Abstract
Biosurfactant-mediated oil recovery may be an economic approach for recovery of significant amounts of oil entrapped in reservoirs, but evidence that biosurfactants can be produced in situ at concentrations needed to mobilize oil is lacking. We tested whether two Bacillus strains that produce lipopeptide biosurfactants can metabolize and produce their biosurfactants in an oil reservoir. Five wells that produce from the same Viola limestone formation were used. Two wells received an inoculum (a mixture of Bacillus strain RS-1 and Bacillus subtilis subsp. spizizenii NRRL B-23049) and nutrients (glucose, sodium nitrate, and trace metals), two wells received just nutrients, and one well received only formation water. Results showed in situ metabolism and biosurfactant production. The average concentration of lipopeptide biosurfactant in the produced fluids of the inoculated wells was about 90 mg/liter. This concentration is approximately nine times the minimum concentration required to mobilize entrapped oil from sandstone cores. Carbon dioxide, acetate, lactate, ethanol, and 2,3-butanediol were detected in the produced fluids of the inoculated wells. Only CO2 and ethanol were detected in the produced fluids of the nutrient-only-treated wells. Microbiological and molecular data showed that the microorganisms injected into the formation were retrieved in the produced fluids of the inoculated wells. We provide essential data for modeling microbial oil recovery processes in situ, including growth rates (0.06 ± 0.01 h−1), carbon balances (107% ± 34%), biosurfactant production rates (0.02 ± 0.001 h−1), and biosurfactant yields (0.015 ± 0.001 mol biosurfactant/mol glucose). The data demonstrate the technical feasibility of microbial processes for oil recovery.
Oil is an essential source of energy and one of the main factors that drive the economic development of the world (16). Current oil production technologies recover only about one-third to one-half of the oil originally present in an oil reservoir (16, 26, 37). The exploitation of oil resources in existing mature reservoirs is essential for meeting future energy demands as the number of oil-exporting countries dwindles. A key to exploiting this untapped resource is to overcome the capillary forces that entrap oil in small pores within the reservoir. Enhanced oil recovery (EOR), i.e., the use of heat, chemicals such as surfactants, microbial processes, and miscible gas injection (15), has the potential to recover a significant portion of this entrapped oil. However, oil recovered by EOR constitutes less than 10% of the total amount of oil produced in the United States (http://www.fe.doe.gov/programs/oilgas/eor/). Interfacial tension between the hydrocarbon and aqueous phases is largely responsible for trapping the hydrocarbon in the porous matrix, and a reduction of several orders of magnitude in interfacial tension is needed for hydrocarbon mobilization (1, 15, 47). To achieve large reductions in interfacial tension, surfactant concentrations significantly above that needed to form micelles (i.e., the critical micelle concentration) are required (7, 41, 42). The high chemical costs have prevented the widespread use of surfactants for enhanced oil recovery.
Microbially enhanced oil recovery (MEOR) processes employ the use of microbial metabolites, such as biosurfactants, to lower interfacial tension between brine and oil and hence mobilize entrapped oil (2, 8, 24, 32). Several biosurfactants, in particular the lipopeptides made by Bacillus species, generate the low interfacial tensions between the hydrocarbon and the aqueous phases required to mobilize residual hydrocarbon (25, 30). MEOR has several advantages compared to other EOR processes in that it does not consume large amounts of energy, as do thermal processes, nor does it depend on the price of crude oil, as do many chemical processes (2, 31, 32). MEOR can also be cost-effective, since microbial products can be produced from inexpensive, renewable resources, and several MEOR processes have been shown to produce incremental oil for about $19 per m3 ($3 per barrel) (6, 9, 31, 32). Nevertheless, microbial processes have always been viewed with considerable skepticism for a number of reasons. First, the lack of quantitative information regarding the reaction rates, stoichiometries, product concentrations, and yields needed to simulate the performance of microbial processes makes it difficult to extrapolate the results for a given field test to those for other reservoirs (31, 32). Second, it is not clear whether microbial processes can generate the necessary metabolites in sufficient quantities and at rates needed to mobilize entrapped oil in oil reservoirs (10, 31, 32). Third, technical performances in many field trials have been inconsistent (14, 23). Last, it is unclear whether the microbial strains used as inocula actually grow and metabolize in the reservoir (31, 32).
Here, we show in a well-controlled field experiment that biosurfactants are produced in situ in amounts sufficient to mobilize substantial amounts of entrapped oil. We provide, for the first time, data on in situ product concentrations and yields, rates of growth, substrate utilization, and metabolite formation and an excellent carbon mass balance. This quantitative information shows that biosurfactant-mediated oil recovery is technically feasible and will facilitate the use of computer simulations to determine the efficacy of MEOR in different reservoirs.
MATERIALS AND METHODS
Preparation of the inoculum.
Two halotolerant biosurfactant producers, Bacillus strain RS-1 and Bacillus subtilis subsp. spizizenii strain NRRL B-23049, were used as the inoculum. Both strains grow in medium with 5% NaCl. Bacillus strain RS-1 and Bacillus subtilis subsp. spizizenii strain NRRL B-23049 were each grown in 200 ml of medium E (48, 49). When the culture reached the late exponential phase of growth, it was used to inoculate a 10-liter carboy of the same medium, which was incubated at 37°C for 48 h. Agitation and aeration were maintained by using glass gas dispersion tubes with fritted cylinders (Fisher Scientific, Inc.). The cells were concentrated by using a tangential membrane flow system (0.45-μm-pore-size filter) (Millipore, Bedford, MA) to yield on average 2 liters of cell concentrate from each 10-liter carboy. The concentrated cells were stored at 4°C. One liter of cell concentrate was used to inoculate a 132-liter tank with the following components (in grams per liter of tap water): dibasic potassium phosphate (1.2 g/liter), monobasic potassium phosphate (0.23 g/liter), sucrose (8.6 g/liter), sodium chloride (8.6 g/liter), sodium nitrate (0.86 g/liter), and Saccharomyces cerevisiae extract (0.86 g/liter). The medium was prepared septically due to the lack of facilities on site, and tanks were incubated for approximately 48 h at an ambient temperature.
Field experiment.
