000024.1 |
| c | DGGE | Citrobacter freundii | 1 | Citrobacter freundii | 93 | AF458082.1 |
| j | DGGE | Unidentified | 6 | Kocuria halotolerans, Rothia spp. | 100, 100 | DQ979377, DQ822568 |
| b′ | DGGE | Propionibacterium acidipropionici, Kocuria spp. | 4 | Kocuria rhizophila | 100 | AF542072.1 |
Lactococcus lactis (band b′) appeared as a relatively major band, as observed from the TTGE profile (Fig. 1). The presence of L. lactis was confirmed in all tested samples using a species-specific PCR assay (Table 3). In most cases, subspecies belonging to the same species (e.g., Lactococcus lactis subsp. lactis and L. lactis subsp. cremoris) could not be differentiated by the TTGE approach (40). Specific PCR tests were then applied to determine which one was more dominant in the Domiati cheese samples. L. lactis subsp. lactis was detected in all 11 samples, whereas L. lactis subsp. cremoris was detected in only three samples (Table 3). The presence of L. lactis subsp. lactis was also confirmed by sequencing of the corresponding bands (Table 4).
Many other bacterial species, belonging to different genera (e.g., Lactococcus, Lactobacillus, Leuconostoc, Enterococcus, Streptococcus, Pseudomonas, and Acinetobacter), were identified as subdominant or minor species in Domiati cheese (Fig. 1). Most of the band assignments using the species database were confirmed by specific PCR tests and/or by cloning and sequencing strategies (Tables 3 and 4). These complementary tests were very useful for identifying precisely the bacteria at the species level. For example, band g was identified as Lactobacillus plantarum and L. pentosus by the reference database and also by band sequencing (Fig. 1 and Table 4), whereas, using the specific PCR tests, L. plantarum was absent (Table 3). Consequently, it supposed that this band might correspond to Lactobacillus pentosus.
Biodiversity among high-G+C-percentage species present in Domiati cheese.
The DGGE profile for Domiati cheese samples (Fig. 2) included 31 different bands. The species database allowed the identification of only 21 bands, whereas the other 10 (Fig. 2, bands d, e, j, q, t, v, x, c′, d′, and e′) could not be identified. The major bands (Fig. 2) putatively identified were those of Kocuria kristinae or Brevibacterium linens (band a′) and Propionibacterium acidipropionici or Kocuria spp. (band b′). A species-specific PCR assay failed to identify B. linens (Table 3), thus making it likely that band a′ was amplified from K. kristinae. Sequencing of the V3 region fragment of band b′ identified K. rhizophila as the species (Table 4). Some other frequent bands of moderate intensity identified Klebsiella pneumoniae (band a), Arthrobacter spp./Brachybacterium tyrofermentans (band s), Corynebacterium variabile (band g), and Propionibacterium freudenreichii (band z) as the corresponding species. Other bacterial species were identified by the DDGE approach (Fig. 2) and confirmed, in some cases, Escherichia coli (band b), Citrobacter freundii (band c), L. casei (band m), and B. casei (band y) (Table 3 and 4).
Enumeration of different bacterial groups.
Several selective media were used to enumerate the different bacterial groups present in Domiati cheese samples (Table 2). The highest bacterial counts (ranging between 7.1 and 7.6 log10 CFU/g) were obtained with MSA medium, which is generally selective for the staphylococcal populations. Total coliforms were recorded as the lowest number of cells (2.1 to 3.8 log10 CFU/g). The counts of salt-tolerant flora ranged between 6.4 and 7.1 log10 CFU/g. In general, the counts of coccal lactic acid bacteria (LAB) (Lactococcus, Streptococcus, Leuconostoc, and Enterococcus), as presented in Table 2, were higher than those of rod members of LAB (mesophilic and thermophilic lactobacilli).
DISCUSSION
Traditional cheeses like Domiati are widely consumed regionally and also contribute to the cultural heritage. To date, little was known about the bacterial communities responsible for fermentation and ripening of Domiati cheese. This is due in part to the sole use of culture-dependent methods for bacterial identification, which provide relatively poor discrimination of species present in a product. In particular, media used for isolation might fail to monitor bacteria that cannot multiply outside the cheese environment (40).
