Abstract
p53 triggers cell cycle arrest and apoptosis through transcriptional regulation of specific target genes. We have investigated the effect of p53 activation on the proteome using 2D gel electrophoresis analysis of mitomycin C-treated HCT116 colon carcinoma cells carrying wild-type p53. Approximately 5,800 protein spots were separated in overlapping narrow-pH-range gel strips, and 115 protein spots showed significant expression changes upon p53 activation. The identity of 55 protein spots was obtained by mass spectrometry. The majority of the identified proteins have no previous connection to p53. The proteins fall into different functional categories, such as mRNA processing, translation, redox regulation, and apoptosis, consistent with the idea that p53 regulates multiple cellular pathways. p53-dependent regulation of five of the up-regulated proteins, eIF5A, hnRNP C1/C2, hnRNP K, lamin A/C, and Nm23-H1, and two of the down-regulated proteins, Prx II and TrpRS, was examined in further detail. Analysis of mRNA expression levels demonstrated both transcription-dependent and transcription-independent regulation among the identified targets. Thus, this study reveals protein targets of p53 and highlights the role of transcription-independent effects for the p53-induced biological response.
Keywords: proteomics, transcription factor, cancer
The p53 tumor suppressor regulates transcription of specific target genes. The levels of p53 are tightly controlled under normal conditions by the MDM2 protein that is up-regulated by p53 and targets p53 for proteasome-mediated degradation. p53 accumulates in response to cellular stress, e.g., DNA damage, oncogene activation, and hypoxia. Activated p53 induces or represses transcription of genes involved in cell cycle arrest, DNA repair, apoptosis, and senescence, all of which are important for maintenance of genomic stability and/or elimination of incipient tumor cells. Among the p53-induced genes, p21 is an effector of cell cycle arrest, whereas Bax, PUMA, and others are mediators of p53-dependent apoptosis (1). p53-mediated transrepression of Bcl-2, the IGF1 receptor, and telomerase (hTERT) is also important for the apoptotic response (2–4). Transcriptional transactivation occurs through p53 binding to specific motifs in target genes, whereas transrepression probably involves interactions with other proteins. p53 can also promote cell death by transcription-independent mechanisms (5). Indeed, p53 can translocate to mitochondria and interact with antiapoptotic proteins to facilitate apoptosis (6).
Close to half of all human tumors carry mutant p53 (see http://p53.free.fr and www-p53.iarc.fr). p53 inactivation leads to inappropriate regulation of p53 target genes and failure to trigger p53-dependent responses. p53 null mice are prone to spontaneous lymphomas and sarcomas at an early age (7). Transcriptional activation by p53 is critical for p53-induced apoptosis, which serves as an important barrier to tumor development (8, 9).
To reach a better understanding of the p53 pathway and p53-dependent tumor suppression, it is essential to learn more about the function of known p53 target genes and also identify novel p53 targets. Several studies have addressed p53-dependent gene expression by microarray analysis (10, 11). These studies have shown that p53 can induce a broad range of target genes that are involved in multiple cellular processes in addition to cell cycle arrest and apoptosis, and many genes are repressed by p53 (12). A recent study identified novel p53 target genes by using a ChIP-based approach (13). Consistent with these studies, >4,800 genes were found to contain at least one p53 motif (14). Thus, p53 can potentially regulate many genes, although not necessarily all in the same cell in response to a given type of stress.
So far, studies of p53-dependent gene expression patterns have been performed at the mRNA level. However, because the function of protein-coding genes is carried out by the protein product, it is important to study p53-dependent expression at the protein level. This may reveal novel p53 targets, including targets that are regulated by transcription-independent mechanisms, for example through posttranslational modifications and/or altered protein stability. Here, we present analysis of the p53-regulated proteome, using HCT116 colon carcinoma cells carrying wild-type p53 and isogenic p53 null cells (15), and narrow range 2D gel electrophoresis (2DE) and mass spectrometry. We identify p53 targets involved in such processes as mRNA processing, translation, metastasis, and apoptosis.
Results
Activation of p53 and Downstream Targets by Mitomycin C (MMC).
