Abstract
Some psychrotrophic lactic acid bacteria (LAB) are specific meat spoilage organisms in modified-atmosphere-packaged (MAP), cold-stored meat products. To determine if incoming broilers or the production plant environment is a source of spoilage LAB, a total of 86, 122, and 447 LAB isolates from broiler carcasses, production plant air, and MAP broiler products, respectively, were characterized using a library of HindIII restriction fragment length polymorphism (RFLP) patterns of the 16 and 23S rRNA genes as operational taxonomic units in numerical analyses. Six hundred thirteen LAB isolates from the total of 655 clustered in 29 groups considered to be species specific. Sixty-four percent of product isolates clustered either with Carnobacterium divergens or with Carnobacterium maltaromaticum type strains. The third major product-associated cluster (17% of isolates) was formed by unknown LAB. Representative strains from these three clusters were analyzed for the phylogeny of their 16S rRNA genes. This analysis verified that the two largest RFLP clusters consisted of carnobacteria and showed that the unknown LAB group consisted of Lactococcus spp. No product-associated LAB were detected in broiler carcasses sampled at the beginning of slaughter, whereas carnobacteria and lactococci, along with some other specific meat spoilage LAB, were recovered from processing plant air at many sites. This study reveals that incoming broiler chickens are not major sources of psychrotrophic spoilage LAB, whereas the detection of these organisms from the air of the processing environment highlights the role of processing facilities as sources of LAB contamination.
Contamination of modified-atmosphere-packaged (MAP) meat products with specific psychrotrophic lactic acid bacteria (LAB) may result in rapid spoilage during the expected shelf life (4, 9, 11, 16) and is thus a major concern of the modern meat industry. Due to the ability of these LAB to grow at refrigerated temperatures and survive the elevated levels of CO2 in the modified atmospheres, they can proliferate to become a major portion of the microbial population of MAP meat products, whereas the growth of aerobic spoilage bacteria and Enterobacteriaceae is restricted (11, 24). The typical meat spoilage changes caused by LAB are an off-odor, an off-taste, gas or slime formation, and/or discolorations (4, 11, 13, 24). These sensory defects become evident after the spoilage LAB population has remained in the stationary-growth phase (22). Usually, LAB levels above 107 CFU/g are then detected. LAB are not equal in their meat spoilage potentials. The genera associated with quality deterioration in MAP fresh meats include Carnobacterium, Lactobacillus, and Leuconostoc (11, 24). While spoilage LAB populations in MAP beef, pork, and cooked meat products have been well documented and also reviewed in the literature (11, 14, 24, 29, 30), few publications have dealt with LAB levels and species in retail MAP poultry. These studies have focused mainly on marinated products due to their high LAB counts (8, 31) and the unexpected, rapidly developing gaseous type of spoilage caused by Leuconostoc gasicomitatum (4).
There are no data on sources of specific spoilage LAB in poultry slaughter and processing. Although incoming live broiler chickens introduce vast indigenous LAB populations to slaughterhouses, little is known about the LAB species variety associated with broilers. While the LAB in chicken gastrointestinal and respiratory tracts have been reported to belong mainly to the genera of Lactobacillus, Enterococcus, and Streptococcus (15, 17, 25), no research has focused on showing if psychrotrophic meat spoilage LAB belong to the microbiome of broilers. Since some spoilage LAB have been associated with meat products from certain animal species, the animal has been considered to be the source of contamination. The temperature of the chicken intestine (body temperature, 41 to 42°C) is too high to support the colonization of the intestinal mucosa by psychrotrophic LAB, which usually do not grow at temperatures exceeding 37°C (4, 20, 21, 23). However, it is not known if broiler skin, feathers, or mucous membranes of lower temperatures harbor psychrotrophic spoilage LAB.
The aim of this study was to determine whether the skin or mucous membranes of broilers harbor psychrotrophic LAB or whether, alternatively, products become contaminated with these bacteria during further processing stages. For the latter possibility, we chose monitoring of airborne LAB, since roof-attached hanging conveyers expose carcasses to airborne contamination. To allow the comparison between LAB in broilers, the processing environment, and products, psychrotrophic LAB from cold-stored, nonmarinated, late-shelf-life products were also characterized.
MATERIALS AND METHODS
Study design for carcass and air sampling in a broiler processing facility.
Sampling of broiler carcasses and processing plant air was conducted in a large-scale commercial processing facility, manufacturer B, manufacturing all the different types of products included in this study. This facility was divided into two major sections: the slaughterhouse, where the broilers were killed, defeathered, eviscerated, and chilled, and the processing plant section, where carcasses were processed further (i.e., cut, boned, and packaged). Carcasses were sampled during four visits to the scalding and defeathering area of the slaughterhouse section. Samples were collected at two sites situated before and after the scalding tank prior to the defeathering and evisceration operations. Broiler carcasses hanging on the moving conveyer line were sampled by cutting pieces from the neck skin and oropharynx and by plucking single feathers from breast area. Of the feathers, only the feather shaft was cut for analysis; the rest was discarded. Air samples were collected during three visits to a routine operation in the processing plant section. Following the slaughtering operations, whole eviscerated carcasses were air chilled and graded for weight and quality in a preselection room (preprocessing area) before entering the main processing area. The final processing stages for boneless products occurred in a separate room (boned-product packaging). The processing line was highly automated, and manual handling of certain products was limited to trimming, boning, and packaging steps.