The combination of molecular tools (i.e., PCR-TTGE, PCR-DGGE, species-specific PCR assays, and cloning/sequencing analyses) allowed us to identify 46 different bacterial species that are present in Domiati cheese. Most of the bacteria were first presumably identified by the assignation of the TTGE/DGGE bands to a complex species database (40, 41). But the limitation of this method concerns the species comigrations. Despite sequence differences, melting temperatures of comigrated V3 fragments are similar, and thus, they migrate at the same position in denaturing gels (37). To confirm species assignments or to distinguish between comigrating species, species-specific PCR tests and/or cloning and sequencing were performed. This strategy proved to be very useful to more precisely identify the bacterial species. According to the frequencies and intensities of the bands on TTGE and DGGE gels (TTGE/DGGE methods are considered semiquantitative techniques [53] and generally, band intensities reflect the relative proportion of each species in the total bacterial population [39]), we could differentiate the bacterial population of Domiati cheeses into three groups. (i) The first is composed of dominant bacterial species (they are almost identified in each sample), e.g., Leuconostoc mesenteroides, Lactococcus garvieae, Aerococcus viridans, Lactobacillus versmoldensis, Pediococcus inopinatus/Macrococcus caseolyticus, and Lactococcus lactis subsp. lactis. The bacteria of this group may play the main role in the fermentation and organoleptic properties of Domiati cheeses because of their common presence in the tested samples. (ii) The second group is composed of frequently encountered bacterial species. These bacteria belonged to both LAB species, e.g., Leuconostoc citreum, Lactobacillus casei, Lactobacillus johnsonii, Staphylococcus thermophilus, E. faecalis, E. faecium/Leuconostoc pseudomesenteroides, and to non-LAB species, e.g., Vibrio spp., K. kristinae, C. variabile, K. rhizophila, Arthrobacter spp./B. tyrofermentans, plus numerous species of coagulase-negative staphylococci. (iii) The third group consists of occasionally encountered bacterial species, e.g., Lactococcus raffinolactis, Acinetobacter lwoffii, Staphylococcus lentus, S. chromogenes, Enterobacter cloacae, and Klebsiella oxytoca. The bacteria of the second and third groups may have a secondary activity in the fermentation process of Domiati cheeses, but the origins and potential roles of these as well as the bacteria of the first group would need further investigation.
Domiati cheese manufacture involves several technological steps, including natural fermentation of milk, salting, renneting, and ripening (pickling) in brine or salted whey solutions. The salt added to cheese milk plays an important role during manufacturing by favoring or inhibiting bacterial growth (the salt content in our cheese samples ranged between 5.46 and 9.50% of wet weight). Many of the identified bacterial species could be recognized as partially or totally salt-tolerant bacteria, e.g., Leuconostoc mesenteroides, Lactobacillus versmoldensis (up to 14% salt tolerance) (28), Lactococcus lactis subsp. lactis, Staphylococcus saprophyticus, K. rhizophila, K. halotolerans, Brachybacterium tyrofermentans (up to 16% salt tolerance) (48), K. kristinae, and Microbacterium gubbeenense. In some cases, the identified species were directly related to a marine environment, e.g., Lactococcus garvieae (which often occurs in dairy environments [3, 12, 23] and is considered one of the major pathogens responsible for fish mortality [44]), Aerococcus viridans, Vibrio spp., and marine sediment bacterium. As previously described (47), our results indicate that salt-tolerant and marine bacteria may play a role in the ripening process of Domiati cheese. Other studies of red-smear soft cheeses have demonstrated that a fraction of cheese flora was composed of microorganisms related to a marine environment (22, 32).
This study allows us to clarify reports of the occurrence of staphylococcal species in Domiati cheese, as numerous species have been identified by molecular tools. These results reflected the high bacterial counts using MSA medium (7.1 to 7.6 log10 CFU/g). Previous studies focused on the occurrence of Staphylococcus aureus because of its important role in food poisoning. Our results indicate a relative predominance of S. sciuri and S. saprophyticus (both coagulase-negative staphylococci) in Domiati cheese. Fortunately, S. aureus was detected in only one cheese sample, indicating either that good sanitation procedures were applied during Domiati cheese manufacturing or that S. aureus is not a good competitor with other bacterial species (36). This study also revealed the presence of other coagulase-negative staphylococci: S. simulans and S. chromogenes were identified for the first time in Domiati cheese. These species are common causes of subclinical mastitis (27, 29). However, it is notable that S. simulans produces lysostaphin, a cell wall-degrading enzyme that lyses practically all known staphylococcal species (45), and was recently developed for its bactericidal antistaphylococcal effects (49). Its presence might actually control development of populations of susceptible staphylococci in Domiati cheese. Other bacterial species known to cause mastitis were also identified (e.g., Streptococcus parauberis, Staphylococcus epidermidis, Citrobacter freundii, and Serratia marcescens) but were detected as minor species.