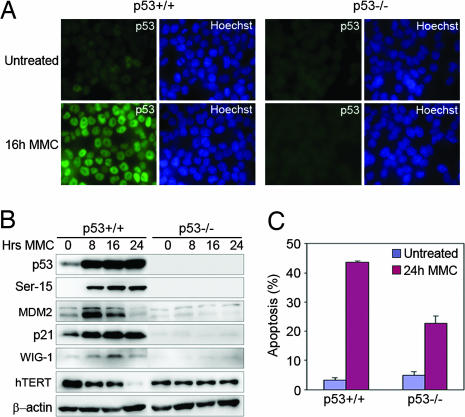
To activate endogenous p53 we treated HCT116 colon carcinoma cells with 10 μg/ml MMC, a DNA-damaging agent that cross-links DNA, preventing separation of the two DNA strands. Immunostaining showed p53 induction after 8 h of incubation with MMC in the wtp53+/+ cells (Fig. 1 A). To confirm activation of p53 target genes, we analyzed MMC-treated p53+/+ and p53−/− cells by Western blotting. Induction of p53 and p53 phosphorylation at Ser-15, was observed in the p53+/+ but not in the p53−/− cells. We also detected up-regulation of p21 and MDM2, two classical p53 targets, and the p53 target Wig-1 (Fig. 1B). In addition, expression of the p53-repressed human telomerase reverse transcriptase (hTERT) gene was decreased in the p53+/+ cells (Fig. 1B). FACS analysis confirmed a higher fraction of cell death in the p53+/+ cells (Fig. 1C).
Fig. 1.
Activation of p53 and induction of apoptosis in MMC-treated HCT116 cells. (A) p53 immunostaining of HCT116 p53+/+ and p53−/− cells after treatment with 10 μg/ml MMC for 24 h. (B) Western blot analysis of MMC-treated HCT116 p53+/+ and p53−/− cells showing induction of p53 and regulation of several p53 targets. (C) Apoptosis was assessed by propidium iodide (PI) staining and FACS analysis.
Identification of 55 Differentially Expressed Proteins by Mass Spectrometry.
Total protein from MMC-treated p53+/+ or p53−/− HCT116 cells at time points 0, 8, 16, and 24 h was subjected to 2DE. In the four matchsets (pH 3.9–5.1; 4.7–5.9; 5.5–6.7; and 6.3–8.3) up to 5,789 protein spots were separated and detected by PDQuest software (Bio-Rad, Hercules, CA) analysis (data not shown). Matchsets II (pH4.7–5.9) and III (pH5.5–6.7) contained 2/3 of all separated spots. We found 115 protein spots that showed statistically significant differences in expression levels (P < 0.05; Mann–Whitney test). Statistically significant changes were defined as a minimum 1.5-fold increase or decrease in expression level over at least two time points in comparison with the protein expression level in the p53 null control cells. To assess reproducibility, the correlation coefficients among three replicate gels were calculated. The average r value was 0.89 (range: 0.86–0.93), indicating high-quality 2DE gels as well as stable and reproducible culture and treatment conditions. We identified 55 p53-regulated polypeptides by mass spectrometry [supporting information (SI) Table 2]. Forty-two of these proteins were up-regulated and 13 were down-regulated in a p53-dependent manner. MMC-induced proteins showed up to 100-fold increase or more in the p53+/+ cells, whereas their expression levels were mostly unchanged or even decreased in the p53 null cells. Among the down-regulated proteins, expression was reduced by a factor of up to 10 in the p53+/+ cells, with no induction or in some cases a slight increase in the p53−/− cells (SI Table 2). Some of the identified proteins are listed in Table 1.
Table 1.
Identified proteins showing p53-dependent regulation
| Protein | Expression, fold change |
Putative binding site | |||||
|---|---|---|---|---|---|---|---|
| p53+/+ |
p53−/− |
||||||
| 8 h | 16 h | 24 h | 8 h | 16 h | 24 h | ||
| Up-regulated | |||||||
| Caspase 3 | 4.19 | 6.41 | — | — | — | — | Yes |
| eIF5A | 1.94 | 11.26 | 13.01 | — | — | 1.42 | Yes |
| hnRNP C1/C2 | 2.18 | 2.58 | 3.49 | — | — | — | No |
| hnRNP K | 7.67 | 8.3 | — | — | — | — | Yes |
| Lamin A/C | 3.69 | 67.2 | 133.7 | 3.7 | 11.1 | 18.0 | Yes |
| Nm23-H1 | 1.46 | 2.21 | 1.38 | — | — | — | Yes |
| Nucleophosmin | 1.49 | 1.66 | — | — | — | — | Yes |
| TAT-binding protein 1 | — | 1.48 | 13.43 | — | — | — | Yes |
| Thioredoxin | 1.43 | 8.11 | 11.03 | 1.3 | 1.7 | 1.77 | No |
| 14-3-3 σ | — | 2.06 | 6.07 | — | — | — | Yes |
| Down-regulated | |||||||
| hnRNP K, isoform a | −1.54 | −3.69 | −10.28 | — | — | — | Yes |
| hnRNP C1/C2 a | — | −1.67 | −4.96 | — | 1.32 | 1.39 | No |
| PP2A | −1.5 | −2.3 | — | — | — | — | Yes |
| Prx II | — | −1.56 | −2.85 | — | — | — | No |
| TrpRS | −1.3 | −1.9 | −2.71 | — | — | — | Yes |
Values indicate fold change in expression levels compared with untreated cells. —, no significant change.