Enrichment of carcass samples for specific spoilage LAB.
Two separate selective enrichment procedures were performed to target the analysis at the meat-spoilage-associated LAB. Vancomycin was added to MRS broth to select for leuconostocs, which have been identified as spoilage organisms of marinated MAP poultry (4, 31). Members of the genus Leuconostoc are intrinsically resistant to vancomycin (2), whereas in Finland many other LAB, such as fecal enterococci, are generally susceptible to vancomycin (27). A total of 31 carcass samples, of which 5, 10, and 16 were from neck skin, oropharynges, and feather shafts, respectively, were enriched separately in MRS broth (Difco, Detroit, MI) supplemented with 10 μg/ml vancomycin (Sigma, St. Louis, MO) and incubated at 25°C for 16 h. If no growth occurred, incubation was prolonged for as long as 5 days. In addition, prior to the experiment, the ability of Leuconostoc gasicomitatum to grow in vancomycin-containing MRS broth was checked with four strains.
Another selective enrichment, for psychrotrophic LAB in general, was performed. Thirty-four carcass samples, of which 4, 10, and 20 were from neck skin, oropharynges, and feathers, respectively, were placed separately in MRS broth and incubated at 6°C for 38 days. After psychrotrophic incubation, the tubes were transferred to 25°C to show if any LAB were present. From each enriched broth showing growth, a loopful (10 μl) was streaked onto MRS agar (Oxoid, Basingstoke, United Kingdom) and incubated in an anaerobic atmosphere (sealed jars with AnaeroGen sachet; Oxoid) at 25°C for 5 days to produce individual colonies. Generally, one colony was streaked to purity on MRS plates. If clearly different colony morphologies were observed, more colonies were picked. Each colony was purified by subculturing in MRS broth and streaking onto MRS agar. For DNA isolation, the strains were inoculated into MRS broth to produce cell mass.
Sampling of airborne LAB.
Airborne LAB were sampled using two Reuter centrifugal air samplers (RCS sampler; Biotest AG, Dreieich, Germany) and a sampling time of 8 min (320 liters of air). The air-sampling sites associated with carcass processing were categorized as follows: the preprocessing area, the main processing area, and the room for boned-product packaging. A total of 43 air samples were taken, of which a total of 6, 29, 6, and 2 were from the preprocessing area, the main processing area, the room for boned-product packaging, and the corridor adjacent to the processing areas, respectively. Samples from air-chilling and processing areas were taken near the railed carcasses or conveyors, avoiding drips from the carcasses or carcass portions. The RCS samplers were swabbed with 70% ethanol between measurements, and airborne LAB on each site were sampled on a fresh MRS agar strip. The strips were incubated under an anaerobic atmosphere at 25°C for 5 days. CFU were counted, and the LAB levels were reported as CFU per square meter of air sampled. A total of 122 colonies, 1 to 5 from each strip, were randomly picked and cultured pure for DNA isolation.
Sampling and enumeration of LAB in broiler products at the retail level.
Table 1 shows the numbers of packages analyzed, the product types included in the study, and their manufacturer-defined shelf lives. Branded products from three large-scale broiler product manufacturers (A, B, and C) were purchased from retail stores during a 3-month period to obtain products possessing typical quality for the retail market level. Both skin-on and skinless broiler meat products, representing the main types manufactured, were included (i.e., skin-on leg and breast cuts, boned and skinned breast fillet cutlets and strips, and minced breast meat). All manufacturers had applied an anaerobic modified atmosphere with 80% CO2 and 20% N2 for packaging. The packages were stored at 6°C until analysis.
TABLE 1.
LAB levels in MAP broiler meat products of three manufacturersa
| Manufacturer | Leg cuts
|
Breast cuts
|
Skinned fillet cutlets
|
Skinned fillet strips
|
Minced meat
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLb | No.c | LAB levels (CFU/g) | SL | No. | LAB levels (CFU/g) | SL | No. | LAB levels (CFU/g) | SL | No. | LAB levels (CFU/g) | SL | No. | LAB levels (CFU/g) | |
| A | 10 | 2 | 1.1 × 108-1.2 × 108 | 10 | 8 | 1.4 × 104-7.9 × 107 | 10 | 6 | 2.7 × 106-3.3 × 108 | ||||||
| B | 10 | 5 | 1.0 × 106-3.6 × 107 | 10 | 5 | 1.0 × 107-1.3 × 108 | 10 | 3 | 3.3 × 107-1.2 × 108 | 10 | 5 | 1.0 × 106-1.9 × 108 | 7 | 5 | 2.1 × 106-1.2 × 107 |
| C | 7 | 5 | 1.9 × 105-4.2 × 106 | 8 | 5 | 1.7 × 104-1.5 × 106 | |||||||||
Products were analyzed on the last day of the manufacturer-defined shelf life.
SL, manufacturer-defined shelf life (in days).
Number of packages analyzed.