A push-pull test was conducted on five oil production wells in the Bebee field (Section 19, T5N, R5E; Pontotoc City, OK) that produce from the same formation (a Viola limestone). Two wells received an inoculum of the Bacillus strains and nutrients, two wells received just nutrients, and one well received an equivalent volume of formation water and served as the negative control. Each well that was inoculated received 396 liters of Bacillus strain RS-1 and 264 liters of B. subtilis subsp. spizizenii NRRL B-23049. The nutrient package consisted of 79.5 kg of glucose; 7.9 kg of sodium nitrate; 19.9 g of magnesium sulfate; 2.0 g each of manganese sulfate, zinc sulfate, and iron sulfate; 0.2 g each of copper sulfate, aluminum potassium sulfate, boric acid, and sodium molybdate; 0.1 g of sodium selenate; and 0.6 g of nickel chloride per well. Each well that received nutrients also received fluorescein (125 g) and sodium bromide (2 kg). Fluorescein was used to detect visually (by a green signal) the production of the slug and to determine when to start and stop sampling. Bromide served as the conservative tracer to account for dilution in the reservoir. The nutrients, inoculum, and tracers were mixed with formation water (∼8,000 liters) by circulation supplied by the pump truck. The formation water was obtained from a storage tank located near the production wells. Each well received an initial injection (preflush) of ∼1,600 liters (10 barrels) of formation water and an injection of ∼8,000 liters (50 barrels) of the treatment (nutrients and cells, nutrients only, or formation water), followed by an injection of ∼8,000 liters (50 barrels) of formation water as a postflush treatment, to make a total of ∼17,500 liters (110 barrels) of fluids injected per well. The postflush treatment was used to move the nutrient package 1.2 to 2.4 m into the formation. After injection of the treatment package, production from each well was stopped for 108 h to allow time for growth and metabolism to occur in the formation. After this incubation period, the pump was started and samples from each of these wells were collected with time.
Flow meters were attached to the tubing of three of the five wells to measure the volume of fluids produced. The total volume produced was recorded when each sample was collected. Since all the wells were set to pump at almost the same rate, the volume of fluid produced during a given time interval in the two wells that did not have flow meters was estimated from the average volume produced in the three wells that had the flow meters attached.
Sampling.
Samples for chemical and microbiological analyses were collected on several occasions prior to treatment and for a 32-hour period after production recommenced after the 108-hour incubation period. Each sample was collected in a 2-liter glass bottle that was allowed to overflow to minimize contact with air. The temperature of the sample was immediately recorded with a handheld probe. A sample for chemical analyses was filtered through a 0.45-μm membrane filter to remove particulate material and oil. The remainder of the unfiltered sample was used for measurement of oil-spreading activity and for microbiological enumerations. All samples were stored on ice until analyzed. The analyses for pH, conductivity, nitrate, nitrite, ammonium, alkalinity, oil spreading, and surface tension were completed on site within 2 hours after sample collection. For the other analyses, the samples were transported back to the laboratory and stored at 4°C until analyzed. All measurements for each sample were done in duplicate unless otherwise indicated.
Detection of biosurfactant production.
The biosurfactant activities of the unfiltered samples were measured by using the oil-spreading technique (33, 49). The diameter of the clear zone on the oil surface was measured in triplicate for each sample. Biosurfactant activity was defined as the diameter of the clearing on the oil surface in centimeters. The surface tensions of the filtered samples were measured with a Du Nuoy ring tensiometer (Fisher Scientific Inc., Hampton, NH) calibrated with water as the high-surface-tension standard and isopropanol as the low-surface-tension standard (25, 30). The lipopeptide biosurfactant was quantified by using high-performance liquid chromatography (HPLC) with a reversed-phase C18 column (250-mm length by 1.5-mm inside diameter [ID]) and 60% acetonitrile in water as the mobile phase (48). Twenty microliters of 1:4 and 1:2 dilutions of filtered samples was injected onto the column. The retention times for the biosurfactant were 2, 2.3, and 3.1 min, corresponding to three different fatty acid tails of the lipopeptide. The peak areas of the three peaks were added together, and the concentration was calculated from standard curves prepared in a similar manner with surfactin (Sigma Chemical Co., St. Louis, MO) and the highly purified lipopeptides (48) produced by each of the two microorganisms, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049. The standard curve was linear up to 500 mg/liter of the lipopetide. The lipopeptides detected in the samples had the same retention times as the highly purified biosurfactant obtained from cultures of each inoculum strain.
Fermentation analyses.
A modified orcinol-H2SO4 method was used to determine the amount of glucose in each sample (45). Acetate, ethanol, and 2,3-butanediol were measured by using gas chromatography with an 80/120 Carbopack B-DA*/4% Carbowax 20 M (2-m length by 2-mm ID) glass column (Sigma Chemical Co., St. Louis, MO). Helium was used as the carrier gas at a flow rate of 24 ml/min (18, 22). The injector temperature was 200°C; the flame ionization detector was set at 180°C. The column temperature was kept constant at 155°C for 3.5 min and then increased to 180°C at 30°C/min. The temperature was then held at 180°C for 10 min. One microliter of the sample diluted in 30 mM oxalic acid was injected onto the column. Oxalic acid was added to the samples in order to condition the column according to the manufacturer's recommendation. Lactate was measured by using HPLC with an Alltech Prevail organic acid column (250-mm length by 1.5-mm ID) (Alltech Associates, Deerfield, IL) and 25 mM KH2PO4 (pH 2.5) as the mobile phase according to the manufacturer's instructions. A 50-μl aliquot of a 1:10 dilution of the sample was used. The above-mentioned metabolites were identified and quantified by comparison of their retention times and peak areas, respectively, with those of known standards.
To test whether the two Bacillus strains produced the same fermentation products from glucose as those detected in the produced fluids of the inoculated wells, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049 were each grown anaerobically in duplicate serum bottles containing 75 ml formation water supplemented with glucose, sodium nitrate, and metals at the same concentration as that used for the field experiment. The cultures were incubated at 37°C without shaking for 48 h. A three-series most-probable-number (MPN) technique (see below) was used to enumerate the number of viable cells immediately after inoculation and after 48 h of incubation.
Other chemical analyses.
The pHs were measured for the filtered samples by using a handheld pH/conductivity meter (EXTECH Instruments, Waltham, MA). Nitrate, nitrite, ammonium, and alkalinity (in mg/liter) were measured colorimetrically by using Hach kits (Hach Chemical Co., Loveland, CO) according to the manufacturer's instructions. The alkalinity values and pHs were used to calculate the CO2 concentrations in the samples by using the Henderson-Hasselbach equation, pH = pKa + log (value for ionized species/value for un-ionized species). Bromide in filtered samples was analyzed by using liquid chromatography with an analytical anion exchange column (IonPac AS4A-SC, 4 by 250 mm; Dionex Corporation, CA) (39). The concentrations of bromide in the samples were calculated from the standard curve of NaBr.
Microbiological analysis.
A three-series MPN technique was used to enumerate total heterotrophic bacteria, spore-forming bacteria, and halotolerant bacteria in unfiltered samples from both the injected and the produced fluids. The procedure was modified for use of 96-well plates. Three columns of wells were used for each sample, which was serially 10-fold diluted from 10−1 to 10−5.