Expression Changes of Seven Identified Proteins.
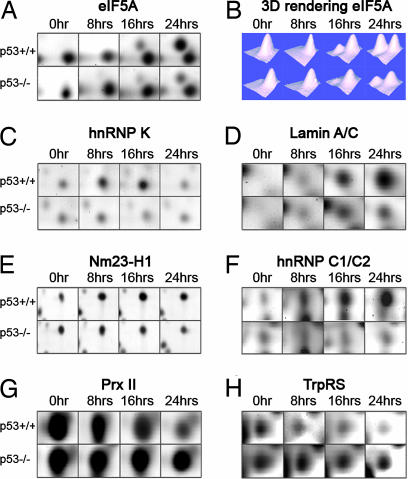
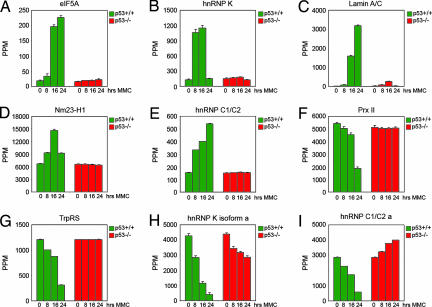
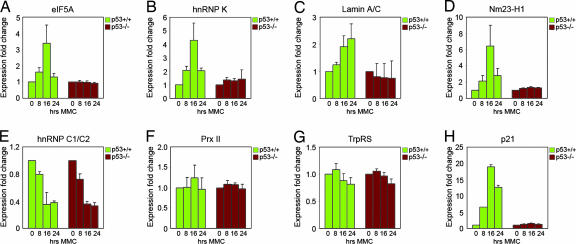
The expression changes of five identified up-regulated spots, eukaryotic translation factor 5A (eIF5A), heterogenous nuclear ribonucleoprotein C1/C2 (hnRNP C1/C2), heterogenous nuclear ribonucleoprotein K (hnRNP K), lamin A/C, and metastasis-inhibitor factor Nm23-H1 and two identified down-regulated spots, peroxiredoxin II (Prx II) and tryptophanyl-tRNA synthetase (TrpRS), are illustrated in Fig. 2. The 3D rendering function of the PDQuest software was applied to visualize protein spots and minimize misinterpretation of two or multiple overlapping spots as one single protein, as shown for eIF5A (Fig. 2B). The optical density of all spots from all matchsets and from three individual experiments was calculated and compiled into histograms showing protein expression at the different time points (Fig. 3 A–I). We detected a >10-fold p53-dependent increase in the levels of the eIF5A protein at 24 h in comparison with the levels at 0 h. hnRNP K levels were markedly induced already at 8 h, remained high at 16 h, and declined to levels comparable to those in untreated cells at 24 h. Lamin A/C was induced from almost undetectable levels before treatment to robust levels of expression at 16 h and even higher levels at 24 h. Nm23-H1 levels reached a peak at 16 h, whereas hnRNP C1/C2 was expressed at the highest levels at 24 h. Levels of the Prx II and TrpRS proteins were gradually reduced down to 25–30% of the levels in untreated cells at 24 h. In all cases, we observed no major expression changes in the p53 null cells (Fig. 3 A–G). Furthermore, we identified two down-regulated spots as hnRNP K isoform a and hnRNP C1C/2 a (Fig. 3 H and I). These spots may represent specific isoforms of these proteins and/or different mobility because of posttranscriptional modifications (see Discussion).
Fig. 2.
p53-dependent expression of seven identified proteins. (A, C–H) Gel sections show differential expression of the indicated protein spots at four time points. (B) PDQuest software 3D rendering analysis of the eIF5A spots.
Fig. 3.
p53-regulated protein expression. Optical density of the indicated spots was quantified by PDQuest software analysis. The graphs show the mean value of three independent gel runs.
Verification of p53-Dependent Regulation at the Protein and RNA Levels.