On the sell-by day (±1 day), 22-g samples were aseptically weighed and homogenized in 198 ml of 0.1% peptone water. Prior to microbiological analyses, the sample LAB were enumerated from serial 10-fold dilutions on MRS agar after incubation of the plates under anaerobic conditions at 25°C for 5 days. To characterize the prevailing LAB, 10 colonies were randomly picked from the plates of the highest dilutions showing growth and were purified by subculturing on MRS medium.
After microbiological sampling, the pH of each sample homogenate was measured using a WTW-530 digital pH meter (Wissenschaftliche-Technische Werkstätten, Weilheim, Germany).
Isolation of DNA, restriction endonuclease analysis, and restriction fragment length polymorphism (RFLP) of the 16 and 23S rRNA-encoding genes for LAB species identification.
Cells harvested from 1 to 2 ml of MRS broth culture were used for DNA analyses. DNA was isolated by the guanidium thiocyanate method of Pitcher et al. (28) as modified by Björkroth and Korkeala (5) with combined lysozyme and mutanolysin (Sigma). Restriction endonuclease treatment of 8 μg of DNA was done by using the HindIII restriction enzyme (New England Biolabs, Beverly, MA) as recommended by the manufacturer. DNA fragments were separated by agarose gel electrophoresis and the resulting fingerprint patterns transferred to a nylon membrane via Southern blotting using a vacuum blotting device (Vacugene; Pharmacia, Uppsala, Sweden). Ribotyping was performed using a cDNA probe reverse transcribed (with avian myeloblastosis virus reverse transcriptase; Promega, Madison, WI) from 16 and 23S rRNA and digoxigenin labeled with a DIG DNA labeling kit (Roche Molecular Biochemicals, Mannheim, Germany) as described by Blumberg et al. (10). Membranes were hybridized at 58°C overnight, and the digoxigenin-labeled fragments (ribopatterns) were detected as recommended by Roche Molecular Biochemicals.
LAB database and numerical pattern analysis.
The HindIII ribopatterns were compared to the corresponding patterns in the previously established LAB database of the Department of Food and Environmental Hygiene, University of Helsinki, Helsinki, Finland. This database comprises patterns of all relevant food-associated LAB in the genera Aerococcus, Carnobacterium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, and Weissella (3, 4, 5, 6, 7, 9, 19, 21, 26, 31). It utilizes 16 and 23S rRNA gene HindIII RFLP patterns of more than 300 type and reference strains as operational taxonomic units in numerical analyses. The isolates are identified based on the locations of type and reference strains within the clusters. The reliability of the clusters for distinguishing between different species has been evaluated in several polyphasic taxonomy studies of LAB (8, 18-21).
For the numerical analysis, the ribopatterns were scanned using a Hewlett-Packard (Boise, ID) ScanJet 4c/T scanner. The patterns were normalized based on the mobility of standards, and a similarity matrix was created using the BioNumerics (version 4.1) software package (Applied Maths, Sint-Martens-Latem, Belgium). The similarity between all pairs was expressed by Dice coefficient correlation, and UPGMA (unweighted-pair group method using arithmetic averages) clustering was used for the construction of the dendrogram. Based on the use of internal controls in the database, a pattern optimization and a band position tolerance of 0.5 and 1.5, respectively, were allowed.
16S rRNA gene sequence analysis of representative strains from the major LAB groups.
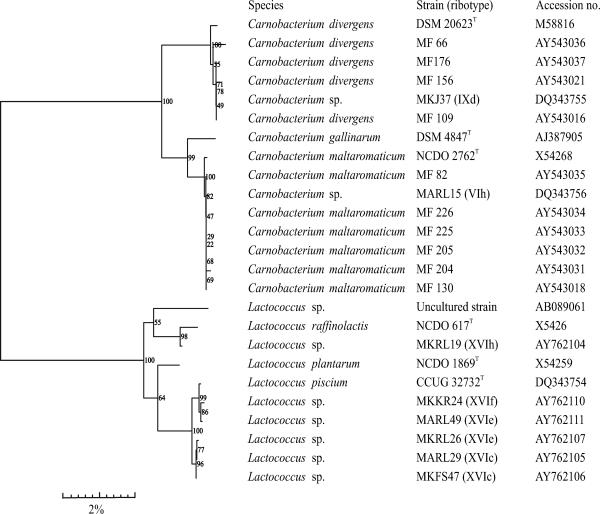
To confirm/obtain the genus-level identification of the three major RFLP clusters, of which two were considered to be carnobacteria and one was unknown, the 16S rRNA genes of eight isolates representing six different ribopatterns (ribotypes VIh and IXd from the carnobacterial clusters and ribotypes XVIc, XVIe, XIVf, and XVIh from the unknown cluster [Fig. 1 ]) were sequenced. Numerical analyses of the ribopatterns considered unknown had not resulted in clustering with any of the type strains. DNA was isolated as for the RFLP analysis. The nearly complete (at least 1,400 bases sequenced) 16S rRNA gene was amplified by PCR with the universal primer pair F8-27 (5′-AGAGTTTGATCCTGGCTGAG-3′) and R1541-1522 (5′-AAGGAGGTGATCCAGCCGCA-3′). The purified (QIAquick PCR purification kit; QIAGEN, Venlo, Netherlands) PCR product was sequenced from both directions by Sanger's dideoxynucleotide chain termination method using primers F19-38 (5′-CTGGCTCAGGAYGAACGCTG-3′), F926 (5′-AACTCAAAGGAATTGACGG-3′), R519 (5′-GTATTACCGCGGCTGCTG-3′), and R1541-1522. Samples were run in a Global IR2 sequencing device with e-Seq (version 2.0) software (LiCor, Lincoln, NE) according to the manufacturer's instructions. The consensus sequences of these strains (created with AlignIR software; LiCor) and of representative strains belonging to the same phylogenetic group (retrieved from GenBank [http://www.ncbi.nlm.nih.gov] using BLASTN 2.2.6 [1]) were aligned, and a phylogenetic tree (Fig. 2) was constructed based on the neighbor-joining method using the BioNumerics (version 4.5) software package (Applied Maths, Sint-Martens-Latem, Belgium).
