Abstract
The yeast Saccharomyces cerevisiae is an attractive host for the production of heterologous proteins. However, low-yield production of many proteins (from micrograms to milligrams/liter) leaves considerable room for optimization. By engineering the yeast cell via traceable genome-wide libraries, genes that can enhance protein expression level because of their roles in protein transcription, translation, folding, and trafficking processes can be readily identified. This report details a novel approach that combines yeast cDNA overexpression libraries with yeast surface display to allow the rapid flow cytometric screening of engineered yeast for gene products that improve the display of heterologous proteins. After optimization of the screening conditions, a genome-wide scan yielded five yeast gene products that promoted increased display levels of a single-chain T-cell receptor (scTCR). The display-enhancing genes included those coding for cell wall proteins (CCW12, CWP2, and SED1), a ribosomal subunit protein (RPP0), and an endoplasmic reticulum-resident protein (ERO1). Under the premise that yeast surface display levels could be used as a predictor of secretion efficiency, each display-enhancing gene product was tested for its ability to affect secretion levels of multiple scTCR and single-chain antibodies (scFv). All of the selected yeast gene products were shown to promote increased secretion of active protein (1.5-fold to 7.9-fold), with CCW12 and ERO1 being the most generalizable enhancers of scFv/scTCR secretion.
Difficulties related to heterologous protein production often limit the development of industrial and therapeutic protein products (7). The yeast Saccharomyces cerevisiae frequently serves as a reasonable host for heterologous protein expression, since this eukaryote contains much of the cellular machinery necessary to process mammalian proteins, while also being a “generally regarded as safe” organism that is easily cultured. Yet with all of these purported advantages, heterologous protein expression in yeast, in many instances, is far from optimal, with yields as low as micrograms per liter (11a, 35). However, yeast protein production capacity in general terms is much higher, given the ability to secrete certain proteins at levels approaching a gram per liter (1, 24).
The wide range of expression levels for different protein products raises the important question as to whether cell- or protein-based factors are limiting expression. Protein engineering of a desired product has been employed to increase yeast secretion levels of various heterologous proteins, including insulin precursor (14), barley α-amylase 2 (6), and single-chain T-cell receptor (scTCR) (33). However, this approach is highly protein specific, introduces the possibility of deleterious functional and immunogenic alterations, and provides no guarantee of success. As an alternative to engineering of the protein product, the host cell can be altered. Often, the host cell is subjected to multiple rounds of random mutagenesis and selection to provide for the desired increases in protein production. Although this approach can be successful, identification of the actual genetic alterations leading to increased production levels is difficult, even with the use of gene microarray analyses. As a contrasting approach, the folding and secretion apparatus of yeast can be rationally tuned by overexpression or deletion of target genes thought to play a role in protein secretion. A nonexhaustive list of examples includes the overexpression of heavy chain binding protein (BiP), yielding increased production of single-chain antibodies (scFv) (35), and scTCR (33); overexpression of PDI, increasing yields of scFv (35), granulocyte colony-stimulating factor (44), and acid phosphatase (30); and deletion of the Golgi apparatus-resident calcium ATPase gene, PMR1, increasing the production yields of prochymosin (10) and propapain (28). However, this semirational approach requires preliminary knowledge of potential gene targets. In addition, many of the genetic manipulations prove to be protein specific (3, 41), and owing to the discrete sampling approach of this methodology, only a limited subset of yeast sequence space has been investigated. This presents a problem as a comprehensive understanding of the molecular players of the secretory pathway does not yet exist. For example, the endoplasmic reticulum (ER)-associated unfolded protein response has been shown to specifically regulate the expression of 381 genes that are involved in functions ranging from transcription, folding, and posttranslational modification to vesicular trafficking (39), and successful mining of these pathways using a “molecule at a time” approach, although possible, is not desirable.
Yeast gene libraries that allow either overexpression or deletion of each of the approximately 6,000 yeast gene products are available and could allow secretion analysis on a genome-wide scale that could prove beneficial for discovering multiple gene products that improve the secretory processing of proteins (4, 43). However, identification of improved yeast secretion strains requires quantitative measurement of secreted proteins by methods such as Western blotting or enzyme-linked immunosorbent assay (ELISA) that tend to be prohibitive on a genome-wide scale. An alternative method that is suitable for high-throughput single clone analysis is yeast surface display. Yeast surface display is accomplished by fusion of the protein of interest to an endogenous yeast protein that is shuttled through the secretory pathway and “displayed” on the yeast cell surface. Importantly, it was recently demonstrated that the surface display level of a series of mutant scTCR proteins correlated well with soluble secretion levels (33, 34) and suggested that yeast surface display level could be used as a readout for secretion efficiency. Thus, capture of the secreted fusion protein on the surface of the cell of origin would provide a genotype-phenotoype linkage between the engineered yeast cell and protein production level. Combined with quantitative flow cytometric sorting of displaying yeast, the improved secretion strains could be evaluated on a single-cell basis in rapid fashion. This study demonstrates that the yeast surface display-gene library approach was successful in identifying improved secretion strains provided an appropriate selection pressure was used, and several yeast genes that could not have been predicted a priori to impact expression were identified.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains and plasmids used for this study along with their sources are detailed in Table 1. Surface display data for the scFv and scTCR were obtained by transformation of pCT-OX26, pCT-7/15, pCT-LWHI, or pCT-4420His6 into the following yeast strains: EBY100 (rat PDI [rPDI] or BiP), AWY100 (yeast PDI [yPDI]), AWY101, or AWY102. EBY100 is the S. cerevisiae yeast surface display strain (2). AWY100 was developed by changing the selectable marker for the tandem integrated AGA1 cassette from URA3 to LEU2. AWY101 and AWY102 were also generated in this study from YVH10 and BJ5464, respectively, by integrating GAL1-AGA1 in tandem with endogenous AGA1, as in the creation of EBY100. The plasmids pMAL5.1 (rPDI), pGAL-KAR2LEU (BiP), and pCT37 (yPDI) were used to increase the copy number of the corresponding folding assistants. Secreted scFv and scTCR expression data were obtained by expressing pRS-GALOX26, pRS-GALTLWHI, pRS-GALT7/15, or pRS-4420His6 in either BJ5464 or YVH10. When necessary for yeast strains harboring multiple plasmids, open reading frames were transferred to plasmid backbones possessing different auxotrophic markers. Control strains were created by transformation with null plasmids containing the identical nutritional marker (pRS-314, pRS-315, or pRS-316) (36). All yeast transformations were performed by the lithium acetate method (8) and grown in minimal medium (2% dextrose, 0.67% yeast nitrogen base) buffered at pH 6.0 with 50 mM sodium phosphate and containing either 1% Casamino Acids (SD-CAA; lacking tryptophan and uracil) or 2× SCAA amino acid supplement (SD-SCAA, 190 mg/liter Arg, 108 mg/liter Met, 52 mg/liter Tyr, 290 mg/liter Ile, 440 mg/liter Lys, 200 mg/liter Phe, 1260 mg/liter Glu, 400 mg/liter Asp, 480 mg/liter Val, 220 mg/liter Thr, 130 mg/liter Gly, lacking leucine, tryptophan, and uracil). Leucine (200 mg/liter), tryptophan (20 mg/liter), and uracil (20 mg/liter) were supplemented when necessary for proper auxotrophic selection. Induction of protein display and secretion was performed in the same medium, with the dextrose substituted for by 2% galactose. Fresh transformants were used in all experiments.
