Abstract
Hepatocyte growth factor (HGF), the ligand for the receptor tyrosine kinase Met, is secreted as single chain pro-HGF that lacks signaling activity. Pro-HGF acquires functional competence upon cleavage between R494 and V495, generating a disulfide-linked α/β-heterodimer, where the β-chain of HGF (HGF β) has a serine protease fold that lacks enzymatic activity. We show that, like serine proteases, insertion of the newly formed N terminus in the β-chain is critical for activity, here by allosterically stabilizing interactions with Met. The HGF β crystal structure shows that V495 inserts into the “activation pocket” near the Met binding site where the positively charged N terminus forms a salt bridge with the negatively charged D672, and the V495 side chain has hydrophobic interactions with main- and side-chain residues. Full-length two-chain HGF mutants designed to interrupt these interactions (D672N, V495G, V495A, G498I, and G498V) displayed <10% activity in Met receptor phosphorylation, cell migration, and proliferation assays. Impaired signaling of full-length mutants correlated with >50-fold decreases in Met binding of the low-affinity HGF β domain alone bearing the same mutations and further correlated with impaired N-terminal insertion. Because high-affinity binding resides in the HGF α-chain, full-length mutants maintained normal Met binding and efficiently inhibited HGF-mediated Met activation. Conversion of HGF from agonist to antagonist was achieved by as little as removal of two methyl groups (V495A) or a single charge (D672N). Thus, although serine proteases and HGF have quite distinct functions in proteolysis and Met signal transduction, respectively, they share a similar activation mechanism.
Keywords: allosteric regulation, signal transduction, receptor tyrosine kinase, trypsin, cancer
Hepatocyte growth factor (HGF), also known as scatter factor, is the ligand for the receptor tyrosine kinase Met and induces biological activities such as cell migration, branching morphogenesis, and proliferation, which are essential for tissue repair and embryonic development (1–4). Upon misregulation of HGF/Met signaling, cells can undergo invasive growth, resulting in tumor progression and metastasis. These observations have lead to a wide variety of antagonist approaches of HGF/Met signaling, which may ultimately result in therapeutic benefit (5).
HGF, a member of the plasminogen-related growth factor family, is a disulfide-linked α/β-heterodimer consisting of an N-terminal domain (PAN module; apple domain) followed by four kringle domains in the α-chain and a trypsin-like serine protease domain in the β-chain (6, 7). It is secreted from mesenchymal cells as a single-chain precursor (pro-HGF), which undergoes proteolytic cleavage at R494–V495 to generate the biologically active form of the ligand. Serine proteases capable of catalyzing pro-HGF activation in vitro include HGF activator (8), matriptase (9), hepsin (10, 11), Factor XIIa (8), Factor XIa (12), and plasma kallikrein (12). Both pro-HGF and two-chain HGF bind Met tightly; however, only two-chain HGF induces Met signaling (13–15).
The activation cleavage of HGF is reminiscent of the activation cleavage of serine protease zymogens to their enzymatically active forms (16, 17). Upon cleavage of the peptide bond between residues [c15] and [c16] (chymotrypsinogen numbering in brackets throughout) of the zymogen, there are large conformational changes of the so-called “activation domain,” three surface-exposed loops designated the [c140], [c180], and [c220] loops, and the newly formed N terminus (18). This concerted rearrangement results in the formation of a catalytically competent active site region. Previously, we demonstrated that pro-HGF activation leads to the formation of a Met binding region that corresponds to the active site and activation domain of serine proteases (19, 20). The functional importance of the β-chain of HGF (HGF β) interacting with Met is illustrated by the markedly reduced Met signaling of HGF mutants bearing amino acid changes in this contact region (19). Thus, although HGF β lacks the essential Asp–His–Ser catalytic triad found in all serine proteases, it still possesses structural features akin to serine proteases based on its tertiary structure.