FIG. 1.
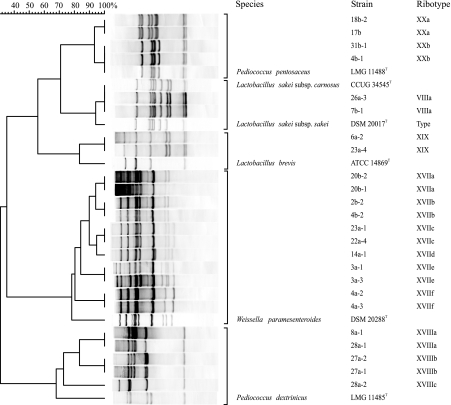
HindIII ribopatterns of LAB from modified-atmosphere-packaged poultry meat products and of product-associated LAB sampled from processing plant air. Products were analyzed at the end of shelf life. The numerical analysis of pattern similarities is presented as a dendrogram and converted to percentages for convenience. Bands on the left have high molecular masses of <23 kbp; those on the right have low molecular masses of >1,000 bp. The footnote a) indicates genus-level identification based on 16S rRNA gene sequence analysis.
FIG. 2.
Phylogenetic tree based on homologies of almost-entire 16S rRNA gene sequences (at least 1,450 bp) of lactococci, carnobacteria, and eight isolates representing the main riboclusters obtained in the study. Bootstrap probability values from 500 trees that were resampled are given at the branch points.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the following eight strains (with accession numbers in parentheses) have been deposited in the GenBank nucleotide sequence database: Carnobacterium sp. strains MKJ37 (DQ343755) and MARL15 (DQ343756) and Lactococcus sp. strains MKRL19 (AY762104), MARL29 (AY762105), MKFS47 (AY762106), MARL49 (AY762107), MKKR24 (AY762110), and MARL49 (AY762111).
RESULTS
LAB detected in carcasses by selective enrichment procedures.
The vancomycin enrichment procedure of the 31 broiler carcass samples resulted in subjecting a total of 86 LAB isolates to numerical analysis of their HindIII ribopatterns. Table 2 shows the species distribution of those 85 LAB isolates that clustered together with type strains. The different patterns obtained and a dendrogram based on pattern similarity are presented in Fig. 3. The species detected were Weissella paramesenteroides (36 isolates), Pediococcus pentosaceus (33 isolates), Pediococcus dextrinicus (7 isolates), Lactobacillus sakei (6 isolates), and Lactobacillus brevis (3 isolates), whereas Leuconostoc spp. were not recovered. One isolate could not be identified by the database. The LAB species detected by vancomycin enrichment have been reported either to be intrinsically glycopeptide resistant or to be resistant to high vancomycin levels (2, 12). However, with the exception of L. sakei, which is sometimes associated with the production of ropy slime in cooked meat products, these species are not considered to be meat spoilage LAB.
TABLE 2.
Numbers and species distribution of 85 LAB isolates obtained from broiler skin, feather shaft, and oropharyngeal samplesa
| Species | Ribotype | No. of isolates obtained from:
|
|||||
|---|---|---|---|---|---|---|---|
| Prescalding samples
|
Postscalding samples
|
||||||
| O | N | F | O | N | F | ||
| Lactobacillus brevis | XIX | 1 | 2 | ||||
| Lactobacillus sakei | VIIIa | 3 | 2 | 1 | |||
| Pediococcusdextrinicus | XVIIIa | 2 | 1 | 1 | |||
| XVIIIb | 2 | ||||||
| XVIIIc | 1 | ||||||
| Pediococcuspentosaceus | XXa | 9 | 4 | 8 | |||
| XXb | 5 | 1 | 4 | 2 | |||
| Weissellaparamesenteroides | XVIIa | 3 | |||||
| XVIIb | 2 | ||||||
| XVIIc | 7 | 2 | 8 | ||||
| XVIId | 2 | 5 | 2 | ||||
| XVIIe | 2 | ||||||
| XVIIf | 3 | ||||||
Samples were enriched in MRS broth supplemented with 10 μg/ml vancomycin. Identification was based on a 16S and 23S rRNA gene HindIII RFLP database. O, oropharyngeal samples; N, neck skin samples; F, feather shaft samples.
FIG. 3.
HindIII ribopatterns of LAB obtained by vancomycin enrichment from broiler skin, oropharynges, and feathers. The numerical analysis of pattern similarities is presented as a dendrogram and converted to percentages for convenience. Bands on the left have high molecular masses of <23 kbp; those on the right have low molecular masses of >1,000 bp.