TABLE 1.
Strains and plasmids used in this study
| Yeast strain or plasmid | Genotype or gene | Display or secretion type | Source or reference |
|---|---|---|---|
| Strains | |||
| EBY100 | MATaAGA1::GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Display | 2 |
| AWY100 | MATaAGA1::GAL1-AGA1::LEU2 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Display | This study |
| AWY101 | MATα AGA1::GAL1-AGA1::URA3 PDI1::GAPDH-PDI1::LEU2 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Display | This study |
| AWY102 | MATα AGA1::GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Display | This study |
| BJ5464 | MATα ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Secretion | 12 |
| YVH10 | MATα PDI1::GAPDH-PDI1::LEU2 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | Secretion | 30 |
| Plasmids | |||
| pCT-4420His6 | Aga2p-4420 | Both | This study |
| pCT-OX26 | Aga2p-OX26 | Both | This study |
| pCT-7/15 | Aga2p-7/15 | Display | 13 |
| pCT-LWHI | Aga2-LWHI | Display | 33 |
| pRS316-4420His6 | 4420 | Secretion | This study |
| pRS-GALT7/15 | 7/15 | Secretion | 34 |
| pRS-GALTLWHI | LWHI | Secretion | 33 |
| pRS316-GALOX26 | OX26 | Secretion | 9 |
| pGAL-KAR2LEU | BiP (Kar2p) | Both | 29 |
| pMAL5.1 | Rat PDI | Both | 16 |
| pCT37 | Yeast PDI | Both | 38 |
Yeast surface display and library screening.
For all surface display experiments, yeast cells were grown for 1 to 2 days with shaking at 30°C in SD-SCAA medium, and these starter cultures were subsequently diluted uniformly to an optical density at 600 nm (OD600) of 0.1 and regrown in SD medium. When the cultures reached an OD600 of 1 to 2, protein surface display was induced by changing to SG medium, and cultures were placed at the appropriate induction temperature (20, 30, or 37°C) for 16 to 18 h. This growth and induction method yielded optimal reproducibility in display levels between replicates and between independent experiments. Then, 2 × 106 yeast cells were collected and washed with 500 μl phosphate-buffered saline (PBS)-bovine serum albumin (BSA) (PBS at pH 7.4, with 1 mg/ml BSA) prior to immunolabeling for detection and flow cytometry. Surface-displayed scFv were detected by antibody labeling with the anti-c-myc epitope tag antibody 9E10 (1:100; Covance), while scTCRs were detected with the conformationally specific 1B2 monoclonal antibody (19) (10 μg/ml) for 30 min at 4°C. All samples were washed with 500 μl PBS-BSA and subsequently labeled with anti-mouse immunoglobulin G conjugated to phycoerythrin (1:35; Sigma) for 30 min at 4°C. After being washed with 500 μl PBS-BSA, samples were resuspended in 750 μl PBS-BSA and analyzed on a Becton Dickinson FACSCalibur benchtop flow cytometer.
The cDNA overexpression library, a kind gift of Haoping Liu, consists of a pool of CEN-based plasmids, each with a single yeast open reading frame under the control of the GAL1 promoter. The plasmid library had been previously utilized in studies to analyze growth and cell cycle effects of overexpressed yeast genes (17, 37). Because the library was created from yeast mRNAs whose levels in the cell will vary for different genes, a minimum of 50,000 variants must be evaluated to ensure complete coverage of the yeast transcriptome (17). After transformation of the plasmid library into AWY100 yeast cells already transformed with the 7/15 scTCR plasmid, the yeast overexpression library contained approximately 2.3 × 105 individual clones and was screened for yeast having increased levels of 7/15 scTCR surface display (1B2 labeling). The library was oversampled by screening 8 × 106 clones using a Becton Dickinson FACSVantage SE flow cytometric sorter at the University of Wisconsin Comprehensive Cancer Center. In the first round of sorting, the top ∼1% of the displaying cells were recovered and subpooled. Those subpools exhibiting the most enrichment were then carried to the next round. Subsequent rounds were performed with multiple sorting gates between 0.1 and 2%, depending on the display distribution observed. The use of various gating percentages for rounds 2 to 4 helped ensure successful enrichment of the clones exhibiting improved display because relatively low absolute increases in display were being observed (1.2- to 2.5-fold). Cultures were maintained to include at least 10 times the size of the library (or subsequent sorted pools) at all times. After the fourth round of enrichment, the entire sorted population was plated on nutritionally selective plates and individual clones were analyzed by flow cytometry.
Identification and recovery of cDNA from yeast.
Clones identified as exhibiting increased 7/15 surface display via flow cytometry were spotted onto SD-SCAA plates and grown overnight. Approximately 0.2 μl cells was resuspended in 30 μl 0.2% sodium dodecyl sulfate (SDS). Cells were lysed by incubating samples at −80°C for 2 min, 95°C for 2 min, −80°C for 2 min, and 95°C for 5 min. Approximately 2 μl of the lysed cells was added as the DNA template to a 50-μl PCR mixture containing 0.4% Triton X-100 with Platinum Taq polymerase (Invitrogen) and the M13 primers (17). PCR products were run on a 1% agarose gel, and bands corresponding to the cDNA inserts were visually identified.
Plasmids having uniquely sized cDNA inserts were recovered from the yeast with the Zymoprep II yeast plasmid miniprep kit (Zymo Research). DH5α cells (Invitrogen) were used to amplify the recovered plasmid DNA and the cDNA inserts sequenced at the University of Wisconsin Biotechnology Center, utilizing the 5′-TACTTCTTATTCCTCTACCG-3′ primer to obtain the forward sequence and the T7 primer (17) for the reverse sequence. To confirm that the increased display efficiency was the result of the harbored overexpression plasmid, the parent AWY100 strain was freshly cotransformed with each recovered cDNA-containing plasmid and pCT-7/15 and analyzed by flow cytometry as described above.
Heterologous protein secretion and activity analyses.