In the present study, several bacterial species (never previously isolated by culturing from Domiati cheese) were detected for the first time in Domiati cheese samples, e.g., LAB (Lactococcus garvieae, A. viridans, Pediococcus inopinatus, Lactobacillus versmoldensis, L. johnsonii, Leuconostoc citreum, and S. parauberis), and non-LAB (Pseudomonas putida, Staphylococcus sciuri, S. chromogenes, S. simulans, Macrococcus caseolyticus, Citrobacter freundii, Corynebacterium variabile, Microbacterium. gubbeenense, Propionibacterium freudenreichii, K. rhizophila, K. halotolerans/Rothia spp., and marine sediment bacterium). These results confirm interest in using the molecular methods for exhaustive and precise identification of complex microbial biodiversity occurring in artisanal cheeses such as Domiati.
The considerable bacterial biodiversity found in Domiati cheeses may explain the heterogeneous production of cheeses available in Egyptian markets. It is expected that each bacterial species present in Domiati cheese may contribute in some way to the ripening process but would vary according to its tolerance toward the probably highly variable cheese production conditions (e.g., salt concentrations, pH, ripening temperatures, and type of pickling solutions [whey or brine]).
Although traditional Domiati cheese is made from raw milk without the addition of any starters, most of the bacteria identified as dominant belonged to LAB. Nevertheless, improvements in Domiati cheese manufacture may be necessary to obtain a safe and homogenous product. This would require the systematic use of clean raw materials, controlled manufacturing steps, and selection of the appropriate LAB for the ripening process. The origin and role of the salt-tolerant and marine bacteria present in Domiati cheese should be further investigated. Finally, the application of culture methods will be valuable for the isolation of “positive” bacteria that characterize Domiati cheese for future use as cheese starter cultures.
Acknowledgments
This research was carried out during the scientific mission granted by AUF (Agence universitaire de la Francophonie).
We are very grateful to Alexandra Gruss for reading and correcting the English of the manuscript and also to Marina Aigle for very useful technical assistance. G. El-Baradei thanks Pierre Pery for support and encouragement; colleagues at UBLO and VIM Units, INRA, Jouy-en-Josas, France; S. E. El-Rakshy, M. H. Ragab, and the director of the Middle East Bureau (AUF); and the teams in Alexandria, Egypt, and Beirut, Lebanon.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Abou-Donia, S. A. 1986. Egyptian Domiati soft white pickled cheese. N. Z. Dairy Sci. Technol. 21:167-190. [Google Scholar]
- 2.Aly, S., and E. A. Galal. 2002. Effect of milk pretreatment on the keeping quality of Domiati cheese. Pakistan J. Nutr. 1:132-136. [Google Scholar]
- 3.Blaiotta, G., O. Pepe, G. Mauriello, F. Villani, R. Andolfi, and G. Moschetti. 2002. 16S-23S rDNA intergenic spacer region polymorphism of Lactococcus garvieae, Lactococcus raffinolactis and Lactococcus lactis as revealed by PCR and nucleotide sequence analysis. Syst. Appl. Microbiol. 25:520-527. [DOI] [PubMed] [Google Scholar]
- 4.Cibik, R., E. Lepage, and P. Talliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 5.Coeuret, V., S. Dubernet, M. Bernardeau, M. Gueguen, and J. P. Vernoux. 2003. Isolation, characterization and identification of lactobacilli focusing mainly on cheeses and other dairy products. Lait 83:269-306. [Google Scholar]
- 6.Coppola, S., G. Blaiotta, D. Ercolini, and G. Moschetti. 2001. Molecular evaluation of microbial diversity occurring in different types of Mozzarella cheese. J. Appl. Microbiol. 90:414-420. [DOI] [PubMed] [Google Scholar]
- 7.de los Reyes-Gavilán, C., G. K. Y. Limsowtin, P. Tailliez, L. Sechaud, and J.-P. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Effat, B. A., I. M. Hosny, and N. M. Dabiza. 2000. Occurrence of Aeromonas hydrophila and its growth in Egyptian soft cheese. Egypt. J. Dairy Sci. 28:1-12. [Google Scholar]
- 9.El-Baradei, G., H. Mahmoud, and I. El-Sayed. 1999. Metachromatic agar-diffusion as a simple method for detection of staphylococcal thermonuclease in Domiati cheese. Alex. J. Agric. Res. 44:143-149. [Google Scholar]
- 10.El-Baradei, G., J. C. Ogier, A. Delacroix-Buchet, and P. Pery. 2004. Analysis of bacterial communities of Egyptian dairy products using molecular fingerprinting tools. II. Mish cheese, p. 35-37. In M. H. Abd El-Salam, A. M. Kholif, S. M. Abdou, and S. El-Shibiny (ed.), Proceedings of the 9th Egyptian Conference for Dairy Science and Technology. Egyptian Society of Dairy Science, Cairo, Egypt.