With the cold enrichment, none of the 34 carcass samples incubated in MRS broth at 6°C resulted in growth in 38 days. However, after the psychrotrophic incubation, the tubes were transferred to 25°C, and they all showed growth. A total of 30 LAB isolates originating from these tubes were subjected to numerical analysis, resulting in clustering of 24 isolates in groups considered species specific. The species detected were Enterococcus faecalis (10 isolates), Enterococcus faecium (7 isolates), Lactococcus garviae (3 isolates), Enterococcus raffinosus (1 isolate), Lactobacillus curvatus (1 isolate), Pediococcus acidilactici (1 isolate), and W. paramesenteroides (1 isolate). Six isolates could not be identified by the RFLP database.
Airborne LAB in the processing environment.
Airborne LAB levels showed great variability depending on the area sampled. The highest counts were detected inside the air-chilling area (>344 CFU/m3), while the counts obtained from the other areas were generally very low (<30 CFU/m3) or the growth was undetectable. Of the 122 LAB isolates randomly picked from the air sample strips, 45% (55 of 122) represented species also detected in the late-shelf-life broiler products. The number and species distribution of these LAB are presented in Table 3, and the corresponding ribopatterns are shown in Fig. 1. In addition, air sampling yielded a total of 67 isolates representing various ribotypes not associated with the products. Thirteen of the latter isolates were assigned to the species Enterococcus malodoratus (3 isolates), Enterococcus pseudoavium (1 isolate), Leuconostoc citreum (3 isolates), Leuconostoc lactis (3 isolates), Leuconostoc pseudomesenteroides (2 isolates), and P. acidilactici (1 isolate). Numerical analysis of the ribopatterns obtained from a total of 54 isolates did not result in species-specific clusters by the RFLP LAB database.
TABLE 3.
Numbers and species distribution of 55 airborne LAB isolates associated with productsa
| Species | Ribotype | No. of LAB isolates
|
|||
|---|---|---|---|---|---|
| Preprocessing area | Main processing area | Boned-product packaging area | Corridor | ||
| Leuconostocgasicomitatum | Ib | 4 | |||
| Streptococcus parauberis | IVb | 2 | |||
| IVc | 2 | 1 | |||
| Carnobacteriummaltaromaticum | VIa | 1 | 6 | 1 | 1 |
| VIb | 2 | ||||
| VIc | 2 | ||||
| VIf | 1 | ||||
| Carnobacteriumdivergens | IXa | 1 | 2 | 1 | 2 |
| Lactobacillus curvatus | X | 2 | |||
| Enterococcusraffinosus | XIIb | 1 | |||
| Enterococcusfaecium | XIII | 6 | 7 | 1 | |
| Streptococcus alactolyticus | XV | 1 | |||
| Lactococcus spp. | XVIa | 1 | |||
| XVId | 1 | ||||
| XVIe | 1 | ||||
| XVIf | 1 | 1 | |||
| XVIh | 1 | 2 | |||
| Total | 13 | 31 | 7 | 4 | |
Identification was based on a 16 and 23S rRNA gene HindIII RFLP database.
LAB levels and species detected in the products.
Table 1 presents the ranges of LAB levels detected in the different types of products. In the products of manufacturers A and B, LAB populations had reached levels (>107 CFU/g) generally indicative of early spoilage changes in meat products (4, 8, 22) in 28 of the 39 packages studied. In the products of manufacturer C, with shorter manufacturer-defined shelf-lives, the LAB levels were 1 to 2 log units lower than those in the products of the other two manufacturers. The pH of the skin-containing leg and breast cuts ranged from 6.2 to 6.4 and from 5.8 to 6.2, respectively. In skinned products made of broiler breast fillets and fillet strips, the pH ranged from 5.8 to 6.1. The highest pH values, ranging from 6.4 to 6.5, were measured in minced meat.
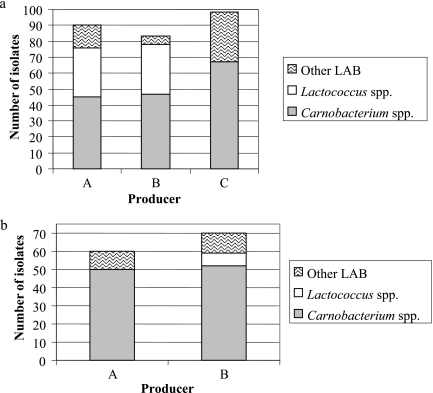
Table 4 shows the species division of the 447 product-associated LAB isolates based on the RFLP database analysis. The dendrogram presenting clustering of the representative HindIIIribopatterns of these isolates is shown in Fig. 1. Carnobacterium divergens and Carnobacterium maltaromaticum were clearly the most abundant LAB identified in both skin-containing and skinless products of all three manufacturers (Table 4; Fig. 4), with the exception of L. curvatus, which was detected along with C. divergens in minced meat. Forty-three percent of all product isolates (191 of 447 isolates) were identified as C. divergens and 21% (96 of 447 isolates) as C. maltaromaticum. In harmony with the RFLP results, the phylogenetic tree deduced from the 16S rRNA gene sequences (Fig. 2) located the two strains MKJ37 and MARL15, representing the C. divergens and C. maltaromaticum RFLP clusters, respectively, in the same branches as the C. divergens and C. maltaromaticum type and reference strains.