Cultures were inoculated in SD-CAA and allowed to grow for 1 to 2 days at 30°C prior to dilution to a uniform OD600 of 0.1 and regrowth for 3 days to an OD600 of 8 to 10. Protein expression was then induced by switching to nutritionally selective SG medium (plus 1 mg/ml BSA) and placing the cultures at 20, 30, or 37°C for 3 days. Although we have previously shown 3 days to be the most general approach for maximum production (35), the growth time (1 or 3 days) and induction time (1 or 3 days) were varied for the CCW12-overexpressing, 4-4-20 system. CCW12 overexpression similarly enhanced secretion levels relative to the wild type in all cases. In terms of absolute expression levels, the 3-day-3-day system and the 1-day-3-day system were similar, while the 1-day-1-day system was substantially lower, as expected. Therefore, throughout this study, we continued to employ the 3-day-3-day system. Cell-free culture supernatants were resolved on a 12.5% polyacrylamide-SDS gel and transferred to nitrocellulose membranes. In the case of LWHI or Aga2p-scFv supernatants, samples were first deglycosylated prior to SDS-polyacrylamide gel electrophoresis (PAGE) (endo-β-N-acetylglucosaminidase H; New England Biolabs). The membranes were probed with either anti-c-myc antibody for scFv samples (9E10; 1:3,000) or anti-tetra-His antibody for LWHI scTCR samples (0.2 μg/ml; QIAGEN). All membranes were probed with a horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (1:2,000; Sigma), followed by enhanced chemiluminescence detection with the Amersham ECL system. Western blot films of various exposure times were analyzed with ImageJ (NIH) to determine band intensities. The slope of the intensity versus exposure time curve in the unsaturated, linear region was then utilized to determine relative protein concentrations and, hence, secretion levels. Lack of significant cell lysis was determined by probing cell supernatants for the endogenous intracellular glyceraldehyde-3-phosphate dehydrogenase (G3PDH) yeast protein. Yeast supernatants for both wild-type and CCW12-overexpressing cells were loaded onto SDS-PAGE gels along with purified G3PDH protein (Sigma) and cell lysate. After being transferred to nitrocellulose, the membrane was probed with a mouse anti-yeast G3PDH antibody (1:500; Chemicon) detected via ECL as described previously, and exposed for 35 min. No signal was detected, and based on the sensitivity of the assay, it was determined that at the very maximum, less than 0.1% of the total heterologous protein in the supernatants could be derived from cell lysis (11a). All statistics presented in the text were determined by two-tailed unpaired Student's t test.
The 7/15 scTCR secretion levels were detected via ELISA as the anti-tetra-His Western blot was not sensitive enough due to low 7/15 secretion levels. The ELISA also served as an activity assay since the 1B2 antibody used in detection recognizes a nearly identical epitope to that of the native peptide-major histocompatibility complex (MHC) and has proven to be a high-affinity surrogate for peptide MHC (19, 33). In addition, where indicated, LWHI activity was also evaluated by ELISA to correlate increases in protein activity with increases in total protein as assessed by Western blotting. To perform the ELISA, wells of a Nunc-Immuno 96-well Maxisorp plate (Nunc) were coated with the anti-tetra-His antibody (2.7 μg/ml; QIAGEN) overnight at 4°C. After being blocked for 2 h with 400 μl PBS-BT (PBS at pH 7.4 with 1 mg/ml BSA and 1 ml/liter Tween 20), wells were washed four times with 250 μl PBS-BT. Various dilutions of culture supernatants were applied for 1 h, and after four rounds of washing with PBS-BT, biotinylated 1B2 (5 μg/ml) was applied for 30 min. After being washed four more times with PBS-BT, streptavidin-HRP (1:1,000; Amersham) was added for 30 min and followed by another four washes. Samples were developed with the tetramethylbenzidine two-component microwell peroxidase substrate kit (Kirkegaard and Perry Laboratories), and the reaction was halted with 2 M H3PO4. Absorbance at 450 nm was measured, and appropriate predilution of samples ensured that only those data in the linear range and at similar signal intensities were considered in the analysis. The slope of the absorbance versus concentration curve was used to determine the relative amount of scTCR in each sample.
To confirm that the increases in 4-4-20 secretion determined by Western blotting also corresponded to increases in active 4-4-20 secretion, fluorescein-binding assays were performed. Fifteen microliters of biotin-coated polystyrene bead suspension (FluoSpheres biotin-labeled microspheres; Invitrogen) was incubated with 600 μl of BlockAid blocking solution (Invitrogen) and sonicated for 5 min. Ten microliters of a NeutrAvidin-fluorescein conjugate (5 mg/ml; Pierce) was then added, and the mixture was incubated at 25°C with shaking for 1 h. After being washed three times with 500 μl PBS-BSA, 10 μl of yeast supernatant containing the 4-4-20 scFv was applied to the fluorescein antigen-coated beads for 1 h at 25°C with shaking. The beads were collected by centrifugation, and the liquid was removed (depleted supernatant, inactive fraction). The beads were then resuspended in 20 μl of 100 mM fluorescein sodium salt (Sigma) for 30 min at 25°C with shaking. The excess free fluorescein competed with the fluorescein-labeled beads to release the bound 4-4-20 scFv (active fraction). Nonspecific binding of 4-4-20 scFv to the polystyrene beads was analyzed by following the same protocol but labeling the beads with NeutrAvidin (Pierce), which lacks the conjugated fluorescein. Samples of the original yeast supernatant, the depleted supernatant, and the active fraction were analyzed by quantitative Western blotting as described previously.
RESULTS
Method for screening engineered yeast for increased protein production using yeast surface display.
The quantitative level of protein display on the surface of yeast was used as a proxy screening variable for improved secretion strains. A library of yeast display strains was created by transforming the yeast surface display strain with a cDNA overexpression library (17). The resultant library contains engineered yeast strains that harbor two plasmids, each under the galactose-inducible GAL1-10 promoter. One plasmid contains an expression cassette that directs surface display of the heterologous protein of interest via fusion to the Aga2p mating protein that self-assembles to the cell wall-anchored Aga1p protein. The second contains a yeast cDNA and mediates overexpression of an endogenous yeast protein. The yeast library was amplified in glucose to prevent growth rate and expression bias effects and then was switched to induction medium containing galactose (Fig. 1). Upon induction, the protein of interest was displayed on the yeast surface, with the protein products of the different yeast cDNA plasmids harbored by each cell causing increased or decreased protein display levels or no change in protein display levels.
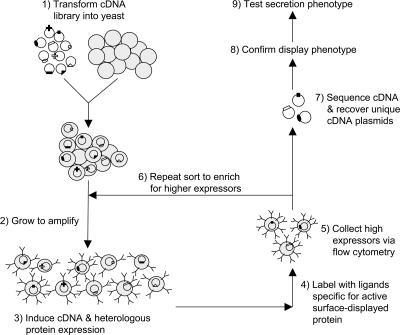
FIG. 1.
Schematic of method for identification of yeast genes that elevate the yeast surface display levels of a target protein. (Step 1) Prior to introduction of the plasmid-based cDNA library, yeast cells were transformed with a plasmid containing the protein of interest (scFv or scTCR) such that after step 1, each cell contained two galactose-inducible plasmid constructs. (Step 2) Cells were amplified in glucose-based media, and (step 3) protein expression was induced in galactose-based media. Step 3 was performed under normal 20°C induction conditions or at elevated temperatures of 30°C and 37°C as described in the text. After step 3, yeast cells had various levels of surface-displayed protein as a result of the induced genetic alteration (via the cDNA plasmid). (Step 4) The yeast cells were then probed for active surface-tethered protein and analyzed by flow cytometry. (Step 5) Using FACS, the cells exhibiting higher fluorescence, and thus elevated levels of surface-displayed protein, were isolated. (Step 6) The enriched population was then cycled through the selection process again for further enrichment. (Step 7) The display-enhancing genes were then identified and (step 8) the display-enhancing phenotype was confirmed. (Step 9) Finally, the display-enhancing cDNAs were tested for their effects on protein secretion.