The physiological properties of Bacillus species, e.g., biofilm formation (17), sporulation (17), and halotolerance (32), allowed the use of specific media and/or manipulations for the MPN analysis as follows. Bacillus biofilm growth medium (BBGM) (17) was used to enumerate heterotrophic bacteria and to promote biofilm formation since biosurfactant production has been associated with biofilm formation in Bacillus species (5, 19, 44). Since Bacillus species are known to sporulate (17), a portion of each sample was heat treated at 85°C for 20 min and then diluted in BBGM to estimate the number of spore-forming bacteria. Finally, to estimate the number of halotolerant bacteria and to avoid underestimation of Bacillus species due to heat treatment, plate count broth (Difco Laboratories Inc., Detroit, MI) with 5% NaCl was used. To estimate the number of biosurfactant producers, 5 μl of sterile crude oil was added to the surface of the medium in all wells where growth was observed and the dissipation of the oil drop was noted. The MPNs of biosurfactant producers in the samples were estimated from those wells where the oil dissipated on the surface. Published tables (www.fsis.usda.gov/Ophs/Microlab/Appendix2.02.pdf) were used to calculate the MPNs.
Samples (50 μl) from the injected and produced fluids of each well were patch inoculated onto blood agar plates and incubated overnight at 37°C. Multiple blood agar plates were subsequently streaked from the growth patch. Resulting beta-hemolytic (12) clearing zones were picked and streak purified until pure isolates were obtained. The beta-hemolytic isolates with the same colony morphologies as those of Bacillus strain RS-1 or Bacillus subtilis subsp. spizizenii NRRL B-23049 were used for further culture-dependent molecular analyses (see below).
Molecular analysis.
Formation water samples (2 liters) before and after treatment were vacuum filtered on a polyethersulfonate membrane with a 90-mm diameter and a 0.2-μm pore size (VWR, West Chester, PA) after oil separation. Membranes containing the microorganisms were cut and used for culture-independent DNA extraction. Cells were lysed by rapid thawing and bead beating to shear the membrane into small pieces (typically less than 5 mm), followed by mixing with stool lysis buffer (QIAGEN, Valencia, CA), vortexing for 2 min, and incubating at 95°C for 10 min in a water bath. Using a modified QIAamp DNA stool mini kit protocol (QIAGEN, Valencia, CA), DNA was extracted from lysed cells and amplified in a Taq DNA PCR, using degenerate primers designed to hybridize with the gyrA gene sequences of a variety of Bacillus strains (13, 40). PCR products were gel extracted (QIAGEN), pGEM cloned (Promega), and plasmid prepped (QIAGEN). Four clones from each gyrA amplicon were sequenced by the Oklahoma Medical Research Foundation (OMRF; Oklahoma City, OK). Sequences were analyzed by DNAMAN multiple sequence alignment (Lynnon Biosoft) with the sequences obtained for Bacillus strain RS-1 (GenBank accession number DQ995270) and Bacillus subtilis subsp. spizizenii strain NRRL B-23049 (GenBank accession numbers DQ995271 and AF272020).
Cell templates from isolates obtained on blood agar plates (see above) were utilized as a source of culture-dependent DNA for 16S rRNA gene (34, 35) and gyrA analyses as well as repetitive extragenic palindromic (REP)-PCR (21, 46). Three clones of each gene from each isolate were prepared and analyzed as described above. Consensus sequences were obtained by multiple sequence alignment and compared with those from other isolates and the strains used as the inoculum. The GenBank accession numbers for the 16S rRNA gene (551 bp) of Bacillus strain RS-1 and Bacillus subtilis subsp. spizizenii strain NRRL B-23049 are DQ995269 and AF074970, respectively. REP-PCRs were resolved first on a 1% agarose gel and subsequently on a 5% polyacrylamide gel with 0% denaturant at 75 V for 3.5 h at 60°C.
Calculation of total recoveries.
The total amount of glucose utilized (CST) in a well was calculated from the equation 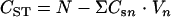 , and the total amount of each metabolite produced (CPT) was calculated from the equation
, and the total amount of each metabolite produced (CPT) was calculated from the equation 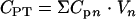 , where N is the number of moles of glucose injected in the well,
, where N is the number of moles of glucose injected in the well, 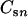 or
or 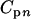 is the molar concentration of glucose or metabolites, respectively, in the produced fluid of the nth sample collected from the well, and Vn is the volume of fluid produced during the time interval between the collection of samples n − 1 and n from the well. The total MPN of cells (CXT) was calculated from the equation
is the molar concentration of glucose or metabolites, respectively, in the produced fluid of the nth sample collected from the well, and Vn is the volume of fluid produced during the time interval between the collection of samples n − 1 and n from the well. The total MPN of cells (CXT) was calculated from the equation 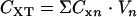 , where
, where 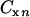 is the MPN per liter of cells in the produced fluid of the nth sample, and Vn is the volume of fluid produced during the time interval between the collection of samples n − 1 and n.
is the MPN per liter of cells in the produced fluid of the nth sample, and Vn is the volume of fluid produced during the time interval between the collection of samples n − 1 and n.
Bromide was used as a conservative tracer to estimate the amount of dilution of the nutrient package by dispersion in the formation (4, 39). The bromide recovery factor was calculated for each of the three wells that had total volumetric flow meters attached. The average bromide recovery factor from the three wells was 1.09 (24% variation), which indicated that little dispersion or adsorption of the tracer occurred. The values for CST, CPT, and CXT from these three wells were divided by the corresponding bromide recovery factors obtained for those wells. For the other two wells, the average bromide recovery factors were used. The corrected numbers of moles of glucose utilized and moles of end products were used for percent carbon recovery calculations.
RESULTS
Preinjection data.
Samples were collected from all the wells 1 and 2 weeks prior to the treatment. The biosurfactant and common bacterial fermentation end products, such as acids and alcohols, were not detected in any of the wells prior to treatment (Table 1) . In addition, we did not detect any viable bacteria at any dilution used for the MPN enumerations (Table 1).
TABLE 1.