- 11.El-Baradei, G., A. Delacroix-Buchet, P. Pery, and J. C. Ogier. 2005. Identification of bacterial communities of Egyptian Karish cheese using molecular fingerprinting tools. Egypt. J. Dairy Sci. 33:25-34. [Google Scholar]
- 12.El-Baradei, G., A. Delacroix-Buchet, P. Pery, and J. C. Ogier. 2005. Occurrence of Lactococcus garvieae in four types of Egyptian cheeses by specific polymerase chain reaction assay. Egypt. J. Dairy Sci. 33:35-41. [Google Scholar]
- 13.El-Baradei, G., A. Delacroix-Buchet, and J. C. Ogier. 2006. Identification of bacterial ecosystems of Egyptian Ras cheese using molecular biology fingerprinting tools, p. 89-90. In V. Juillard and J.-C. Piard (ed.), Proceedings of 14ème colloque du Club des Bactéries Lactiques (CBL), Mai 2006. CBL, Paris, France.
- 14.El-Gendy, S. M., A. F. El-Erian, and O. M. El-Demerdash. 1980. Morphological characters and nutritional requirements of isolated dairy Arthrobacters. Assiut Vet. Med. J. 7:161-174. [Google Scholar]
- 15.El-Soda, M., M. El-Ziney, S. Awad, G. Osman, N. Omran, G. Gamal, N. Ezzat, and H. El-Shafei. 2003. A culture collection of lactic acid bacteria isolated from raw milk and traditional Egyptian dairy products. Egypt. J. Dairy Sci. 31:217-226. [Google Scholar]
- 16.El-Zayat, A. I., A. Goda, E. El-Bagoury, J.-P. Dufour, S. Collin, and M. Osman. 1995. Bacteriological studies on Domiati cheese. Egypt. J. Dairy Sci. 23:239-247. [Google Scholar]
- 17.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 18.Ercolini, D., P. J. Hill, and C. E. R. Dodd. 2003. Bacterial community structure and location in Stilton cheese. Appl. Environ. Microbiol. 69:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ercolini, D., G. Mauriello, G. Blaiotta, G. Moschetti, and S. Coppola. 2004. PCR-DGGE fingerprints of microbial succession during a manufacture of artisanal water buffalo mozzarella cheese. J. Appl. Microbiol. 96:263-270. [DOI] [PubMed] [Google Scholar]
- 20.Fahmy, T. K., and L. M. Youssef. 1978. Incidence of micrococci in Domiati cheese. Agric. Res. Rev. 56:145-148. [Google Scholar]
- 21.Fahmy, T. K., and L. M. Youssef. 1978. Incidence of streptococci and lactobacilli in Domiati cheese. Agric. Res. Rev. 56:149-152. [Google Scholar]
- 22.Feurer, C., F. Irlinger, H. E. Spinnler, P. Glaser, and T. Vallaeys. 2004. Assessment of the rind microbial diversity in a farmhouse-produced vs a pasteurized industrially produced soft red-smear using both cultivation and rDNA-based methods. J. Appl. Microbiol. 97:546-556. [DOI] [PubMed] [Google Scholar]
- 23.Flórez, A. B., and B. Mayo. 2006. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 110:165-171. [DOI] [PubMed] [Google Scholar]
- 24.Forsman, P., A. Tilsala-Timisjarvi, and T. Alatossava. 1997. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 143:3491-3500. [DOI] [PubMed] [Google Scholar]
- 25.Furet, J. P., P. Quenee, and P. Talliez. 2002. Identification des bactéries lactiques par PCR quantitative. Sci. Aliments 22:33-44. [Google Scholar]
- 26.Guarneri, T., L. Rossetti, and G. Giraffa. 2001. Rapid identification of Lactobacillus brevis using the polymerase chain reaction. Lett. Appl. Microbiol. 33:377-381. [DOI] [PubMed] [Google Scholar]
- 27.Jarp, J. 1991. Classification of coagulase-negative staphylococci isolated from bovine clinical and subclinical mastitis. Vet. Microbiol. 27:151-158. [DOI] [PubMed] [Google Scholar]
- 28.Krockel, L., U. Schillinger, C. M. Franz, A. Bantleon, and W. Ludwig. 2003. Lactobacillus versmoldensis sp. nov., isolated from raw fermented sausage. Int. J. Syst. Evol. Microbiol. 53:513-517. [DOI] [PubMed] [Google Scholar]
- 29.Kudinha, T., and C. Simango. 2002. Prevalence of coagulase-negative staphylococci in bovine mastitis in Zimbabwe. J. S. Afr. Vet. Assoc. 73:62-65. [DOI] [PubMed] [Google Scholar]