Because the protein of interest is displayed on the cell surface, it is accessible to epitope-specific antibodies. When followed by fluorescent secondary antibodies, the yeast cells were sorted on a single-cell basis using flow cytometry to rapidly provide quantitative data corresponding to protein display levels. Thus, yeast cells exhibiting higher levels of fluorescence were isolated from the library population via fluorescence-activated cell sorting (FACS) (Fig. 1). Subsequently, these desirable clones were amplified in glucose-based media and the cycle was repeated to purify yeast clones that have elevated levels of surface display. Then, individual clones were tested for their level of protein display, and those containing cDNA that enhances display were sequenced for identification. Unique cDNA clones were then retransformed into the parent display strain to confirm the phenotype and eliminate yeast mutation or epigenetic phenomena as causes of the observed display increases. Finally, the cDNAs that improved surface display were tested for their effects on protein secretion. As described earlier, the selection strategy was designed based on previous findings that surface display levels of engineered scTCR proteins correlated well with secretion levels of these proteins (33, 34). Since this relationship was crucial to the success of the selection strategy, we first tested whether or not the correlation between display level and secretion level holds when the yeast cell, rather than the protein, is engineered.
Testing the correlation between display and secretion for engineered yeast.
Previous studies have revealed that certain yeast genes, such as those coding for BiP and PDI, when overexpressed result in increased secretion of proteins such as the anti-fluorescein 4-4-20 scFv (35) and the an anti-transferrin receptor scFv (OX26) (9). We therefore wished to test whether or not display levels of strains engineered to overexpress BiP and PDI would correlate with the observed increases in secreted levels. Since secretion of scFv is maximal in yeast when expression is induced at 20°C (11a, 42, 44) and the goal of these screens was to maximize protein production, the presence or absence of a correlation was evaluated at this temperature. Yeast display and secretion strains were engineered to overexpress yeast BiP (Kar2p), yeast PDI (by plasmid [pCT37] and by integration [YVH10]), and rat PDI, and display and secretion were induced at 20°C. Surface display data were obtained by flow cytometry and compared with secreted expression levels measured by Western blotting (Fig. 2). Both approaches employed the c-myc epitope tag present near the carboxy terminus of each construct. Although secreted scFv was generally increased from yeast strains overexpressing BiP and PDI (1.4- to 2.1-fold for OX26 and 1.5- to 5.8-fold for 4-4-20), the surface display of these two proteins was unchanged by BiP and PDI overexpression (the lone display increase was 4-4-20 with plasmid-based PDI overexpression 1.2-fold) (Fig. 2). Since the expected correlation between secretion and display of engineered yeast strains did not materialize in these small-scale experiments, it was hypothesized that the presence of the Aga2p fusion partner required for scFv surface display negated the effects of the folding assistants by altering the secretory processing of the scFv.
FIG. 2.
Fusion to Aga2p eliminates the effects of folding assistants on secretion and display levels for the (A) 4-4-20 and (B) OX26 scFvs. Relative expression levels represent surface display levels of Aga2p-scFv measured by flow cytometry, secreted scFv levels measured by Western blotting, and secreted Aga2p-scFv levels measured by Western blotting. Because of variations in plasmid markers and the required yeast cell strains, each engineered cell was normalized to its appropriate wild-type background. The wild type depicted in these plots was that corresponding to the wild-type system that yielded the highest standard error. Each data point represents triplicate display and secretion samples. Single and double asterisks represent P < 0.05 and P < 0.07, respectively. WT, wild type; rPDI, rat PDI; BiP, yeast BiP/Kar2p; pCT37, plasmid-based yeast PDI overexpression; YVH10, integration-based yeast PDI overexpression.
Therefore, to test this hypothesis we analyzed the effect of the ER-resident folding assistants on secretion of the Aga2p-scFv fusion protein. Without high-level expression of the Aga1p cell wall anchor, Aga2p-scFv fusions are secreted from the cell rather than being displayed on the yeast surface (11). In this way, the Aga2p-scFv constructs were secreted from the same strain as that used for secretion of the unfused scFv products, and secretion levels were analyzed by Western blotting. Figure 2 indicates that the Aga2p-scFv fusions were not secreted at higher levels in the presence of BiP or PDI overexpression. (The lone exception was 4-4-20 with integrated PDI [2.2-fold].) This contrasts dramatically with the effects that folding assistants have on the secretion of unfused scFv (Fig. 2), confirming that fusion to Aga2p obviates the positive effects that chaperones have on scFv processing. Therefore, Aga2p, and not scFv, determines display efficiency for these proteins.
Since the abovementioned experiments tested only BiP and PDI effects, it was possible that these folding assistants were simply special cases that were not responsive in a display format. Thus, we tested the effects of Aga2p fusion on the display responses for the entire yeast genome using the selection procedure outlined in Fig. 1 with 20°C induction at step 3. Yeast overexpression libraries were screened for factors that increased the display of three different proteins, 4-4-20 and two scTCR (7/15 and LWHI). The proteins were chosen because they have been previously produced in yeast as fully active proteins and because they represent a range in display and secretion competence. The 4-4-20 scFv has secreted levels of 1 mg/liter (35), while the 7/15 and LWHI scTCR are closely related mutants that differ in both display and secretion efficiency by approximately 20-fold (7/15, 0.1 mg/liter, versus LWHI, 2 mg/liter) (33). Neither the 4-4-20 nor the scTCR screens yielded cDNA-dependent improvements in display levels after four rounds of sorting (data not shown). Combined with the data regarding the lack of BiP and PDI effects on display, the inability to recover improved clones from the genome-wide libraries of engineered yeast for three different proteins prompted us to conclude that unlike the correlation between display and secretion for an engineered protein, display effects generated by engineering the yeast cell were masked by fusion to Aga2p.
Screening of yeast cDNA library under selection pressure.