Preinjection chemical and microbial analyses for all the wellsa
| Well group | Temp (°C) | pH | Glucose (μg/liter) | Nitrate (mg/liter) | Ammonium (mg/liter) | Bromide (μM) | Alkalinity (mg/liter) |
|---|---|---|---|---|---|---|---|
| Inoculatedb | 27.9 ± 1.2 | 7.54 ± 0.55 | 4.22 ± 5.7 | 11.3 ± 2.4 | 8.12 ± 3.75 | 1.62 ± 0.02 | 745 ± 34 |
| Nutrient only treatedb | 27.8 ± 3 | 7.4 ± 0.38 | 4.2 ± 4.3 | 11.8 ± 2.4 | 6.25 ± 1.4 | 1.64 ± 0.03 | 710 ± 66 |
| Negative controlc | 28.8 ± 0.3 | 7.6 ± 0.43 | 1.97 ± 1.34 | 9.5 ± 3 | 7.5 ± 5 | 1.62 ± 0.01 | 720 ± 280 |
Acetate, lactate, ethanol, 2,3-butanediol, nitrite, the biosurfactant, total heterotrophic bacteria, and biosurfactant producers were not detected in any of the samples before injection. The detection limits for acetate, ethanol, and butanediol were 0.1 mM. The detection limit for lactate was 0.2 mM. The biosurfactant detection limit was 10 mg/liter.
Numbers are averages ± standard deviations for four replicates (two samples collected for each of two wells within a 1-week interval).
Numbers are averages ± ranges for two samples collected within a 1-week interval.
Evidence that the injected strains were maintained in the inoculated wells.
MPN analysis using the BBGM showed biosurfactant producers in the injected fluids of both the inoculated wells and the nutrient-only-treated wells (Table 2). The total number of biosurfactant producers (CXT) in the injected fluids was 1 order of magnitude higher for the inoculated wells than for the nutrient-only-treated wells (4.2 × 1010 compared to 3.7 × 109) (Table 2). No spore-forming microorganisms were detected in either the injected or the produced fluids of the nutrient-only-treated wells or the negative control. Since Bacillus species are known to sporulate (17), the absence of spore-forming microorganisms in the nutrient-only-treated wells indicates that the biosurfactant producers that were introduced into these wells were probably not Bacillus species but microorganisms present in the storage tank. On the other hand, in the inoculated wells, spore-forming, biosurfactant-producing microorganisms were both introduced in and retrieved from the injected and produced fluids, respectively (Table 2). The CXT values for spore-forming biosurfactant producers in the injected and produced fluids of the inoculated wells were not significantly different (Table 2). These data indicate that biosurfactant producers in the inoculum maintained viability but did not grow during the incubation period.
TABLE 2.
MPN analysis of total and spore-forming heterotrophic bacteria and biosurfactant producers in the injected and produced fluidsa
| Well group | Log MPN (CXT) of indicated organism in indicated fluidb
|
|||
|---|---|---|---|---|
| Injected
|
Produced
|
|||
| Heterotrophic bacteria | Biosurfactant producers | Heterotrophic bacteria | Biosurfactant producers | |
| Inoculated | ||||
| Total | 15 ± 0.01 | 10.3 ± 0.6 | 14.6 ± 0.4 | -c |
| Spore formers | 11.2 ± 0.6 | 11.2 ± 0.6 | 10.3 ± 0.2 | 10.3 ± 0.2 |
| Nutrient only treated | ||||
| Total | 11.5 ± 2.3 | 9.6 ± 0.6 | 12.1 ± 0.8 | - |
| Spore formers | - | - | - | - |
| Negative control | ||||
| Total | 9.15 ± 2.3 | - | 9.7 ± 0.11 | - |
| Spore formers | - | - | - | - |
BBGM was used for the MPN analysis.
Numbers are average log10 total MPNs ± standard deviations for four independent determinations except for the negative control, where numbers are averages ± ranges for two independent determinations.
-, no growth was detected in the MPN medium.
Although data in Table 2 suggest that biosurfactant-producing Bacillus species were present only in the produced fluids of the inoculated wells and not in those of the nutrient-only-treated wells, it was essential to eliminate any underestimation of Bacillus MPN due to the heat treatment. Plate count broth medium modified to contain 5% salt was used for the MPN analysis to select for the halotolerant Bacillus species (Table 3). Although halotolerant biosurfactant producers were detected in the produced fluids of the nutrient-only-treated wells, the total MPN (CXT) of these microorganisms was 53-fold lower than that in the injected fluids of the same wells. On the other hand, MPN data showed that the total MPN (CXT) of halotolerant biosurfactant producers in the produced fluids of the inoculated wells was about 1.5 times that present in the injected fluids of the same wells.
TABLE 3.
MPN analysis of halotolerant heterotrophic bacteria and biosurfactant producers in the injected and produced fluidsa
| Well group | Log MPN (CXT) of indicated organism in indicated fluidb
|
|||
|---|---|---|---|---|
| Injected
|
Produced
|
|||
| Heterotrophic bacteria | Biosurfactant producers | Heterotrophic bacteria | Biosurfactant producers | |
| Inoculated | 12 ± 0.3 | 11.9 ± 0.3 | 15 ± 0.5 | 12.2 ± 0.05 |
| Nutrient only treated | 12 ± 0.45 | 10.6 ± 1.4 | 13.7 ± 0.3 | 9.8 ± 0.3 |
| Negative control | 12.5 ± 0.4 | 9.4 ± 0.4 | 11.1 ± 0.04 | -c |
Plate count broth supplemented with 5% NaCl was used for the MPN analysis.
Numbers are average log10 MPNs ± standard deviations for four independent determinations except for the negative control, where numbers are averages ± ranges for two independent determinations.
-, no growth was detected in the MPN medium.
The MPN data suggest the absence of Bacillus species (no spore-forming, biosurfactant-producing microorganisms) and the presence of non-spore-forming, halotolerant biosurfactant producers in the nutrient-only-treated wells. The latter were probably introduced into the wells with the treatment (e.g., from the storage tank formation water) but were not maintained. On the other hand, MPN data from the inoculated wells indicate the survival of Bacillus species (spore-forming, halotolerant, biosurfactant-producing microorganisms) in these wells.
Colonies with the same morphologies as those of the two strains used as the inoculum, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049, were retrieved on blood agar plates from both the injected and the produced fluids of the inoculated wells but not from the injected and produced fluids of the wells that received only nutrients or the negative-control well. The 16S rRNA and gyrA gene sequences of these clones were 100% similar to those of Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049.
Culture-independent gyrA amplicons were obtained with DNA extracted from produced fluids of the inoculated wells and not from DNA extracted from produced fluids of the wells that received only nutrients or the control well that received brine. The gyrA amplicons were not detected with DNA extracted from produced fluids of the inoculated wells collected prior to treatment. The resulting sequences of the gyrA clones obtained were 99.72% similar to those of Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049.
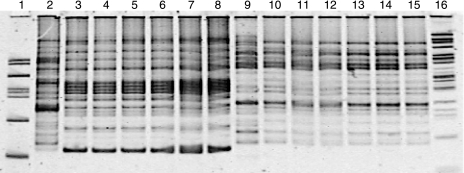
REP-PCR reactions (21, 46) on blood agar plates containing isolates from the injected and produced fluids of the inoculated wells showed patterns similar to those of the inoculum strains, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049 (Fig. 1).