- 30.Lee, H. J., S. Y. Park, and J. Kim. 2000. Multiplex PCR-based detection and identification of Leuconostoc species. FEMS Microbiol. Lett. 15:243-247. [DOI] [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (ribosomal database project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maoz, A., R. Mayr, and S. Scherer. 2003. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 69:4012-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martineau, F., F. J. Picard, L. Grenier, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT Trial. J. Antimicrob. Chemother. 46:527-534. [DOI] [PubMed] [Google Scholar]
- 34.Mauriello, G., L. Moio, A. Genovese, and D. Ercolini. 2003. Relationships between flavoring capabilities, bacterial composition, and geographical origin of natural whey cultures used for artisanal water-buffalo mozzarella cheese manufacture. J. Dairy Sci. 86:486-497. [DOI] [PubMed] [Google Scholar]
- 35.Mayeux, J. V., W. E. Sandine, and P. R. Elliker. 1962. A selective medium for detecting Leuconostoc in mixed-strain starter cultures. J. Dairy Sci. 45:655. [Google Scholar]
- 36.Mossel, D. A. A., and P. Van Netten. 1990. Staphylococcus aureus and related staphylococci in foods: ecology, proliferation, toxinogenesis, control and monitoring. Soc. Appl. Bacteriol. Symp. Ser. 19:S123-S145. [DOI] [PubMed] [Google Scholar]
- 37.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer, G. 1999. DGGE/TTGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 39.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogier, J.-C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis, Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogier, J.-C., V. Lafarge, V. Girard, A. Rault, V. Maladen, A. Gruss, J-Y. Leveau, and A. Delacroix-Buchet. 2004. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:5628-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu, Z. Y., M. Dobos, G. K. Limsowtin, and I. B. Powell. 2002. Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the gram-positive bacterial genus Lactococcus. J. Appl. Microbiol. 93:353-361. [DOI] [PubMed] [Google Scholar]
- 43.Randazzo C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravelo, C., B. Magariños, S. López-Romalde, A. Toranzo, and J. L. Romalde. 2003. Molecular fingerprinting of fish-pathogenic Lactococcus garvieae strains by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 41:751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recsei, P., A. Gruss, and R. P. Novick. 1987. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc. Natl. Acad. Sci. USA 84:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riffon, R., K. Sayasith, H. Khalil, P. Dubreuil, M. Drolet, and J. Lagace. 2001. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J. Clin. Microbiol. 39:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roushdy, I. M., M. A. Ehrmann, and R. F. Vogel. 1998. Molecular identification and characterization of halo-tolerant lactic acid bacteria isolated from soft pickled “Domiati” cheese. Adv. Food Sci. 20:40-45. [Google Scholar]
- 48.Schubert, K., W. Ludwig, N. Springer, R. M. Kroppenstedt, J. P. Accolas, and F. Fiedler. 1996. Two coryneform bacteria isolated from the surface of French Gruyere and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. Int. J. Syst. Bacteriol. 46:81-87. [DOI] [PubMed] [Google Scholar]
- 49.von Eiff, C., J. F. Kokai-Kun, K. Becker, and G. Peters. 2003. In vitro activity of recombinant lysostaphin against Staphylococcus aureus isolates from anterior nares and blood. Antimicrob. Agents Chemother. 47:3613-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, V. L., B. C. Tatford, X. Yin, S. C. Rajki, M. M. Walsh, and P. LaRock. 1999. Species-specific detection of hydrocarbon utilizing bacteria. J. Microbiol. Methods 39:59-78. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X., R. L. Kilmer, and A. Muhammad. 2003. A descriptive analysis of Egypt and Saudi Arabia who import United States dairy products, monograph MGTC 03-8. International Agricultural Trade and Policy Center, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL.
- 53.Zoetendal, E. G., C. T. Collier, S. Koike, R. I. Mackie, and H. R. Gaskins. 2004. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 134:465-472. [DOI] [PubMed] [Google Scholar]