TABLE 4.
Number and species distribution of 447 LAB isolates from MAP broiler products of manufacturers A, B, and C analyzed at the end of their manufacturer-defined shelf livesa
| Species | Ribotype | No. of LAB isolates in the following products from the indicated manufacturer:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Skin-containing products
|
Skinned and boned breast fillet products
|
Total | |||||||||
| Breast cuts
|
Leg cuts
|
Strips and cutlets
|
Minced meat (B) | ||||||||
| A | B | C | A | B | C | A | B | ||||
| Leuconostocgasicomitatum | Ia | 1 | 1 | 2 | |||||||
| Ib | 2 | 2 | |||||||||
| Ic | 1 | 3 | 4 | ||||||||
| Leuconostocgelidum | II | 1 | 1 | ||||||||
| Leuconostoccarnosum | IIIa | 2 | 2 | ||||||||
| IIIb | 1 | 1 | 2 | ||||||||
| Streptococcus parauberis | IVa | 1 | 1 | 2 | |||||||
| Lactococcusgarviae | V | 1 | 1 | ||||||||
| Carnobacteriummaltaromaticum | VIa | 4 | 13 | 11 | 2 | 17 | 1 | 1 | 13 | 4 | 66 |
| VIb | 1 | 2 | 2 | 5 | |||||||
| VIc | 1 | 1 | 2 | 1 | 5 | ||||||
| VId | 1 | 1 | |||||||||
| VIe | 2 | 2 | |||||||||
| VIf | 1 | 2 | 6 | 4 | 1 | 14 | |||||
| VIg | 1 | 1 | |||||||||
| VIh | 2 | 2 | |||||||||
| Lactobacillus oligofermentans | VIIa | 1 | 2 | 3 | |||||||
| VIIb | 2 | 1 | 3 | ||||||||
| VIIc | 1 | 5 | 2 | 8 | |||||||
| Lactobacillus sakei | VIIIa | 3 | 7 | 3 | 13 | ||||||
| VIIIb | 5 | 1 | 6 | ||||||||
| VIIIc | 3 | 3 | |||||||||
| Carnobacteriumdivergens | IXa | 22 | 1 | 5 | 3 | 12 | 40 | 30 | 15 | 128 | |
| IXb | 2 | 2 | |||||||||
| IXc | 8 | 8 | 3 | 1 | 24 | 7 | 8 | 59 | |||
| IXd | 2 | 2 | |||||||||
| Lactobacillus curvatus | X | 1 | 2 | 3 | 1 | 6 | 13 | ||||
| Lactobacillus fuchuensis | XI | 1 | 1 | ||||||||
| Enterococcusraffinosus | XIIa | 2 | 2 | ||||||||
| Enterococcusfaecium | XIII | 2 | 2 | ||||||||
| Lactococcusraffinolactis | XIV | 1 | 1 | ||||||||
| Streptococcus alactolyticus | XV | 1 | 1 | ||||||||
| Lactococcus spp. | XVIb | 1 | 1 | ||||||||
| XVIc | 11 | 1 | 12 | ||||||||
| XVId | 7 | 1 | 2 | 10 | |||||||
| XVIe | 5 | 10 | 9 | 2 | 5 | 31 | |||||
| XVIf | 2 | 5 | 3 | 10 | |||||||
| XVIg | 3 | 1 | 1 | 5 | |||||||
| XVIh | 1 | 1 | 1 | 3 | |||||||
| XVIi | 2 | 2 | |||||||||
| Unknown I | UI | 1 | 1 | ||||||||
| Unknown II | UII | 1 | 1 | ||||||||
| Unknown III | UIII | 7 | 2 | 9 | |||||||
| Unknown IV | UIVa | 2 | 2 | ||||||||
| UIVb | 1 | 1 | |||||||||
| Total no. of strains | 74 | 35 | 49 | 17 | 46 | 50 | 60 | 70 | 46 | 447 | |
Identification was based on a 16 and 23S rRNA gene HindIII RFLP database.
FIG. 4.
(a) Proportions of the major LAB groups in modified-atmosphere-packaged skin-containing broiler leg and breast cuts manufactured by three plants, A, B, and C. Products were analyzed on the last day of the manufacturer-defined shelf life. (b) Proportions of the major LAB groups in modified-atmosphere-packaged skinless breast fillet products of two manufacturers, A and B. Products were analyzed on the last day of the manufacturer-defined shelf life.
The third major LAB group (74 of 447 isolates) distinguished by the RFLP database was formed by unidentified isolates possessing nine different ribopatterns. Based on the phylogenetic analysis of the 16S rRNA gene sequences of six isolates, they were considered to belong to the genus Lactococcus. Five of them (ribotypes XVIc to -f) were located in a branch (Fig. 2) containing Lactococcus piscium type strain CCUG 32732, and one (ribotype XVIh) clustered together with Lactococcus raffinolactis type strain NCFB 617. Lactococci were recovered mainly from skin-containing products (Table 4). From the products of manufacturer C, only two lactococcal isolates were obtained. This manufacturer differed from the other two by setting shorter shelf lives for its products, which also showed lower LAB levels than the products of the two other manufacturers (Table 1).