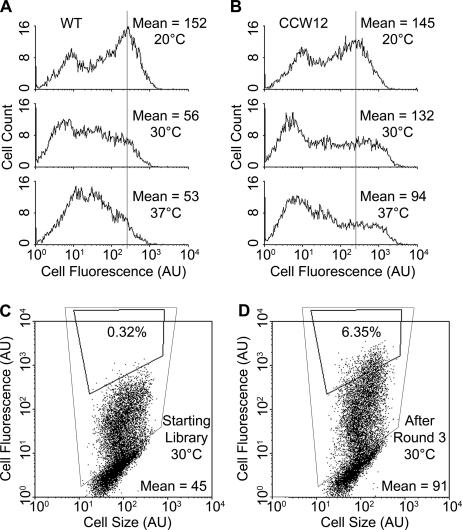
Since recovery of yeast proteins that enhance display was not possible under normal induction conditions (20°C), a selection pressure was applied with the idea of making the fusion partner, rather than Aga2p, the dominant determinant of display efficiency. Of the four proteins studied here, the 7/15 scTCR is particularly poorly processed by the yeast cell at elevated temperatures (33). When surface display induction temperature was raised from 20°C to 30°C and 37°C, the surface display levels of scTCR dropped substantially (Fig. 3A), while the display of Aga2p alone was not dramatically affected (data not shown). Thus, it appeared that selection at elevated induction temperatures would allow isolation of yeast proteins that could overcome the defective processing and attenuated display of 7/15. In contrast, even though secretion levels of 4-4-20 and OX26 are decreased at elevated induction temperatures (9, 35), the display levels were not significantly altered by induction at elevated temperatures (data not shown), indicating that Aga2p was still serving as the determinant of display efficiency for these scFvs. Thus, we chose to use 7/15 as our bait to select cDNAs that recover the deficit in display at elevated temperatures. Yeast cells displaying 7/15 scTCR were transformed with the cDNA overexpression library, induced at either 30°C or 37°C, and sorted for four rounds using the conformationally sensitive 1B2 antibody that is a surrogate for the natural peptide-major histocompatibility complex and thus serves as a probe for display of active scTCR protein (19, 33) (Fig. 1, with selection pressure at step 3). At the conclusion of the third sorting round, the mean cellular fluorescence level (scTCR display level) of the sorted pool was twofold higher than that of both the starting library and the wild-type display strain (Fig. 3C and D), indicating the enrichment of clones with improved 7/15 display. After a final round of sorting to purify the improved clones, 29 total clones from the 30 and 37°C sorts were tested and 28 of these led to 1.4- to 2.5-fold increased surface display at the elevated sort temperatures (data not shown). In contrast, the 28 clones were also tested at the 20°C display induction temperature and only 12 showed higher expression ranging from 1.2- to 2.3-fold (data not shown). All individual clones were subjected to whole-cell PCR to recover the overexpressed cDNA, and those genes that possessed inserts of unique nucleotide lengths were sequenced for identification. The overexpressed genes leading to increased surface display at their respective elevated induction temperatures were identified as CCW12, CWP2, RPP0, SED1, and ERO1 (Table 2). With the lack of diversity in the pool sorted at 30°C that contained mostly CCW12 clones, four clones from the earlier third round of sorting at 30°C were tested as well. Only three of these clones showed increased 7/15 surface display when induced at 30°C, with two of the clones being identified as CCW12, while a third was homologous to RPL6A/B. The products of CCW12, CWP2, and SED1 are all cell wall-associated proteins (21, 32, 42), while the products of RPP0 and RPL6A/B are constituents of the ribosome (22, 25). ERO1 localizes to the ER and provides PDI with oxidizing equivalents for disulfide bond rearrangement (26, 40).
FIG. 3.
Identification of yeast genes that increase 7/15 scTCR surface display by using elevated induction temperatures. (A) Histograms depicting the effects of increasing induction temperature (20, 30, and 37°C) on 7/15 display. (B) Histograms depicting the effects of CCW12 overexpression on 7/15 display at different induction temperatures (20, 30, and 37°C). (C) Flow cytometric dot plot of 7/15-displaying yeast that harbor the cDNA library (30°C sort). The yeast population that displays active 7/15 is enclosed in the large rectangle, while a sample sort gate is also shown at 0.32% of the total population. (D) The population after the third round of 30°C sorting shows enrichment of clones having higher display levels with the mean of the positive population doubling and the percentage of the total population contained in the original sort gate increasing by almost 20-fold. Reported are geometric means of the population of yeast cells that were displaying protein on the cell surface. This approach makes use of a second epitope tag that allows exclusion of nondisplaying yeast from the analysis (the negative population can be seen in the histograms as a peak with low mean fluorescence of ∼6 to 8). All flow cytometry data were obtained via antibody labeling with the 1B2 activity probe and represent 10,000 yeast cells. Histograms are representative of triplicate samples, and the vertical lines are inserted to facilitate comparison of the fluorescence of the positive display peak.
TABLE 2.
Yeast cDNAs that enhance scTCR display
| Gene | Screen temp (°C) | Frequency |
|---|---|---|
| CCW12 | 30 | 17 |
| SED1 | 30 | 1 |
| CWP2 | 37 | 3 |
| ERO1 | 37 | 4 |
| RPP0 | 37 | 3 |
Effects of display enhancers on scTCR display at normal and elevated temperatures.
Each unique overexpression plasmid listed in Table 2 was recovered from the corresponding yeast clone to confirm the cDNA-based influences on display phenotype. The parent yeast surface display strain (AWY100) was transformed with both the pCT-7/15 display vector and the recovered overexpression plasmids, and display at each induction temperature was evaluated by flow cytometry (Fig. 3A and B and Fig. 4). The mean display level of each engineered yeast strain was normalized to that of the wild-type population at 20°C for comparison purposes. It was found that all genes show statistically significant increases in display levels of active protein for at least one of the elevated induction temperatures compared to the wild-type population at the same temperature (1.2- to 2.2-fold; P < 0.05). Most exhibited improvements in display at both 30°C and 37°C, regardless of what sort they were identified in. The lone exception was RPL6A/B, as it did not prove to increase 7/15 display at any temperature tested and behaved exactly like the wild type (data not shown). Furthermore, at 30°C, CCW12 and RPP0 overexpression recovers 7/15 surface display to the same level as the wild type at 20°C. Only ERO1 overexpression resulted in a statistically significant increase in 7/15 display at 20°C (P < 0.06). Taken together, these results indicate that a 20°C sort would not have identified the majority of the display enhancers, as also indicated by our failed screens at 20°C with 4-4-20, 7/15, and LWHI. Since the screen involved manipulation of cell wall proteins by fusion to the Aga2p protein and surface tethering via the Aga1p protein, CCW12, CWP2, and SED1 could either play a beneficial role in increasing surface display by Aga1p/Aga2p modulation or by improving scTCR processing through the secretory pathway. To test which of these possibilities was most likely, Aga2p was displayed alone without any scTCR fusion. There was no change in Aga2p display in any of the engineered yeast strains at any of the temperatures tested (data not shown), indicating that the observed increases in display in the presence of overexpressed cDNAs were scTCR dependent. This result suggested that the yeast proteins recovered in our screen therefore had potential to increase heterologous protein secretion in addition to improving display levels. Therefore, we opted to test each of the recovered yeast proteins for their effects on the secretion of four heterologous proteins, and in doing so, tested the generality of our hypothesis that display and secretion levels would be coordinately regulated as a result of selection pressure.
FIG. 4.
Effects of overexpressed cDNA on surface display levels of 7/15 scTCR at the various induction temperatures. Samples were analyzed via flow cytometry in the same manner as in Fig. 3. Data represent triplicate cultures whose display levels are normalized to the wild type (WT) at 20°C. Data representing only the positive displaying population were analyzed. The main cause for the appearance of negative display peaks in yeast surface display experiments has been determined to be plasmid stability (27). In no case did the overexpressed cDNA alter the percentage of displaying yeast in the population, indicating that the cDNA did not affect plasmid stability in the displaying strains.
Effects of display enhancers on the secretion of four heterologous proteins.