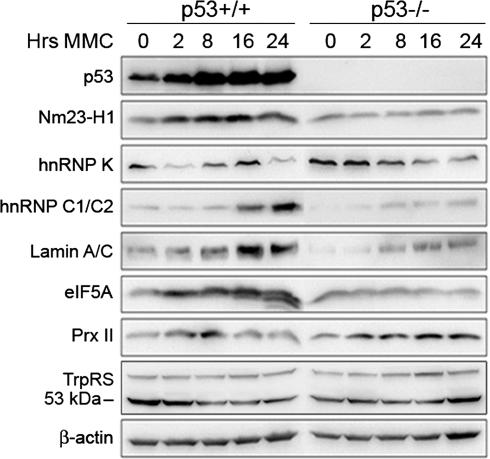
To verify these results, we performed Western blot analysis using antibodies against seven of the identified proteins. The results were largely in agreement with the 2DE data (Fig. 4). However, the induction of hnRNP K and lamin A/C was less pronounced in the Western blot analysis, possibly because of specifically regulated isoforms that appear on the 2D gels. Moreover, we did not detect a substantial down-regulation of Prx II as compared with the levels expressed at 0 h. Western blot analysis revealed a transient increase in Prx II expression at 8 h and low levels at 16 and 24 h in the p53+/+ cells, whereas Prx II levels were increased at all time points upon MMC treatment in the p53 null cells. Interestingly, a faster-migrating eIF5A protein species was detected in the MMC-treated p53+/+ cells (Fig. 4), suggesting that a specific isoform is regulated by p53. Two TrpRS protein species were visualized by Western blotting; whereas the 53 kDa species was down-regulated in the p53+/+ cells, the 70-kDa species did not show any changes in expression (Fig. 4).
Fig. 4.
Western blot analysis of p53-dependent expression of the indicated proteins at the indicated time points in MMC-treated HCT116 p53+/+ and p53−/− cells. β-Actin was used as loading control.
We also examined mRNA levels for each of these proteins by quantitative real-time RT-PCR (Fig. 5 A–G). The known p53 targets p21, MDM2, and Wig-1 were included as positive controls (Fig. 5 H and data not shown). We observed increased mRNA levels for eIF5A, hnRNP K, lamin A/C, and Nm23-H1 as well as all three controls in the MMC-treated p53+/+ cells (Fig. 5 A–D and data not shown), consistent with transcriptional regulation by p53. The mRNA levels showed a good correlation with the protein levels at the different time points. In contrast, mRNA levels for hnRNP C1/C2 were significantly reduced in the MMC-treated cells regardless of p53 status, particularly at the 16- and 24-h time points, indicating that the observed p53-dependent increase in hnRNP C1/C2 protein expression levels was not due to regulation of transcription (Fig. 5E). Similarly, our analysis of Prx II and TrpRS mRNA levels revealed a discordant regulation at the mRNA and protein levels. Prx II mRNA levels did not change, except for a minor increase at 16 h of MMC treatment in the p53+/+ cells, whereas the protein was significantly reduced at 24 h in the p53+/+ cells, according to the 2DE analysis. TrpRS mRNA was only slightly decreased in the MMC-treated cells, indicating that the observed p53-dependent down-regulation occurs at the protein level (Fig. 5 F and G).
Fig. 5.
Transcriptional regulation by p53. (A–G) Real-time PCR analysis of p53-dependent mRNA expression of the indicated genes at the indicated time points in MMC-treated HCT116 p53+/+ and p53−/− cells. (H) The p53 target p21 was included as positive control.
Western blot analysis of two other wild-type p53-carrying human cell lines, MCF7 and U2OS, revealed a similar pattern of protein expression changes in response to MMC treatment, confirming that the observed changes are not a unique feature of the HCT116 cells (SI Fig. 6).
Relationship Between Protein Expression Changes and Apoptosis.
To determine whether the observed changes in expression are related to apoptosis rather than activation of p53 per se, we examined protein levels in MMC-treated HCT116 cells in the presence of the caspase inhibitor zVAD. hnRNP C1/C2, lamin A/C, and Nm23-H1 were all induced in a p53-dependent manner even in the presence of zVAD. Likewise, eIF5A was induced, although to a lesser extent (SI Fig. 7).
We also examined protein expression in HCT116 cells treated with staurosporine (STS), an agent that triggers p53-independent apoptosis. eIF5A, hnRNP C1/C2, lamin A/C, and Nm23-H1 were not induced by STS. These results show that most of the identified proteins tested here are not induced by apoptosis as such but, rather, appear to be direct p53 targets (SI Fig. 8).
Promoter Analysis of the Genes Encoding the Proteins Identified.
We analyzed the promoter region including 5 kb upstream from the transcription start site as well as exon 1 and intron 1 in the genes encoding the identified proteins for p53-binding motifs using the p53MH algorithm (16). Twenty four of the genes encoding up-regulated proteins and 8 of the genes encoding down-regulated proteins contain at least one putative p53-binding site with a minimum similarity of 80% to the p53 consensus binding motif (SI Table 2). These results support the idea that many of the identified p53-induced proteins are indeed bona fide p53 transcriptional targets. These include, for example, eIF5A, hnRNP K, Nm23-H1, Annexin 1, and nucleophosmin (NPM). However, several of the identified genes appear to lack p53-binding motifs, suggesting that they are not classical p53 transcription targets.