In this study, we have investigated whether another paradigm of the serine protease activation domain also applies to HGF. In trypsin-like serine proteases the new N terminus at [c16] inserts into a preformed “activation pocket” and triggers a properly formed active site with an oxyanion hole and the substrate/inhibitor interaction subsites (16, 18). Proper insertion of the N terminus into the activation pocket depends on both hydrophobic and electrostatic interactions. In trypsin, the Ile-16 side chain and the salt bridge formed between Asp-194 and the positively charged N terminus provides 5 and 3 kcal/mol (1 kcal = 4.18 kJ), respectively, of stabilization energy to the activation domain (21). Proper N-terminal insertion is not only critical for the catalytic machinery but also for the interaction with active site inhibitors, such as the binding of bovine pancreatic trypsin inhibitor (BPTI) to trypsin (21).
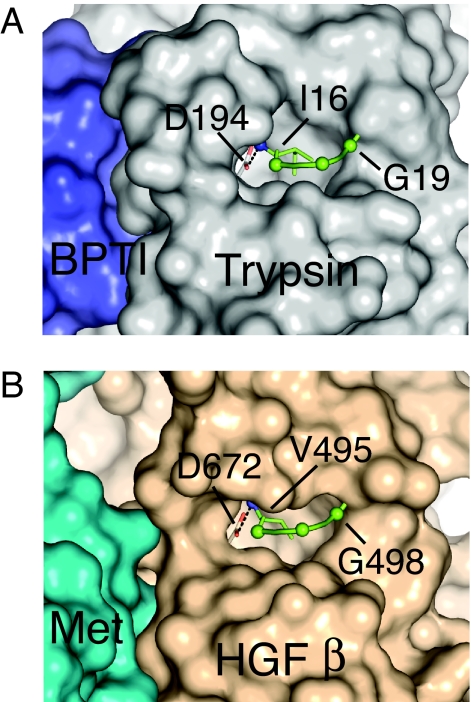
The structure of the complex of BPTI with trypsin further illustrates that the inserted N terminus is not in direct contact with the inhibitor, indicating that active site stabilization by the N terminus must be allosterically driven (Fig. 1). The respective locations of the Met binding site and inserted N terminus in the HGF β-chain are approximately the same as seen in the trypsin/BPTI complex (Fig. 1). Therefore, we hypothesized that an N-terminal insertion in the HGF β-chain is important for stabilizing the Met binding region. Here we provide evidence for the critical importance of electrostatic and hydrophobic interactions of the newly formed HGF β-chain N terminus V495 [c16] with its activation pocket and its stabilizing effect on Met interaction. Thus, although the functions of HGF as a ligand for Met signal transduction and of serine proteases as enzyme catalysts are quite distinct, they share a similar activation mechanism. On the basis of these findings, we were able to convert HGF into a potent receptor antagonist by introducing single amino acid substitutions to impair N-terminal insertion.
Fig. 1.
N-terminal “activation pocket” of trypsin and HGF β. (A) Crystal structure of the trypsin/BPTI complex [Protein Data Bank (PDB) ID code 2FTL]. Trypsin D194 is shown with the carboxylate in light red. The N terminus for I16 of trypsin is shown as a blue sphere. The Cα of V17, G18, and G19 are shown as green spheres. The salt bridge between D194 and I16 is ≈3 Å and is shown as a dotted line. (B) Crystal structure of the HGF β/Met Sema PSI complex (PDB ID code 1SHY). The HGF β carboxylate at D672 [c194] is shown in light red, and the N terminus of V495 [c16] is shown as a blue sphere. The Cα of V496, N497, and G498 are shown as green spheres. The salt bridge between D672 [c194] and V495 [c16] is ≈3 Å and is shown as a dotted line.
Results
Importance of N-Terminal Insertion for Binding of the HGF β-Chain to Met.