FIG. 1.
Polyacrylamide gel of the REP-PCR for identification of Bacillus strains in the injected and produced fluids of the inoculated wells. DNA was extracted from isolates that had colonies with the same morphologies as those of the two strains used as the inoculum (Bacillus strain RS-1 and Bacillus subtilis subsp. spizizenii NRRL B-23049) and used for the PCRs. Lanes 2 and 9 are DNA from B. licheniformis and B. subtilis subsp. spizizenii type strains, respectively. Lanes 3 and 10 are DNA from laboratory-grown Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049, respectively. Lanes 4 to 8 are Bacillus strain RS-1-like isolates, while lanes 11 to 15 are B. subtilis subsp. spizizenii NRRL B-23049-like isolates. Lanes 4 and 11 are DNA from isolates obtained from the tanks used for the inoculation. Lanes 5, 6, 12, and 13 are DNA from isolates obtained from the injected fluids. Lanes 7, 8, 14, and 15 are DNA from isolates obtained from the produced fluids. Lanes 1 and 16 are the DNA ladder.
Collectively, microbiological and molecular data show that the microorganisms injected into the formation were retrieved in the produced fluids of the inoculated wells.
Lipopeptide biosurfactant production.
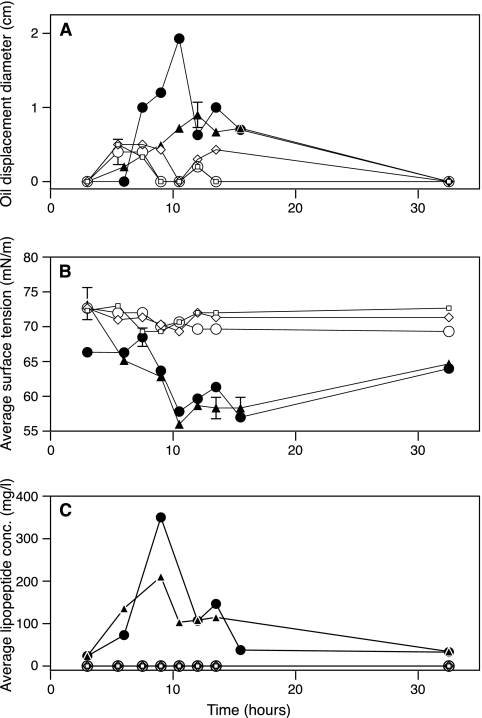
The presence of the biosurfactant in produced fluids was followed over the time of sampling by using three different methods. The oil-spreading technique (33, 49) and surface tension measurement (25) were used to detect the presence of surface-active compounds in the produced fluids of all the wells. Surface activity was observed only in production fluids from the inoculated wells as evidenced by an increase in oil-spreading activity and a decrease in surface tension (Fig. 2A and B). No evidence for surface activity was detected in the production fluids from the nutrient-only-treated wells or the negative-control well (Fig. 2A and B). To determine the natures and the concentrations of the biosurfactant produced in the inoculated wells, HPLC was used (48). The HPLC profile of the samples from the produced fluids of the inoculated wells matched those of the lipopeptide biosurfactants purified from laboratory cultures of the two strains used as the inoculum, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049. The average concentration of the lipopeptide biosurfactant in the produced fluids of the inoculated wells was about 90 mg/liter (a total amount of 7 mol in ∼80,000 liters of produced water) (Table 4 and Fig. 2C). The maximum concentration was as high as 350 mg/liter in the produced fluids of the inoculated wells (Fig. 2C). These maximum concentrations are more than 20 times higher than the critical micelle concentration reported for lipopeptide biosurfactants (ranging from 10 to 20 mg/liter) (33). HPLC analysis of the produced fluids of the nutrient-only-treated wells and the negative-control well showed no lipopeptide biosurfactant production.
FIG. 2.
Evidence for in situ biosurfactant production. The x axis represents the time in hours after the production started from the wells. Closed circles and triangles represent data from the two inoculated wells, open squares and open diamonds represent data from the two nutrient-treated wells, and open circles represent data from the negative-control well. (A) By the oil-spreading technique. Error bars represent standard deviations for three measurements. (B) By surface tension measurement. Error bars represent standard deviations for three measurements. (C) By determination of biosurfactant concentration (conc.) by high-performance liquid chromatography. Error bars represent ranges for duplicate measurements.
TABLE 4.
In situ mass balance and product formation rates and yields compared to those obtained in laboratory culture
| Product | Value for indicated treatment groupa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Inoculated wells
|
Nutrient-only-treated wells
|
Laboratory culture
|
|||||||
| No. of molesb | Rate (h−1)c | Yieldd | No. of molesb | Rate (h−1)c | Yieldd | No. of molesb | Rate (h−1)c | Yieldd | |
| Acetate | 17 ± 7 | 0.04 ± 0.004 | 0.03 ± 0.01 | ND | NA | NA | 0.57 ± 0.03 | 0.04 ± 0.001 | 0.12 ± 0.007 |
| Butanediol | 68 ± 44 | 0.06 ± 0.007 | 0.13 ± 0.08 | ND | NA | NA | 0.54 ± 0.014 | 0.04 ± 0.0005 | 0.11 ± 0.003 |
| Lactate | 40 ± 0.9 | 0.06 ± 0.0002 | 0.08 ± 0.002 | ND | NA | NA | 2.1 ± 0.19 | 0.06 ± 0.001 | 0.42 ± 0.04 |
| Ethanol | 900 ± 50 | 0.08 ± 0.001 | 1.8 ± 0.28 | 130 ± 48 | 0.07 ± 0.003 | 0.36 ± 0.14 | 0.39 ± 0.12 | 0.03 ± 0.007 | 0.08 ± 0.03 |
| CO2 | 640 ± 113 | 0.02 ± 0.001 | 1.3 ± 0.22 | 200 ± 76 | 0.01 ± 0.003 | 0.6 ± 0.21 | 11 ± 0.56 | 0.07 ± 0.001 | 2.3 ± 0.11 |
| Biosurfactant | 6.8 ± 0.67 | 0.02 ± 0.001 | 0.02 ± 0.001 | ND | NA | NA | 0.11 ± 0.05 | 0.05 ± 0.009 | 0.02 ± 0.009 |
| Cellse | 6.7 ± 7.7 | 0.06 ± 0.01 | 0.01 ± 0.02 | 0.24 ± 0.14 | 0.18 ± 0.006 | 0.001 ± 0.0004 | 0.12 ± 0.17 | 0.18 ± 0.06 | 0.03 ± 0.03 |
The numbers of moles of glucose used were 506 ± 118 mol, 360 ± 34 mol, and 4.88 ± 0.0004 mol for the inoculated wells, nutrient-only-treated wells, and laboratory cultures, respectively. Percent carbon recoveries were 106.7 ± 34, 20.7 ± 9, and 94.5 ± 12.8 for the inoculated wells, nutrient-only-treated wells, and laboratory cultures, respectively. ND, not detected; NA, not applicable.