Since 20°C has been shown to be the optimal induction temperature for the secretion of many scFv and scTCR fragments (9, 35), the five yeast proteins were first tested for their effects on 20°C secretion of 7/15 scTCR, LWHI scTCR, 4-4-20 scFv, and OX26 scFv (Fig. 5A and B). Secretion levels of 7/15 scTCR were determined by a high-sensitivity ELISA utilizing a tetra-His capture antibody (intact carboxy terminus) and biotinylated 1B2 (active) as a means of detection. Thus, this assay serves as a direct readout of full-length, active protein. At 20°C, overexpression of CCW12 and ERO1 increased secretion of active 7/15 scTCR protein by 2.5- and 3.2-fold, respectively (Fig. 5B). In contrast, SED1, CWP2, and RPP0 did not reproducibly increase 7/15 secretion titers. Similar to the case with 7/15, 4-4-20 and OX26 scFvs were secreted at higher levels (from 1.6- to 7.4-fold), as assessed by Western blotting, when CCW12 and ERO1 were overexpressed, indicating a generality in the secretion-enhancing effects of the products of these yeast genes (Fig. 5A and B). Overexpression of RPP0 also enhanced the secretion levels of these two scFvs (2.8-fold). To confirm that scFv secretion increases indicated by Western blotting represented increased yields of active protein, anti-fluorescein scFv 4-4-20 activity assays were performed (see Materials and Methods for details) and indicated that the amount of scFv that undergoes a reversible, fluorescein-dependent binding is increased by 4.7-fold in the presence of CCW12 overexpression (compared with 4.1-fold determined by Western blotting). Protein-specific effects were observed as the secretion of LWHI is only affected in the presence of ERO1 overexpression, which leads to a marginal increase in secretion as assessed by Western blotting (1.6-fold). In addition, ERO1-mediated increases in LWHI secretion were confirmed by the ELISA activity assay (1.8-fold) to ensure that Western blotting data were indicative of the increases in secretion levels of active protein. As protein-specific effects suggest, general cell-based phenomena induced by the cDNA were not responsible for the observed increases in secretion. In particular, the cytoplasm-resident G3PDH protein could not be detected in the 4-4-20 supernatants with or without CCW12 overexpressed (data not shown). This finding indicated that cell lysis was not responsible for the increased amount of active protein accumulated in the supernatant.
FIG. 5.
Effects of overexpressed cDNA on secretion levels of scTCRs and scFvs. (A) Representative Western blotting data for 4-4-20 scFv secretion experiments. Triplicate supernatants derived from independent transformants were used to generate the Western blotting signals. Within each temperature condition, the data shown are from the same time point of exposure and can therefore be directly compared. However, these only serve as qualitative comparisons, while quantitative values and associated statistical significance can be found in panels B and C (see Materials and Methods for quantitation details). WT, wild type. (B) Secretion yields after induction at 20°C. (C) Secretion yields after elevated temperature induction. As indicated in panel C, each cDNA overexpression strain was evaluated at the elevated induction temperature at which it was selected. 7/15 secretion levels were detected by quantitative ELISA, while all other protein levels were measured by quantitative Western blotting, as depicted for 4-4-20 in panel A. Activity assays confirmed that increased secretion of 4-4-20 and LWHI expression detected via Western blotting was representative of active protein levels. Single and double asterisks represent P < 0.05 and P < 0.1, respectively, and were reproducibly significant over three independent experiments. Triple asterisks indicate data that were statistically significant (P < 0.05) for the experiment presented, but were not reproducibly significant over multiple experiments. These *** conditions were therefore reported as incapable of increasing secretion levels. Triplicate independent transformants were evaluated for each protein-cDNA combination, and all data were normalized to the wild type at the corresponding temperature.
Since all of the identified yeast genes elicited their most significant effects on surface display at the elevated induction temperatures, secretion using 30 and 37°C induction was also investigated to determine induction temperature effects on secretion levels. Given the sensitivity of 7/15 display as a function of temperature, even though the scTCR is expressed as a fusion to Aga2p, it was not surprising that secreted unfused 7/15 scTCR was not detectable after 30 or 37°C induction, even in the presence of the overexpressed genes (data not shown). Therefore, we also tested the effects of the overexpressed cDNAs and induction temperature on the more stable LWHI scTCR, along with 4-4-40 and OX26 scFvs (Fig. 5C). The first finding was that the elevated temperatures had the general effect of lessening the impact of overexpressed cDNA both by decreasing the magnitude of improvement and by causing fewer cDNAs to have an impact (compare Fig. 5B and C). As described above, activity tests indicated that the increases in secretion reported at the elevated temperatures again represented increases in active protein (4-4-20 with CCW12 at 30°C and LWHI with ERO1 at 37°C). The final two cDNAs tested, SED1 (4-4-20 at 20°C) and CWP2 (4-4-20 at 37°C), had the most limited overall effects on secretion and only modestly boosted secretion for a single scFv under discrete conditions.
Finally, the absolute secretion levels at the different induction temperatures were compared. As observed previously (9, 35), the absolute secretion levels of the scFvs in the wild-type system were not increased by raising the induction temperature from 20°C to 30°C or 37°C. In the presence of cDNA, the only elevated temperature conditions whose absolute scFv production levels exceeded that seen for the same system at 20°C were 4-4-20 with CWP2 at 37°C (1.6-fold increase). In contrast, the optimal secretion temperature for the stable LWHI scTCR in the wild-type system was 30°C (2-fold increase over that seen at 20°C), and only in the presence of ERO1 were the effects of cDNA noticeable at increased temperature (1.6-fold at 37°C). Thus, as a general rule, the 20°C system with overexpressed cDNA (CCW12, ERO1, or RPP0) most often yielded the maximum amount of secreted protein.
DISCUSSION
This study described the mining of a library of engineered yeast strains modified by overexpression of endogenous yeast proteins. It was discovered that although yeast surface display allowed rapid quantitative sorting of the engineered strains, the Aga2p tether masked the effects that the overexpressed yeast proteins had on the scFv or scTCR fusion partner. However, one of the proteins (7/15 scTCR) was particularly sensitive to induction temperature. Thus, under the influence of an elevated temperature that decreased the efficiency of intracellular processing, several yeast strains that promoted increased display and secretion were isolated. The increases were mediated by overexpression of translational components (coded for by RPP0), ER-resident folding assistants (coded for by ERO1), and cell wall proteins (coded for by SED1, CCW12, and CWP2), few of which would likely have been predicted a priori. The increases in heterologous protein secretion were not limited to the screened scTCR, but were also generalizable to additional scTCR and scFv proteins.
Although secreted protein and Aga2p fusion protein destined for display on the cell surface both traverse the same secretory compartments, our observations indicated that the association with the cellular folding machinery, such as BiP and PDI, differed substantially. In particular, increasing the expression levels of the ER-resident BiP and/or PDI had already proven successful in increasing secretion of scFv and scTCR from S. cerevisiae (this work and references 9, 33, and 35). However, BiP and PDI overexpression had no effect on surface display levels. In addition, although an scFv and two scTCR that differ 20-fold in secretion efficiency were put through the initial selection strategy without selection pressure, no overexpressed yeast proteins that could increase display levels were identified. Thus, it appeared that fusion to the Aga2p display scaffold enabled the scFv to bypass the intracellular bottleneck normally encountered by unfused scFv. In addition, even when scFv display was induced at elevated temperatures that normally diminish secretion titers (9, 35), the display levels were not affected, again indicating that Aga2p could dominate the display efficiency of its scFv fusion partner. The consequence of these findings is that screens for engineered yeast cannot be performed under conditions where Aga2p dominates display efficiency.