To investigate the role of the N terminus in the HGF β-chain, we modified residues D672 [c194] and V495 [c16], which form electrostatic (salt bridge) and hydrophobic interactions in the activation pocket, respectively (Fig. 1). D672N eliminates electrostatic interactions, whereas changes at V495 disrupt hydrophobic interactions with residues in the activation pocket. The N terminus was also shortened by one residue in the deletion mutant V495del. Additional changes were made at G498 [c19], which is situated at the entrance to the activation pocket and has main-chain torsion angles (φ and ψ = −125° and −175°, respectively) accessible to other amino acids. Assuming that there are no significant main-chain alterations, the side chains of G498 mutants will project inward toward the pocket occupied by V495 and would interfere with N-terminal insertion. All mutants are summarized in Table 1. Full-length HGF mutants were expressed in Chinese hamster ovary cells, activated by serum, and purified. Pro-HGF mutants were completely cleaved into their two-chain form, indicating that the changes made at the P1′ (residue 495) and P4′ (residue 498) positions did not impair activation cleavage by the serum activator (data not shown).
Table 1.
Relative Met binding of HGF β and HGF mutants and activity of HGF mutants in Met phosphorylation and cell migration assays
| HGF mutant [chymotrypsinogen numbering] | HGF β/Met competition binding, IC50(mut)/IC50(WT) | HGF/Met competition binding, IC50(mut)/IC50(WT) | Stimulation of cell migration of MDA-MB435 cells, % of WT | Maximal Met phosphorylation in A549 cells, % of WT |
|---|---|---|---|---|
| WT | 1 | 1 | 100 | 100 |
| V495A [c16] | >100 | 0.8 ± 0.1 | 6.2 ± 19.4 | 4.5 ± 1.9 |
| V495G [c16] | >100 | 1.7 ± 0.1 | 0.0 ± 13.9 | 1.7 ± 1.2 |
| V495del [c16del] | >100 | 0.7 ± 0.2 | 19.4 ± 7.4 | 16.4 ± 0.9 |
| D672N [c194] | >50 | 2.0 ± 0.8 | −2.0 ± 14.3 | 1.9 ± 1.3 |
| G498A [c19] | 1.0 ± 0.1 | 0.7 ± 0.2 | 77.4 ± 16.3 | 94.8 ± 10.4 |
| G498I [c19] | >100 | 1.1 ± 0.5 | −1.1 ± 10.8 | 5.1 ± 1.4 |
| G498P [c19] | >100 | 1.1 ± 0.02 | 18.2 ± 12.5 | 11.1 ± 1.8 |
| G498V [c19] | >100 | 1.3 ± 0.4 | −1.2 ± 11.9 | 8.2 ± 1.2 |
IC50 for HGF β in the HGF β/Met competition ELISA was 0.8 ± 0.4 μM (n = 14). All HGF β mutants were made in the C604S [c128] background; WT for HGF β refers to C604S [c128]. IC50 for HGF in the two-chain HGF/Met competition ELISA was 3.6 ± 1.8 nM (n = 9). WT, two-chain wild-type HGF; mut, mutant.
The same mutations were also made in just the HGF β-chain itself (HGF β). To examine how changes in the N-terminal insertion peptide influenced interactions with Met, we performed competition binding assays with HGF β mutants and Met–IgG. The results showed that all mutants except G498A displayed >50- to >100-fold reduced binding affinity (Table 1). The dramatic loss of Met binding by these mutants was also confirmed in direct binding ELISAs (data not shown). In contrast to the loss of Met binding for HGF β mutants, full-length HGF mutants still bound Met with essentially the same affinity as WT HGF (Table 1); this is attributed to the high-affinity binding by the HGF α-chain.
Chemical Modification of the HGF β-Chain N Terminus.