The numbers are averages ± standard deviations for four independent determinations. Values were corrected for adsorption using the bromide recovery factor.
The rates were calculated from the equation ln (Ct/C0) = k · t, where Ct is the concentration at time t, C0 is the initial concentration, k is the rate of production, and t (time) was 108 h. The numbers are averages ± standard deviations for four independent determinations.
Yields are shown in mol · mol glu−1 and were calculated from the equation Yp/s = number of moles of product · mol−1 of substrate used. The numbers are averages ± standard deviations for four independent determinations.
The numbers of moles of cells were calculated from the following equation: number of moles of cells = CXT · mass of one cell/molecular weight of the cell, where the mass of a single cell in grams was assumed to be 1E−13 g, and the molecular weight of the cell was calculated from the cell's empirical formula, CH1.8O0.5N0.2.
Glucose utilization and product formation.
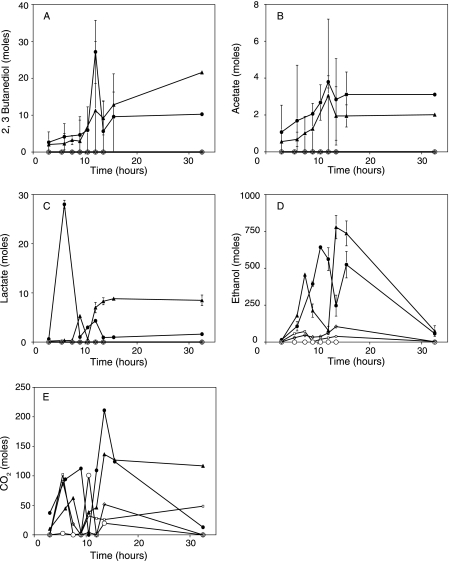
Glucose was the carbon and energy source in our treatments. While complete glucose utilization did not occur, it was clear that large amounts of glucose were used since only 20 to 40% of the glucose added to the nutrient-treated wells (both inoculated and uninoculated) was recovered in the produced fluids and products of microbial metabolism were detected in the produced fluids. The produced fluids of all the nutrient-treated wells (both inoculated and uninoculated) showed increases in alkalinity and carbon dioxide concentrations compared to pretreatment levels, most likely resulting from microbial growth and activity. The produced fluids of the inoculated wells had much higher CO2 concentrations following treatment than those of the nutrient-only-treated wells (Table 4 and Fig. 3E). In addition to CO2, acetate, lactate, ethanol, and 2,3-butanediol were detected in the produced fluids of the inoculated wells (Table 4 and Fig. 3A to D). The percent carbon recovery was 107% ± 34% for the inoculated wells (Table 4). Similar fermentation products from glucose metabolism were obtained by the pure cultures of the strains used as the inoculum, Bacillus strain RS-1 and B. subtilis subsp. spizizenii NRRL B-23049, when the strains were individually grown in formation water supplemented with the nutrients used for well treatments (Table 4). These data argue that the products detected in the produced fluids of the inoculated wells were products of anaerobic glucose metabolism by the inoculum. Although glucose was partially utilized in the wells that received only nutrients, none of the above-mentioned fermentation products were detected, with the exception of CO2 and some ethanol (Fig. 3). We do not know the fate of the remaining glucose carbon in these wells.
FIG. 3.
Total numbers of moles of glucose fermentation products in the produced fluids of the wells. The x axis represents the time in hours after the production started from the wells. Closed circles and triangles represent data from the two inoculated wells, open squares and open diamonds represent data from the two nutrient-treated wells, and open circles represent data from the negative-control well. (A) 2, 3-Butanediol. (B) Acetate. (C) Lactate. (D) Ethanol. (E) CO2.
The detection of 2,3-butanediol, a product often produced by Bacillus species during fermentation (32, 43), and the biosurfactant indicates that we stimulated the microorganisms responsible for biosurfactant production.
DISCUSSION
A major concern with MEOR is whether exogenous microorganisms will be metabolically active in the presence of diverse, natural populations of microorganisms that inhabit oil reservoirs (2, 8, 32). Biosurfactant-mediated oil recovery is even more problematic in that a metabolite, e.g., the biosurfactant, not related to the main energy metabolism of the cell must be produced. The data presented in Fig. 2 and 3 and Table 4 clearly show that the appropriate metabolism was stimulated in the formation and resulted in the production of the biosurfactant and products indicative of a Bacillus fermentation (45). The MPN data indicate that microorganisms physiologically similar to those used as the inoculum (halotolerant, spore-forming biosurfactant producers) were present in high numbers in the produced fluids of the inoculated wells after the incubation period. Molecular characterization of the isolates obtained from the produced fluids of the inoculated wells clearly showed that the same strains used in the inoculum were retrieved from the produced fluids of the inoculated wells. These data provide clear evidence that biosurfactant-mediated oil recovery is technically feasible.
Even though microorganisms exist in the reservoir (as evidenced by growth and glucose utilization in the wells that received only nutrients), the indigenous microorganisms did not prevent the strains used as the inoculum from establishing and metabolizing in the reservoir. The reason for the success of the inoculation procedure might be that the type and amount of nutrients used were more favorable for Bacillus species than for indigenous microorganisms. Pretreatment sampling did not detect the presence of microorganisms in the produced fluids of the wells that were capable of growing in the various media used for enumeration (Table 1). So, it is possible that the injection of nutrients created a niche that allowed the injected strains to be metabolically active. The MPN analysis did not indicate that the biosurfactant producers in the inoculated wells grew, since there was not a significant difference between the total MPN of the spore-forming biosurfactant producers (Table 2) or the halotolerant biosurfactant producers (Table 3) present in the injected fluids and that in the produced fluids. The lack of growth may have been due to a nutrient limitation. Nitrate was used as the nitrogen source, and limiting amounts of nitrate were added to the treated wells to shift the carbon flow from cell mass production to a secondary metabolite production. Previous studies show that nitrogen limitation is associated with an increase in biosurfactant production (11).