Other species detected in the late-shelf-life LAB populations were L. sakei (22 of 447 isolates), Lactobacillus oligofermentans (14 of 447 isolates), L. curvatus (13 of 447 isolates), and L. gasicomitatum (8 of 447 isolates).
DISCUSSION
This study assessed LAB in two stages of broiler product manufacture to determine whether species prevailing in late-shelf-life products are detected in broilers handled at the beginning of slaughter or whether products become contaminated during further processing stages. C. divergens and C. maltaromaticum were prevalent among the LAB populations in MAP broiler products. The other species detected in the products were mainly from the genera Lactococcus (17%) and Lactobacillus (11%). In contrast to the findings of previous studies of marinated fillet products (4, 31), Leuconostoc spp. were rarely (2.9%) recovered.
Based on our results, skin, feathers, and mucous membranes of broilers were not major sources of psychrotrophic LAB associated with late-shelf-life MAP broiler products. The selective enrichment approaches used for recovery of specific spoilage LAB from carcass samples did not, with the exception of L. sakei, detect LAB species associated with meat spoilage. Other LAB detected by the two enrichment procedures belonged neither to species associated with products nor to those considered specific meat spoilage organisms. Moreover, in the cold-enriched carcass samples, no growth was observed in 38 days. However, when these samples were incubated at 25°C after cold enrichment, growth was detected in all samples, and nonpsychrotrophic species, mainly E. faecium and E. faecalis, were recovered. In a previous study (8), these species were commonly detected also in MAP marinated broiler legs of manufacturer B right after packaging but not within the spoilage LAB population after cold storage at 6°C for 17 days. These findings highlight the importance of determining specific spoilage organisms when the role of LAB contaminants in product shelf life stability is evaluated.
In contrast to the broiler carcasses, nearly half of the isolates (45%) obtained from processing plant air represented species associated with retail MAP broiler products and spoilage of MAP poultry in previous studies (4, 8). Carnobacterium spp. and L. gasicomitatum were detected from air sampled during the final processing operations, whereas lactococci were isolated mainly from early processing stages, such as air chilling. Our results show that the processing facilities are a more likely source of psychrotrophic LAB contamination than the broilers. At present, it is not known how these LAB enter the processing environment and how the air becomes contaminated. The products from different manufacturers possessed quite similar LAB diversities. Since manufacturing practices in modern plants are similar, it is tempting to theorize that the plant environment favors the survival of psychrotrophic LAB, resulting in similar in-house populations. On the other hand, product-associated LAB were recovered from corridors adjacent to the poultry meat processing areas, and it is also possible that these organisms are continuously introduced into the processing environment outside the plant as a result of airflows and employer activities.
In conclusion, the spoilage-associated LAB are not introduced to the processing environment or the plant air along with the broilers. However, the common detection of spoilage LAB in air emphasizes the role of processing facilities and manufacturing operations in product contamination. Further studies are needed to examine, and to design strategies to reduce, psychrotrophic LAB contamination during processing.
Acknowledgments
We thank Henna Niinivirta for excellent technical assistance and the collaborating meat manufacturer for participating in this study.
The financial support of the Finnish Funding Agency for Technology and Innovation (decision 440472/03) and the Academy of Finland (project 100479) is gratefully acknowledged.
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Altschul, S. F., and W. Gish. 1996. Local alignment statistics. Methods Enzymol. 266:460-480. [DOI] [PubMed] [Google Scholar]
- 2.Björkroth, J., and W. Holzapfel. 2006. Genera Leuconostoc, Oenococcus and Weissella, p. 267-319. In M. Dworkin (ed.), The prokaryotes: a handbook on the biology of bacteria: Firmicutes, Cyanobacteria, vol. 4, 3rd ed. Springer-Verlag, New York, NY. [Google Scholar]
- 3.Björkroth, J., and H. Korkeala. 1996. rRNA gene restriction patterns as a characterization tool for Lactobacillus sake strains producing ropy slime. Int. J. Food Microbiol. 30:293-302. [DOI] [PubMed] [Google Scholar]
- 4.Björkroth, K. J., R. Geisen, U. Schillinger, N. Weiss, P. De Vos, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2000. Characterization of Leuconostoc gasicomitatum sp. nov., associated with spoiled raw tomato-marinated broiler meat strips packaged under modified-atmosphere conditions. Appl. Environ. Microbiol. 66:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björkroth, K. J., and H. J. Korkeala. 1996. Evaluation of Lactobacillus sake contamination in vacuum-packaged sliced cooked meat products by ribotyping. J. Food Prot. 59:398-401. [DOI] [PubMed] [Google Scholar]
- 6.Björkroth, K. J., and H. J. Korkeala. 1997. Lactobacillus fructivorans spoilage of tomato ketchup. J. Food Prot. 60:505-509. [DOI] [PubMed] [Google Scholar]