Discussion
p53 may regulate downstream targets via both transcription-dependent and transcription-indpendent mechanisms. The transcription-dependent effects can be direct or indirect, i.e., mediated by p53 itself or by one or several transcriptional targets of p53. It is also conceivable that p53 regulates specific proteins directly in a transcription-independent manner by affecting their stability, intracellular localization, and/or activity. This regulation could involve protein–protein interactions and posttranslational modifications such as phosphorylation and ubiquitination. Analysis of p53-regulated protein expression should provide information about both transcription-dependent and transcription-independent regulation. Here, we present a global analysis of the p53-regulated proteome using high-resolution micropH 2DE and mass spectrometry. Close to 5,800 protein spots were separated, and 115 of these spots showed significant p53-dependent changes in expression levels. Thus, ≈2% of all separated protein spots in this study were affected by p53. This result is consistent with previous microarray studies showing that p53 may regulate 2–4% of all human genes (11, 12) and indicates that the number of proteins that are regulated by p53 does not vastly exceed the number of p53-regulated genes. Nonetheless, inhibition of protein synthesis by cycloheximide in a DNA microarray analysis using murine temperature-sensitive p53 resulted in a reduction of the fraction of p53-regulated transcripts from 5.5% to 0.88% (10). This supports the argument that many of the identified p53-regulated genes are indirect targets.
We identified the classical p53 targets 14-3-3σ and caspase-3 (Table 1), confirming that our experimental approach is valid. However, other known p53 targets such as p21, MDM2, Bax, and PUMA, were not among the identified proteins. This is presumably because the resolution and sensitivity of 2DE does not allow detection of all cellular proteins. Detection of a specific protein will depend on abundance, molecular weight, isoelectric point, and other factors.
The p53-regulated proteins identified here belong to different functional categories, including mRNA processing, protein translation, apoptosis, and metastasis, consistent with the notion that p53 regulates multiple cellular pathways. Some of the identified proteins have been identified as p53 transcription targets or otherwise linked to p53, whereas others have no reported p53 connection.
eIF5A was originally described as a translation initiation factor (17), but studies in yeast have indicated that it is not essential for general protein synthesis (18). eIF5A interacts with the general nuclear export receptor CRM1 and functions as a nucleocytoplasmic shuttle protein (19). Overexpression of eIF5A leads to increased expression of p53 targets as well as p53-dependent apoptosis, indicating that it is a positive regulator of p53 (20). We observed a strong p53-dependent up-regulation of eIF5A at 24 h of MMC treatment and identified a p53-binding motif in the first intron (Table 1), suggesting that eIF5A is a bona fide p53 target gene.
The hnRNP K protein is required for p53-mediated transcription of cell cycle checkpoint genes (21). It enhances transcription of oncogenes like c-myc and c-src and is thought to promote cell proliferation, survival, and migration (22). It has also been implicated in chromatin remodeling, mRNA splicing, export, and translation. We identified a p53-binding motif 67 nt upstream of the transcription start site in the hnRNP K promoter (Table 1). Thus, hnRNP K may be a coactivator of p53 that is transcriptionally activated by p53 in a positive feedback loop. Interestingly, we observed a 10-fold down-regulation of the hnRNP K isoform a in the HCT116 wtp53+/+ cells (Fig. 3H). This may be due to reduced stability or altered mobility of this hnRNP K isoform upon p53 activation, perhaps as a result of posttranslational modification. Indeed, hnRNP K is phosphorylated in vivo (23), and its binding to poly(C) is abrogated upon phosphorylation, suggesting that phosphorylation regulates its activity (24).
Similarly, the lamin A/C protein was up-regulated in a p53-dependent manner (Figs. 3C and 4). Lamin A/C proteins encoded by the LMNA gene belong to the type A lamins, which are the major component of nuclear lamina. Mutations in the LMNA gene are associated with the Hutchinson–Gilford progeria syndrome, i.e., premature aging (25). Mice lacking the Zmptse24 protease, which causes prelamin A accumulation, showed activation of the p53 signaling pathway (26). Mice overexpressing a short isoform of p53 or producing a p53-activating carboxy-terminal p53 fragment show signs of early aging (27, 28). This raises the possibility that lamin A/C is somehow involved in the premature aging phenotype associated with p53. The existence of a putative p53-binding site in intron 1 suggests that lamin A/C is a bona fide p53 target.
Nm23-H1 that encodes nucleoside diphosphate kinase (NDPK-A) is up-regulated at the mRNA and protein level upon activation of wild-type p53 (29). It is a negative regulator of cell proliferation and its expression is reduced in metastatic human tumors including melanoma and breast carcinoma. We observed a marked p53-dependent increase in Nm23-H1 expression in the MMC-treated cells, as confirmed by Western blot analysis and real-time RT-PCR (Figs. 4 and 5D). The Nm23-H1 gene contains a putative p53-binding site close to 5 kb upstream of the transcription start site (Table 1).