Sand-packed-column studies have shown that oil recoveries up to 95% occur when the columns are treated with lipopeptide biosurfactants (3, 27, 36, 38). Recent studies have shown that at least 11 mg/liter of a lipopeptide biosurfactant is required to mobilize oil from sandstone cores, and recoveries as high as 40% of entrapped oil were obtained with as little as 38 mg/liter of the lipopeptide biosurfactant (20, 28). In the current field test, the average concentration of lipopeptide biosurfactant in the produced fluids of the inoculated wells was about 90 mg/liter. This concentration is approximately nine times the minimum concentration required to mobilize entrapped oil from sandstone cores (28). These results showed that in situ lipopeptide biosurfactant production indeed meets this important engineering criterion.
Previous laboratory studies that used both sandstone cores and sand-packed columns suggested that 2.2 ml of oil could be recovered per mg of a lipopeptide biosurfactant (29). Based on this information, the 7 mol of lipopeptide biosurfactant recovered from the produced fluids of the two inoculated wells could recover approximately 16 m3 (100 barrels) of oil. The total material expenses for producing 7 mol of biosurfactant were about $164 ($82 per well). The cost of the in situ biosurfactant production process could be as low as $10 per m3 ($1.6 per barrel). Since the main goal of the study was to test whether in situ biosurfactant production is possible, the volume of the reservoir that was contacted was small, and thus, significant amounts of additional oil were not expected. However, the company followed oil production before and after treatment and these data can be used to judge the effectiveness of the process. The lease that had the two inoculated wells, one of the nutrient-only-treated wells, and several other wells showed an average increase in oil production of one barrel of oil per day compared to the oil production rate before treatment. This increase in oil production was maintained for a period of 7 weeks following treatment. On the other hand, the oil production rate of the other well that received only nutrients slightly decreased during the 7 weeks after treatment compared to pretreatment oil production rates. Although the oil production data suggest that the in situ process was not as efficient and cost-effective as predicted from laboratory data, the data still argue for the cost-effectiveness of MEOR compared to the current price of oil (>$65/barrel). However, more definitive results can be obtained if the size of the treatment is scaled up.
Our data show that in situ biosurfactant production is possible and occurred in amounts exceeding the engineering criterion for mobilizing oil from sandstone cores. This work also provides essential data for modeling MEOR processes in situ (Table 4), including growth rates (0.06 ± 0.01 h−1), carbon balances (107% ± 34%), biosurfactant production rates (0.02 ± 0.001 h−1), and biosurfactant yields (0.015 ± 0.001 mol biosurfactant/mol glucose). These data can be used to study computationally microbial activity in subsurface environments, including petroleum reservoirs. We should note that this is the first time that an in situ carbon balance has been obtained for any MEOR process. Overall, the work emphasizes the technical feasibility and cost-effectiveness of MEOR.
Acknowledgments
We thank C. Brackin, M. Brackin, S. Lemons, and other employees of Arrow Oil and Gas, Inc., for the use of the field and assistance in conducting the experiment. We also thank Matthew J. McInerney and K. Woodson for technical assistance.
This work was supported by U.S. Department of Energy contracts DE-FC26-02NT15321 and DE-FC26-04NT15522.
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Austad, T., and K. Taugbol. 1995. Chemical flooding of oil reservoirs. 1. Low tension polymer flood using a polymer gradient in three-phase region. Colloids Surf. A 101:87-97. [Google Scholar]
- 2.Banat, I. M. 1995. Biosurfactants production and possible uses in microbial enhanced oil recovery and oil pollution remediation: a review. Bioresour. Technol. 51:1-12. [Google Scholar]
- 3.Banat, I. M. 1993. The isolation of a thermophilic biosurfactant producing Bacillus sp. Biotechnol. Lett. 15:591-594. [Google Scholar]
- 4.Berkey, J. S., T. E. Lachmer, W. J. Doucette, and R. Ryan Dupont. 2003. Tracer studies for evaluation of in situ air sparging and in-well aeration system performance at a gasoline-contaminated site. J. Hazard. Mater. 98:127-144. [DOI] [PubMed] [Google Scholar]
- 5.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, L. R., A. A. Vadie, and J. O. Stephens. 2002. Slowing production decline and extending the economic life of an oil field: new MEOR technology. SPE Reservoir Eval. Eng. 5:33-41. [Google Scholar]
- 7.Brusseau, M. L., D. A. Sabatini, J. S. Gierke, and M. D. Annable. 1999. Surfactant selection criteria for enhanced subsurface remediation. Adv. Chem. Ser. 725:8-21. [Google Scholar]
- 8.Bryant, R. S., and T. E. Burchfield. 1989. Review of microbial technology for improving oil recovery. SPE Reservoir Eng. 4:151-154. [Google Scholar]
- 9.Bryant, R. S., A. K. Stepp, K. M. Bertus, T. E. Burchfield, and M. Dennis. 1993. Microbial enhanced waterflooding field pilots. Dev. Petrol. Sci. 39:289-306. [Google Scholar]
- 10.Bryant, S. L., and T. P. Lockhart. 2002. Reservoir engineering analysis of microbial enhanced oil recovery. SPE Reservoir Eval. Eng. 5:365-374. [Google Scholar]
- 11.Cameotra, S. S., and R. S. Makkar. 1998. Synthesis of biosurfactants in extreme conditions. Appl. Microbiol. Biotechnol. 50:520-529. [DOI] [PubMed] [Google Scholar]
- 12.Carrillo, P. G., C. Mardaraz, S. J. Pitta-Alvarez, and A. M. Giulietti. 1996. Isolation and selection of biosurfactant-producing bacteria. World J. Microbiol. Biotechnol. 12:82-84. [DOI] [PubMed] [Google Scholar]
- 13.Chun, J., and K. S. Bae. 2000. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Leeuwenhoek 78:123-127. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich, F. L., F. G. Brown, Z. H. Zhou, and M. A. Maure. 1996. Microbial EOR technology advancement: case studies of successful projects. SPE 36746. Society of Petroleum Engineers, Richardson, TX.
- 15.Green, D. W., and G. P. Willhite. 1998. Enhanced oil recovery. Society of Petroleum Engineers, Richardson, TX.