Thus, to overcome the dominant effects of Aga2p and allow the yeast strain engineering approach to identify yeast proteins that can enhance display and secretion, we employed an scTCR protein whose display levels, unlike those of scFv, were particularly responsive to a selection pressure of elevated induction temperature. In this way, five yeast genes that restore or increase display levels of active protein were identified. Although the five yeast genes increased display of the low-stability 7/15 scTCR at the elevated temperatures, only ERO1 overexpression increased display levels at 20°C (1.4-fold increase), again suggesting that the selection pressure was required to select CCW12, RPP0, SED1, and CWP2 from the yeast library due to Aga2p masking effects. Therefore, although it would be ideal to use this system to screen engineered yeast libraries for any heterologous protein of interest, the protein of interest must be responsive to a selection pressure such as elevated induction temperature for the display-based screen to be successful. However, display-based screening with a single protein substrate allowed the identification of five yeast proteins, several of which can serve as fairly generalizable secretion assistants as discussed below.
Although the five yeast genes were selected at higher temperatures, all except CWP2 promoted increased secretion of at least one protein at 20°C, and 20°C proved optimal for the maximum secretion levels. The two scFvs tested behaved similarly to the 7/15 scTCR in that CCW12 and ERO1 could enhance secretion, albeit to different extents. However, unlike 7/15, the scFvs exhibited increased secretion levels in response to RPP0 overexpression. In contrast to the products of these three genes, the ultrastable LWHI scTCR did not respond to any of the overexpressed yeast genes other than showing modest increases with ERO1 overexpression. Taken together, the temperature-dependent display enhancers (CCW12, CWP2, SED1, and RPP0) seem to facilitate secretion of the lower-expression/stability proteins, while secretion of the LWHI protein was unaffected. On the other hand, the lone 20°C display enhancer, ERO1, yielded statistically significant increases in secretion for all proteins tested, indicating that the most general solutions would be those selected under conditions of induction at 20°C. Unfortunately, as discussed above, the presence of Aga2p prevents such direct selections from being successful.
Two of the isolated display enhancers, Ero1p and Rpp0p, are known to function directly in the protein synthesis and folding process and were therefore expected to enhance protein secretion. The Ero1p protein is essential for yeast viability and functions in delivering oxidizing equivalents to folding disulfide-containing proteins through PDI (5, 26, 40). ERO1 is induced by the unfolded protein response and loss of Ero1p results in accumulation of reduced protein in the ER (5, 26). Therefore, since each of the heterologous proteins investigated contains two disulfide bonds, overexpression of Ero1p likely assists in the formation of these disulfide bonds and promotes exit from the ER. For example, overexpression of Kluveromyces lactis ERO1 has led to increased secretion of disulfide-bonded human serum albumin, but not disulfide-free interleukin-1β (18). The P0 protein (Rpp0p) is one of a set of proteins that assemble at the stalk of the large ribosomal subunit in yeast (20, 31), and excess Rpp0p is not normally observed (31). Thus, it may be possible that under conditions of heterologous protein overexpression, the Rpp0p protein may be a limiting component in the ribosomal assembly, and this deficiency in protein translation capacity may be alleviated by overexpression of the Rpp0p protein. Alternatively, Rpp0p may be functioning indirectly as overexpressed Rpp0p has been implicated in alleviating prion formation in yeast by increasing the activity of promoters containing heat shock elements that drive expression of many chaperones and foldases (15).
We initially hypothesized that several of the genes recovered in the library screen, namely CCW12, CWP2, and SED1, might not increase secretion of the unfused 7/15 scTCR, as these genes have cellular functions related to the yeast cell wall. Since the flow cytometry selection process required surface display involving the Aga1p and Aga2p cell wall proteins, the recovered clones could have been the result of “you get what you select for,” and yeast proteins that facilitate Aga1p and/or Aga2p assembly and processing, rather than scTCR processing, could have been selected. However, none of the cell wall proteins, when overexpressed, affected the display of Aga2p lacking the scTCR fusion partner. Thus, it appeared that the cell wall proteins were regulating surface display in an scTCR-dependent manner and may have had a general influence on the secretory processing of scTCR. Indeed, overexpression of the cell wall genes increased both the surface display of 7/15 (CCW12, CWP2, and SED1) and the secretion of 7/15 (CCW12). scFv secretion was also elevated by CCW12 overexpression, and to a lesser extent by CWP2 and SED1. In contrast, the LWHI scTCR was unaffected by cell wall protein expression, indicating protein-specific effects and not a general change in cell physiology.
Each of the cell wall proteins is covalently linked to the cell wall glycan layer after processing as a glycosylphosphatidylinositol-anchored precursor (21, 23, 42). The proteins have generally been implicated in providing cell wall stability and resistance to stresses. For example, CCW12 deletion or overexpression increases the sensitivity to known cell wall perturbants calcofluor white and Congo red (21), deletion of SED1 made stationary-phase cells more sensitive to Zymolyase treatment (32), and deletion of CWP2, like CCW12, increased sensitivity to calcofluor white and Congo red while also increasing the sensitivity of exponentially growing cells to Zymolyase treatment (42). Thus, the stresses imposed by heterologous protein display and secretion may be diminished by overexpression of cell wall proteins. Although further study will be required to elucidate the mechanism whereby the cell wall proteins assist secretion and display, the results of this study clearly point to the cell wall as a novel target for secretion improvement.
Acknowledgments
We thank Haoping Liu for the yeast cDNA overexpression library and David Kranz for the 1B2 hybridoma. Special thanks also goes to Shawn Brueggemeier for his assistance with construction of scFv expression plasmids.