We also identified NPM as a p53-induced protein (Table 1). NPM is an abundant nucleolar phosphoprotein involved in ribosomal protein assembly and transport that shuttles between the nucleus and cytoplasm. NPM binds MDM2 and prevents the interaction between p53 and MDM2, and thereby regulates p53 (30). NPM also binds to p53, causing increased p53 stability and transcriptional transactivation upon stress (31). We identified a p53-binding site 2.4 kb upstream of the transcription start site. This suggests that NPM is a p53-induced protein target that forms a positive feedback loop with p53.
hnRNP C1/C2 proteins are abundant pre-RNA-binding proteins involved in premRNA maturation. The observed p53-dependent induction of hnRNP C1/C2 protein levels suggests that p53 may regulate this process. In contrast to eIF5A, hnRNP K, lamin A/C, and Nm23-H1, hnRNP C1/C2 mRNA levels were markedly reduced upon MMC treatment (Fig. 5E). Thus, hnRNP C1/C2 is not a classical transcriptional p53 target but rather regulated posttranscriptionally. It is conceivable that p53 also regulates other target proteins in a transcription-independent manner. This emphasizes the importance of protein expression profiling as a tool to identify and characterize downstream p53 targets, because transcription-independent targets will not turn up in a microarray analysis. Interestingly, one of the five hnRNP C1/C2 spots in the 2DE analysis, here designated hnRNP C1/C2 a, was down-regulated in the p53+/+ cells (Fig. 3I). This could be due to posttranslational modifications such as phosphorylation and/or small ubiquitin-like modifier modification. Phosphorylation of hnRNP C1/C2 is required for its binding to premRNA, and hyperphosphorylation leads to release of hnRNP C1/C2 from premRNA. Subsequent dephosphorylation by protein phosphatase 2A (PP2A) makes the protein available for the next round of phosphorylation and mRNA processing (32). We found that PP2A is down-regulated upon p53 activation (see below). This could lead to accumulation of hyperphosphorylated hnRNP C1/C2 protein that cannot participate in premRNA processing. It is noteworthy that hnRNP C1/C2 proteins are cleaved by interleukin 1β converting enzyme-like proteases and caspase 3 during apoptosis induced by different stimuli (33, 34). Thus, it is possible that p53-induced apoptosis targets hnRNP C1/C2 for proteolytic cleavage. Of note, caspase-3 was induced at 8 h of MMC treatment (Table 1).
PP2A is a serine–threonine phosphatase with multiple cellular targets. It has antiapoptotic activity that is specifically mediated by the R5/B56 regulatory subunits. Loss of complexes containing the B56 subunit leads to apoptosis in Drosophila (35). The SV40 small-t antigen binds to PP2A and perturbs its activity, contributing to cellular transformation (36). Furthermore, PP2A in complex with polyoma virus small-t antigen (PyST) was shown to inhibit ARF-mediated signals required for p53 activation (37). We detected a 2.3-fold repression of the B56 α-isoform after 16 h of MMC treatment (Table 1). The PP2A gene has a putative p53-binding site in intron 1 but its role in p53-mediated repression of PP2A is unclear.
Another identified p53-repressed target, Prx II, was down-regulated according to our 2DE analysis, but Western blot analysis did not reveal any significant p53-dependent repression. This suggests that p53 down-regulates specific isoforms and/or posttranslationally modified forms of Prx II rather than the entire pool. Prx II regulates cellular senescence and aging. Prx II null mouse embryo fibroblasts showed elevated levels of reactive oxygen species (ROS) and cellular senescence, and Prx II null mice showed signs of accelerated skin aging (38). p53 overexpression induces ROS production, resulting in oxidative degradation of mitochondrial components followed by apoptotic cell death (39). Our results suggest that p53-mediated down-regulation of Prx II has a role in the generation of ROS and p53-dependent apoptosis. This idea is consistent with the observation that mouse Prx V inhibits p53-induced generation of ROS and apoptosis (40). We did not observe any repression of Prx II at mRNA level, indicating that this gene is not a direct transcriptional target of p53.
Aminoacyl-tRNA synthetases play major roles in protein synthesis and catalyze the attachment of specific amino acids to their cognate tRNA but can also participate in diverse cellular processes such as RNA trafficking, rRNA synthesis, apoptosis, angiogenesis, and inflammation. In human cells, TrpRS is expressed as a full-length major form and a truncated form designated mini-TrpRS, because of alternative splicing. Mini-TrpRS was shown to inhibit retinal angiogenesis, whereas full-length TrpRS is thought to function primarily in translation (41). We observed a 2.7-fold down-regulation of full-length TrpRS (MW 53 kDa) at 24 h of MMC treatment in the p53+/+ cells (Table 1). This was confirmed by Western blot analysis, whereas no changes were found by real-time PCR (Figs. 4 and 5G), indicating that p53 down-regulates TrpRS independently of transcription. It is possible that p53 induces posttranslational modifications of TrpRS that leads to down-regulation of either of the protein variants. Evidence suggesting that p53 may inhibit translation by down-regulating TrpRS is previously unreported.