- 16.Hall, C., J. Tharakan, J. Hallock, C. Cleveland, and M. Jefferson. 2003. Hydrocarbons and the evolution of human culture. Nature 426:318-322. [DOI] [PubMed] [Google Scholar]
- 17.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 18.Henis, Y., J. R. Gould, and M. Alexander. 1966. Detection and identification of bacteria by gas chromatography. Appl. Microbiol. 14:513-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofemeister, J., B. Conard, B. Adler, B. Hofemeiser, J. Feesche, N. Kucheryava, G. Steinborn, P. Franke, N. Grammel, A. Zwintscher, F. Leendres, G. Hitzeroth, and J. Vater. 2004. Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol. Gen. Genomics 272:363-378. [DOI] [PubMed] [Google Scholar]
- 20.Knapp, R. M., M. J. McInerney, D. P. Nagle, M. J. Folmsbee, and S. Maudgalya. 2002. Development of a microbially enhanced oil recovery process for the Delaware Childers Field, Nowata County. Okla. Geol. Surv. Circ. 108:193-200. [Google Scholar]
- 21.Koeuth, T., J. Versalovic, and J. R. Lupski. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 5:408-418. [DOI] [PubMed] [Google Scholar]
- 22.Larrson, L., P. A. Mardh, and G. Odham. 1978. Detection of alcohols and volatile fatty acids by head-space gas chromatography in identification of anaerobic bacteria. J. Clin. Microbiol. 7:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar, I. 1991. MEOR field trials carried out over the world during the last 35 years. Dev. Petrol. Sci. 31:485-530. [Google Scholar]
- 24.Li, Q., C. Kang, H. Wang, C. Liu, and C. Zhang. 2002. Application of microbial enhanced oil recovery technique to Daqing oil field. Biochem. Eng. J. 11:197-199. [Google Scholar]
- 25.Lin, S. C., M. A. Minton, M. M. Sharma, and G. Georgiou. 1994. Structural and immunological characterization of a biosurfactant produced by Bacillus licheniformis JF-2. Appl. Environ. Microbiol. 60:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundquist, A. D., D. Cheney, C. L. Powell, P. O'Neill, G. Norton, A. M. Veneman, N. Y. Evans, S. Mineta, S. Abraham, J. M. Allbaugh, C. T. Whitman, J. B. Bolten, M. E. Daniels, L. B. Lindsey, and R. Barrales. 2001. Energy for a new century: increasing domestic energy production. National energy policy report of the National Energy Policy Development Group. U.S. Government Printing Office, Washington, DC.
- 27.Makkar, R. S., and S. S. Cameotra. 1997. Biosurfactant production by a thermophilic Bacillus subtilis strain. J. Ind. Microbiol. Biotechnol. 18:37-42. [Google Scholar]
- 28.Maudgalya, S., M. J. McInerney, R. M. Knapp, D. P. Nagle, and M. J. Folmsbee. 2005. Tertiary oil recovery with microbial biosurfactant treatment of low-permeability Berea sandstone cores. SPE 94213. Society of Petroleum Engineers, Richardson, TX.
- 29.McInerney, M. J., K. E. Duncan, N. H. Youssef, T. Fincher, S. Maudgalya, M. J. Folmsbee, R. M. Knapp, D. R. Simpson, N. Ravi, and D. P. Nagle. 2005. Final report to the U. S. Department of Energy: development of microorganisms with improved transport and biosurfactant activity for enhanced oil recovery. DE-FC-02NT15321 R 02. University of Oklahoma, Norman, OK. http://www.osti.gov/energycitations/servlets/purl/860919-GXQnLc/.
- 30.McInerney, M. J., M. Javaheri, and D. P. Nagle. 1990. Properties of the biosurfactant produced by Bacillus licheniformis strain JF-2. J. Ind. Microbiol. 5:95-102. [DOI] [PubMed] [Google Scholar]
- 31.McInerney, M. J., S. Maudgalya, D. P. Nagle, and R. M. Knapp. 2002. Critical assessment of the use of microorganisms for oil recovery. Recent Res. Dev. Microbiol. 6:269-284. [Google Scholar]
- 32.McInerney, M. J., D. P. Nagle, and R. M. Knapp. 2005. Microbially enhanced oil recovery: past, present, and future, p. 215-237. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington DC.
- 33.Morikawa, M., Y. Hirata, and T. Imanaka. 2000. A study on the structure-function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta 1488:211-218. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 36.Nazina, T. N., D. S. Sokolova, A. A. Grigoryan, Y.-F. Xue, S. S. Belyaev, and M. V. Ivanov. 2003. Production of oil-releasing compounds by microorganisms from the Daqing oil field, China. Microbiology 72:173-178. [PubMed] [Google Scholar]
- 37.Planckaert, M. 2005. Oil reservoirs and oil production, p. 3-19. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, DC.
- 38.Pruthi, V., and S. S. Cameotra. 1997. Short communication: production of a biosurfactant exhibiting excellent emulsification and surface-active properties by Serratia marcescens. World J. Microbiol. Biotechnol. 13:133-135. [Google Scholar]
- 39.Reusser, D. E., J. D. Istok, H. R. Beller, and J. A. Field. 2002. In-situ transformation of deuterated toluene and xylene to benzylsuccinic acid analogues in BTEX-contaminated aquifers. Environ. Sci. Technol. 36:4127-4137. [DOI] [PubMed] [Google Scholar]
- 40.Roberts, M. S., L. K. Nakamura, and F. M. Cohan. 1994. Bacillus mojavenesis sp. nov. distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int. J. Syst. Bacteriol. 44:256-264. [DOI] [PubMed] [Google Scholar]
- 41.Sabatini, D. A., R. C. Knox, J. H. Harwell, and B. Wu. 2000. Integrated design of surfactant enhanced DNAPL remediation. Efficient supersolubilization and gradient systems. J. Contam. Hydrol. 45:95-121. [Google Scholar]
- 42.Salager, J.-L. 1999. Microemulsions., p. 253-302. In G. Bronze (ed.), Handbook of detergents. Part A. Properties. Marcel Dekker, Inc., New York, NY.
- 43.Shariati, P., W. J. Mitchell, A. Boyd, and F. G. Priest. 1995. Anaerobic metabolism in Bacillus licheniformis NCIB 6346. Microbiology 141:1117-1124. [DOI] [PubMed] [Google Scholar]
- 44.Stein, T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56:845-857. [DOI] [PubMed] [Google Scholar]
- 45.Tahzibi, A., F. Kamal, and M. M. Assadi. 2004. Improved production of rhamnolipids by a Pseudomonas aeruginosa mutant. Iran. Biomed. J. 8:25-31. [Google Scholar]
- 46.Versalovic, J., M. Schneider, F. deBruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 47.West, C. C., and J. H. Harwell. 1992. Surfactants and subsurface remediation. Environ. Sci. Technol. 26:2324-2330. [Google Scholar]
- 48.Youssef, N. H., K. E. Duncan, and M. J. McInerney. 2005. Importance of the 3-hydroxy fatty acid composition of lipopeptides for biosurfactant activity. Appl. Environ. Microbiol. 71:7690-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Youssef, N. H., K. E. Duncan, D. P. Nagle, K. N. Savage, R. M. Knapp, and M. J. McInerney. 2004. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 56:339-347. [DOI] [PubMed] [Google Scholar]