- 7.Björkroth, K. J., and H. J. Korkeala. 1997. Use of rRNA gene restriction patterns to evaluate lactic acid bacterium contamination of vacuum-packaged sliced cooked whole-meat product in a meat processing plant. Appl. Environ. Microbiol. 63:448-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Björkroth, K. J., M. Ristiniemi, P. Vandamme, and H. Korkeala. 2005. Enterococcus species dominating in fresh modified-atmosphere-packaged, marinated broiler legs are overgrown by Carnobacterium and Lactobacillus species during storage at 6°C. Int. J. Food Microbiol. 97:267-276. [DOI] [PubMed] [Google Scholar]
- 9.Björkroth, K. J., P. Vandamme, and H. J. Korkeala. 1998. Identification and characterization of Leuconostoc carnosum, associated with production and spoilage of vacuum-packaged, sliced, cooked ham. Appl. Environ. Microbiol. 64:3313-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumberg, H. M., J. A. Kiehlbauch, and I. K. Wachsmuth. 1991. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphisms of rRNA genes. J. Clin. Microbiol. 29:2368-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borch, E., M. L. Kant-Muermans, and Y. Blixt. 1996. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 33:103-120. [DOI] [PubMed] [Google Scholar]
- 12.Danielsen, M., and A. Wind. 2003. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 82:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Egan, A. F. 1983. Lactic acid bacteria of meat and meat products. Antonie Leeuwenhoek 49:327-336. [DOI] [PubMed] [Google Scholar]
- 14.Ercolini, D., F. Russo, E. Torrieri, P. Masi, and F. Villani. 2006. Changes in the spoilage-related microbiota of beef during refrigerated storage under different packaging conditions. Appl. Environ. Microbiol. 72:4663-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Gram, L., L. Ravn, M. Rasch, J. B. Bruhn, A. B. Christensen, and M. Givskov. 2002. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78:79-97. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi, I., H. Hayashidani, K. Kaneko, M. Ogawa, and Y. Benno. 1992. Bacterial flora of the respiratory tracts in chickens with a particular reference to Lactobacillus species. J. Vet. Med. Sci. 54:261-267. [DOI] [PubMed] [Google Scholar]
- 18.Koort, J. 2006. Polyphasic taxonomic studies of lactic acid bacteria associated with non-fermented meats. Ph.D. thesis. Department of Food and Environmental Hygiene, University of Helsinki, Finland.
- 19.Koort, J., T. Coenye, P. Vandamme, A. Sukura, and J. Björkroth. 2004. Enterococcus hermanniensis sp. nov., from modified-atmosphere-packaged broiler meat and canine tonsils. Int. J. Syst. Evol. Microbiol. 54:1823-1827. [DOI] [PubMed] [Google Scholar]
- 20.Koort, J., P. Vandamme, U. Schillinger, W. Holzapfel, and J. Björkroth. 2004. Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int. J. Syst. Evol. Microbiol. 54:1621-1626. [DOI] [PubMed] [Google Scholar]
- 21.Koort, J., A. Murros, T. Coneye, S. Eerola, P. Vandamme, A. Sukura, and J. Björkroth. 2005. Lactobacillus oligofermentans sp. nov., associated with spoilage of modified-atmosphere-packaged poultry products. Appl. Environ. Microbiol. 71:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korkeala, H., T. Alanko, P. Makela, and S. Lindroth. 1989. Shelf-life of vacuum-packed cooked ring sausages at different chill temperatures. Int. J. Food Microbiol. 9:237-247. [DOI] [PubMed] [Google Scholar]
- 23.Korkeala, H., T. Suortti, and P. Makela. 1988. Ropy slime formation in vacuum-packed cooked meat products caused by homofermentative lactobacilli and a Leuconostoc species. Int. J. Food Microbiol. 7:339-347. [DOI] [PubMed] [Google Scholar]
- 24.Korkeala, H. J., and K. J. Björkroth. 1997. Microbiological spoilage and contamination of vacuum-packaged cooked sausages. A review. J. Food Prot. 60:724-731. [DOI] [PubMed] [Google Scholar]
- 25.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyhs, U., J. M. Koort, H. S. Lundstrom, and K. J. Bjorkroth. 2004. Leuconostoc gelidum and Leuconostoc gasicomitatum strains dominated the lactic acid bacterium population associated with strong slime formation in an acetic-acid herring preserve. Int. J. Food Microbiol. 90:207-218. [DOI] [PubMed] [Google Scholar]
- 27.Myllyniemi, A., J. Koppinen, V. Gindonis, and S. Nykäsenoja. 2004. FINRES-Vet 2002-2003: Finnish veterinary antimicrobial resistance monitoring and consumption of antimicrobial agents. National Veterinary and Food Reseach Institute (EELA), Iisalmi, Finland.
- 28.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 29.Sakala, R. M., H. Hayashidani, Y. Kato, T. Hirata, Y. I. Makino, A. Fukushima, T. Yamada, C. Kaneuchi, and M. Ogawa. 2002. Change in the composition of the microflora on vacuum-packaged beef during chiller storage. Int. J. Food Microbiol. 74:87-99. [DOI] [PubMed] [Google Scholar]
- 30.Schillinger, U., and F. K. Lücke. 1986. Lactic-acid bacteria on vacuum packaged meat and their influence on shelf-life. Fleischwirtschaft 66:1515-1520. [Google Scholar]
- 31.Susiluoto, T., H. Korkeala, and K. J. Björkroth. 2003. Leuconostoc gasicomitatum is the dominating lactic acid bacterium in retail modified-atmosphere-packaged marinated broiler meat strips on sell-by-day. Int. J. Food Microbiol. 80:89-97. [DOI] [PubMed] [Google Scholar]