Our analysis of the p53-dependent proteome has allowed identification of 55 putative p53-regulated proteins, many of which are not known p53 targets. These proteins fall into multiple categories. Based on our findings, we can link p53 to mRNA processing (hnRNP C1/C2), translation (TrpRS), redox regulation (Prx II, thioredoxin), apoptosis (caspase-3, eIF5A), cell growth control (14-3-3σ, Nm23-H1), aging (lamin A/C, Prx II), Ras signaling and the MAP kinase pathway (GRB2, MAPK8), proteasomal degradation (TBP-1), phosphatase signaling (PP1 and PP2A), and chaperone activity (Hsp27, Hsp 60). Further work should confirm p53-dependent regulation of the identified proteins and examine p53-dependent regulation at the transcriptional level.
Our findings support the notion that the p53 tumor suppressor regulates cell growth and survival and other cellular processes at multiple levels through diverse and interconnecting pathways and via both transcription-dependent and transcription-independent mechanisms. This study also shows that global proteome analysis is suitable for identification of novel p53 targets and suggests that p53 is involved in posttranslational modification of many proteins. Further analysis of p53-regulated proteins should provide a better understanding of the molecular mechanisms behind p53-mediated tumor suppression. This analysis may also lead to the identification of novel targets for improved cancer therapy.
Materials and Methods
Cells and Cell Culture.
HCT116 p53+/+ and p53−/− colon carcinoma cells were grown as described (4). MMC (Sigma, St. Louis, MO) was added at 10 μg/ml to activate endogenous p53. Staurosporine (100 nM) (Sigma) was added to induce p53-independent cell death. Apoptosis was assessed as outlined (4). Cells were treated with 30 μM ZVAD (MP Biomedicals, Aurora, OH) for 2 h before MMC treatment for inhibition of caspases. Caspase activity was measured by flow cytometry with the CaspaTag Pan caspase kit (Chemicon, Temecula, CA).
Western Blot Analysis and Immunostaining.
Western blot analysis was carried out according to standard procedures by using the following primary antibodies: p53 monoclonal and MDM2 polyclonal from Santa Cruz Biotechnology (Santa Cruz, CA); hTERT monoclonal from Novocastra (Newcastle, U.K.); p21, lamin A/C, and eIF5A monoclonal from Pharmingen (San Diego, CA); β-actin monoclonal from Sigma; and hnRNP C1/C2, Nm23-H1, and Prx II monoclonal and hnRNP K polyclonal from Abcam (Cambridge, MA). Immunofluorescence staining was performed as described (4).
Real-Time PCR.
Reverse transcription of total RNA was performed by using a Reverse Transcription kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Real-time PCR was performed by using the ABI Prism 7500 instrument (Applied Biosystems, Foster City, CA).
2D Gel Electrophoresis.
Isoelectric focusing (IEF) and polypeptide separation (SDS/PAGE) was carried out as described (42). For the first dimension, 250–800 μg of protein per immobilized pH gradient (IPG) strip was loaded and focused in a PROTEAN IEF Cell (Bio-Rad) at ≈60,000 volt-hours. For the second dimension, 10–13% linear SDS/PAGE gradient gels were cast. The IPG strips were applied on top of the SDS/PAGE gels, and electrophoresis was performed for 20 h at 42,000 volt-hours. For image analysis, analytical gels were stained with silver nitrate. Spots on preparative gels were visualized by using Sypro Ruby staining solution (Molecular Probes, Eugene, OR). For more information, see SI Text.
Scanning and Image Analysis.
Scanning was performed as outlined (42). Four matchsets (one for each pH range) were constructed. Protein spots from gels from different time points were matched to spots in the reference gel. Spot intensities were normalized, and the relative staining intensities were determined. Expression changes were determined by statistical analysis with Student's t and Mann–Whitney tests (P < 0.05).
In-Gel Digestion and Mass Spectrometry.
Sypro Ruby-stained protein spots from the 2DE gels were excised by using EXQuest spot cutter (Bio-Rad). Manual in-gel digestion was performed as described (43). Peptide mass fingerprinting was performed in an Ultraflex TOF/TOF instrument (Bruker Daltonics, Billerica, MA). A search for protein identity was carried out with ProFound (http://prowl.rockefeller.edu/profound_bin/). Internal calibration was achieved by analysis of autolytic trypsin cleavage products resulting in an accuracy of ±0.05 Da. Judgement of significance was based on measuring z-value, number of matching peptide masses, and agreement between experimental and theoretical physical properties of the proteins.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein (The Johns Hopkins University, Baltimore, MD) for HCT116 cells and Thierry Soussi for valuable comments. This work was supported by the Swedish Cancer Society (Cancerfonden) and Karolinska Institutet.