This work was funded through an NSF CAREER award (BES-0238864).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Antoniukas, L., H. Grammel, and U. Reichl. 2006. Production of hantavirus Puumala nucleocapsid protein in Saccharomyces cerevisiae for vaccine and diagnostics. J. Biotechnol. 124:347-362. [DOI] [PubMed] [Google Scholar]
- 2.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 3.Butz, J. A., R. T. Niebauer, and A. S. Robinson. 2003. Co-expression of molecular chaperones does not improve the heterologous expression of mammalian G-protein coupled receptor expression in yeast. Biotechnol. Bioeng. 84:292-304. [DOI] [PubMed] [Google Scholar]
- 4.Davydenko, S. G., J. K. Juselius, T. Munder, E. Bogengruber, J. Jantti, and S. Keranen. 2004. Screening for novel essential genes of Saccharomyces cerevisiae involved in protein secretion. Yeast 21:463-471. [DOI] [PubMed] [Google Scholar]
- 5.Frand, A. R., and C. A. Kaiser. 1998. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1:161-170. [DOI] [PubMed] [Google Scholar]
- 6.Fukuda, K., M. H. Jensen, R. Haser, N. Aghajari, and B. Svensson. 2005. Biased mutagenesis in the N-terminal region by degenerate oligonucleotide gene shuffling enhances secretory expression of barley alpha-amylase 2 in yeast. Protein Eng. Des. Sel. 18:515-526. [DOI] [PubMed] [Google Scholar]
- 7.Gerngross, T. U. 2004. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 22:1409-1414. [DOI] [PubMed] [Google Scholar]
- 8.Gietz, R. D., and R. A. Woods. 2006. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 313:107-120. [DOI] [PubMed] [Google Scholar]
- 9.Hackel, B. J., D. Huang, J. C. Bubolz, X. X. Wang, and E. V. Shusta. 2006. Production of soluble and active transferrin receptor-targeting single-chain antibody using Saccharomyces cerevisiae. Pharm. Res. 23:790-797. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 11.Huang, D., and E. V. Shusta. 2005. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol. Prog. 21:349-357. [DOI] [PubMed] [Google Scholar]
- 11a.Huang, D., and E. V. Shusta. 2006. Yeast platform for the production of single-chain antibody-green fluorescent protein fusions. Appl. Environ. Microbiol. 72:7748-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, E. W. 2002. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351:127-150. [DOI] [PubMed] [Google Scholar]
- 13.Kieke, M. C., E. V. Shusta, E. T. Boder, L. Teyton, K. D. Wittrup, and D. M. Kranz. 1999. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc. Natl. Acad. Sci. USA 96:5651-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kjeldsen, T., S. Ludvigsen, I. Diers, P. Balschmidt, A. R. Sorensen, and N. C. Kaarsholm. 2002. Engineering-enhanced protein secretory expression in yeast with application to insulin. J. Biol. Chem. 277:18245-18248. [DOI] [PubMed] [Google Scholar]
- 15.Kryndushkin, D. S., V. N. Smirnov, M. D. Ter-Avanesyan, and V. V. Kushnirov. 2002. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277:23702-23708. [DOI] [PubMed] [Google Scholar]
- 16.Laboissiere, M. C., S. L. Sturley, and R. T. Raines. 1995. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J. Biol. Chem. 270:28006-28009. [DOI] [PubMed] [Google Scholar]
- 17.Liu, H., J. Krizek, and A. Bretscher. 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodi, T., B. Neglia, and C. Donnini. 2005. Secretion of human serum albumin by Kluyveromyces lactis overexpressing KlPDI1 and KlERO1. Appl. Environ. Microbiol. 71:4359-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning, T. C., C. J. Schlueter, T. C. Brodnicki, E. A. Parke, J. A. Speir, K. C. Garcia, L. Teyton, I. A. Wilson, and D. M. Kranz. 1998. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity 8:413-425. [DOI] [PubMed] [Google Scholar]
- 20.Mitsui, K., and K. Tsurugi. 1988. cDNA and deduced amino acid sequence of 38 kDa-type acidic ribosomal protein A0 from Saccharomyces cerevisiae. Nucleic Acids Res. 16:3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mrsa, V., M. Ecker, S. Strahl-Bolsinger, M. Nimtz, L. Lehle, and W. Tanner. 1999. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 181:3076-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton, C. H., L. C. Shimmin, J. Yee, and P. P. Dennis. 1990. A family of genes encode the multiple forms of the Saccharomyces cerevisiae ribosomal proteins equivalent to the Escherichia coli L12 protein and a single form of the L10-equivalent ribosomal protein. J. Bacteriol. 172:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oender, K., M. Loeffler, E. Doppler, M. Eder, S. Lach, F. Heinrich, T. Karl, R. Moesl, H. Hundsberger, T. Klade, P. Eckl, J. R. Dickinson, M. Breitenbach, and L. Koller. 2003. Translational regulator RpL10p/Grc5p interacts physically and functionally with Sed1p, a dynamic component of the yeast cell surface. Yeast 20:281-294. [DOI] [PubMed] [Google Scholar]
- 24.Parekh, R. N., and K. D. Wittrup. 1997. Expression level tuning for optimal heterologous protein secretion in Saccharomyces cerevisiae. Biotechnol. Prog. 13:117-122. [DOI] [PubMed] [Google Scholar]
- 25.Planta, R. J., and W. H. Mager. 1998. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14:471-477. [DOI] [PubMed] [Google Scholar]
- 26.Pollard, M. G., K. J. Travers, and J. S. Weissman. 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1:171-182. [DOI] [PubMed] [Google Scholar]
- 27.Rakestraw, A., and K. D. Wittrup. 2006. Contrasting secretory processing of simultaneously expressed heterologous proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:896-905. [DOI] [PubMed] [Google Scholar]
- 28.Ramjee, M. K., J. R. Petithory, J. McElver, S. C. Weber, and J. F. Kirsch. 1996. A novel yeast expression/secretion system for the recombinant plant thiol endoprotease propapain. Protein Eng. 9:1055-1061. [DOI] [PubMed] [Google Scholar]
- 29.Robinson, A. S., J. A. Bockhaus, A. C. Voegler, and K. D. Wittrup. 1996. Reduction of BiP levels decreases heterologous protein secretion in Saccharomyces cerevisiae. J. Biol. Chem. 271:10017-10022. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, A. S., V. Hines, and K. D. Wittrup. 1994. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Biotechnology 12:381-384. [DOI] [PubMed] [Google Scholar]
- 31.Santos, C., and J. P. Ballesta. 1994. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 269:15689-15696. [PubMed] [Google Scholar]
- 32.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shusta, E. V., P. D. Holler, M. C. Kieke, D. M. Kranz, and K. D. Wittrup. 2000. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 18:754-759. [DOI] [PubMed] [Google Scholar]
- 34.Shusta, E. V., M. C. Kieke, E. Parke, D. M. Kranz, and K. D. Wittrup. 1999. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 292:949-956. [DOI] [PubMed] [Google Scholar]
- 35.Shusta, E. V., R. T. Raines, A. Pluckthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson, L. F., B. K. Kennedy, and E. Harlow. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 98:3946-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tachibana, C., and T. H. Stevens. 1992. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol. Cell. Biol. 12:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 40.Tu, B. P., S. C. Ho-Schleyer, K. J. Travers, and J. S. Weissman. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290:1571-1574. [DOI] [PubMed] [Google Scholar]
- 41.Valkonen, M., M. Penttilä, and M. Saloheimo. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Vaart, J. M., L. H. Caro, J. W. Chapman, F. M. Klis, and C. T. Verrips. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177:3104-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yaver, D. S., M. Lamsa, R. Munds, S. H. Brown, S. Otani, L. Franssen, J. A. Johnstone, and H. Brody. 2000. Using DNA-tagged mutagenesis to improve heterologous protein production in Aspergillus oryzae. Fungal Genet. Biol. 29:28-37. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W., H. L. Zhao, C. Xue, X. H. Xiong, X. Q. Yao, X. Y. Li, H. P. Chen, and Z. M. Liu. 2006. Enhanced secretion of heterologous proteins in Pichia pastoris following overexpression of Saccharomyces cerevisiae chaperone proteins. Biotechnol. Prog. 22:1090-1095. [DOI] [PubMed] [Google Scholar]