Abbreviations
- 2DE
2D gel electrophoresis
- MMC
mitomycin C
- ROS
reactive oxygen species.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700794104/DC1.
References
- 1.Vousden KH, Lu X. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Szekely L, Okan I, Klein G, Wiman KG. Oncogene. 1993;8:3427–3431. [PubMed] [Google Scholar]
- 3.Prisco M, Hongo A, Rizzo MG, Sacchi A, Baserga R. Mol Cell Biol. 1997;17:1084–1092. doi: 10.1128/mcb.17.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman R, Latonen L, Wiman KG. Oncogene. 2005;24:1320–1327. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 5.Caelles C, Helmberg A, Karin M. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 6.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 7.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 8.Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 9.Attardi LD, Lowe SW, Brugarolas J, Jacks T. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan K, Amariglio N, Rechavi G, Jakob-Hirsch J, Kela I, Kaminski N, Getz G, Domany E, Givol D. Oncogene. 2001;20:2225–2234. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- 11.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 12.Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier P, et al. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 13.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wu Q, Qiu P, Mirza A, McGuirk M, Kirschmeier P, Greene JR, Wang Y, Pickett CB, Liu S. J Biol Chem. 2001;276:43604–43610. doi: 10.1074/jbc.M106570200. [DOI] [PubMed] [Google Scholar]
- 15.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 16.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershey JW. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 18.Kang HA, Hershey JW. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 19.Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle MC, Hauber J. J Cell Sci. 1999;112(Pt 14):2369–80. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- 20.Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, Pan X, Man JH, He K, Yu M, et al. J Biol Chem. 2004;279:49251–49258. doi: 10.1074/jbc.M407165200. [DOI] [PubMed] [Google Scholar]
- 21.Moumen A, Masterson P, O'Connor MJ, Jackson SP. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 22.Bomsztyk K, Denisenko O, Ostrowski J. BioEssays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 23.Schullery DS, Ostrowski J, Denisenko ON, Stempka L, Shnyreva M, Suzuki H, Gschwendt M, Bomsztyk K. J Biol Chem. 1999;274:15101–15109. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 24.Dejgaard K, Leffers H, Rasmussen HH, Madsen P, Kruse TA, Gesser B, Nielsen H, Celis JE. J Mol Biol. 1994;236:33–48. doi: 10.1006/jmbi.1994.1116. [DOI] [PubMed] [Google Scholar]
- 25.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, et al. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 27.Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 29.Chen SL, Wu YS, Shieh HY, Yen CC, Shen JJ, Lin KH. Mol Carcinog. 2003;36:204–214. doi: 10.1002/mc.10110. [DOI] [PubMed] [Google Scholar]
- 30.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 31.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 32.Mayrand SH, Dwen P, Pederson T. Proc Natl Acad Sci USA. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse N, Kumar S, Song Q, Strike P, Sparrow L, Dreyfuss G, Alnemri ES, Litwack G, Lavin M, Watters D. J Biol Chem. 1996;271:29335–29341. doi: 10.1074/jbc.271.46.29335. [DOI] [PubMed] [Google Scholar]
- 34.Brockstedt E, Rickers A, Kostka S, Laubersheimer A, Dorken B, Wittmann-Liebold B, Bommert K, Otto A. J Biol Chem. 1998;273:28057–28064. doi: 10.1074/jbc.273.43.28057. [DOI] [PubMed] [Google Scholar]
- 35.Silverstein AM, Barrow CA, Davis AJ, Mumby MC. Proc Natl Acad Sci USA. 2002;99:4221–4226. doi: 10.1073/pnas.072071699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arroyo JD, Hahn WC. Oncogene. 2005;24:7746–7755. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- 37.Moule MG, Collins CH, McCormick F, Fried M. Proc Natl Acad Sci USA. 2004;101:14063–14066. doi: 10.1073/pnas.0405533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han YH, Kim HS, Kim JM, Kim SK, Yu DY, Moon EY. FEBS Lett. 2005;579:4897–4902. doi: 10.1016/j.febslet.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 39.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Kok KH, Chun AC, Wong CM, Wu HW, Lin MC, Fung PC, Kung H, Jin DY. Biochem Biophys Res Commun. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]
- 41.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. Proc Natl Acad Sci USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roblick UJ, Hirschberg D, Habermann JK, Palmberg C, Becker S, Kruger S, Gustafsson M, Bruch HP, Franzen B, Ried T, et al. Cell Mol Life Sci. 2004;61:1246–1255. doi: 10.1007/s00018-004-4049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellman U, Bhikhabhai R. Rapid Commun Mass Spectrom. 2002;16:1851–1859. doi: 10.1002/rcm.805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.