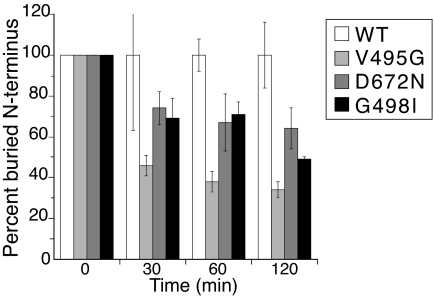
We hypothesized that if the HGF β-chain N terminus is not stably inserted into the activation pocket, then it should be more readily available for chemical modification of the N-terminal primary amine. This phenomenon has been observed previously with Factor VIIa in the presence or absence of its cofactor tissue factor (22). Here, we used an N-hydroxy succinimide reagent to modify any “free” N-terminal residue in three HGF β mutants (V495G, D672N, and G498I) selected because of the lack of Met activation by the corresponding full-length mutants in cell-based assays (see Effects of Impaired HGF β-Chain N-Terminal Insertion on Met-Dependent Signaling). Any reduction in the amount of N-terminal sequence obtained by Edman degradation directly corresponds to the percent of the chemically modifiable “free” N terminus. As shown in Fig. 2, the N terminus for all three mutants was significantly more exposed compared with WT HGF β.
Fig. 2.
Relative extent of structurally buried N terminus of HGF β-chain mutants. The percent of N terminus buried relative to WT is plotted at various quenching times. For comparative purposes, the peak height for each mutant was first normalized relative to itself at t0 (t0 for all proteins = 100%) and then normalized to HGF β (WT) at 30, 60, or 120 min to determine the relative abundance of buried (nonmodifiable) N terminus.
Effects of Impaired HGF β-Chain N-Terminal Insertion on Met-Dependent Signaling.
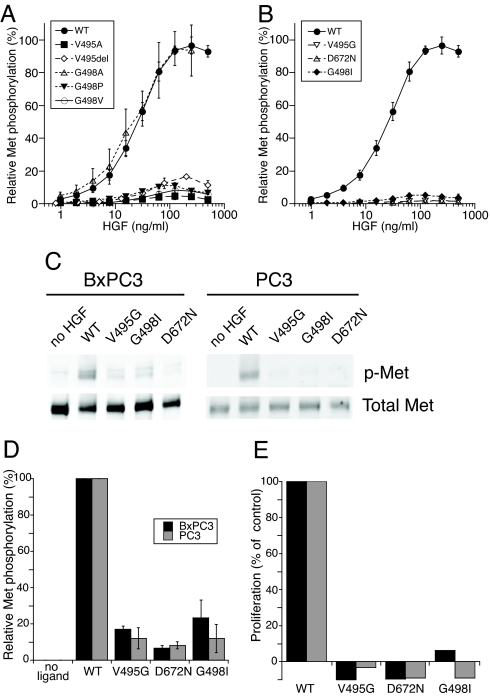
Full-length two-chain HGF mutants showed markedly reduced ability to promote cell migration of breast carcinoma MDA-MB435 cells. Except for the G498A mutant (77% of HGF activity), all mutants had <20% of HGF activity (Table 1). Consistent with their normal Met binding affinity (Table 1), higher concentrations of selected mutants (V495del, V495G, D672N, and G498I) remained ineffective agonists (data not shown), similar to HGF variants with β-chain mutations at the Met binding site (19). In agreement with the migration results, kinase receptor activation (KIRA) assays showed that the HGF mutants also had significantly reduced Met phosphorylation in lung carcinoma A549 cells, except for G498A, which was indistinguishable from HGF (Fig. 3 A and B). There was good correlation between the maximal Met phosphorylation and the cell migration activities of all mutants (Table 1).
Fig. 3.
HGF-dependent Met phosphorylation and cell proliferation. (A and B) Phosphorylation of Met in A549 cells and quantification by KIRA assay was carried out as described in Materials and Methods. (C) Representative immunoblots of phospho-Met from BxPC3 and PC3 cells after stimulation with 100 ng/ml HGF (WT) or HGF mutant. (D) Quantification of immunoblots for phospo-Met normalized to HGF (% of WT). (E) BxPC3 cell proliferation by 0.1 μg/ml HGF (WT) or HGF mutants at 0.1 μg/ml (black bars) or 2.0 μg/ml (gray bars).
Compared with WT, the three most inactive mutants, V495G, D672N, and G498I, had maximal Met phosphorylation activities of 1.7%, 1.9%, and 5.1%, respectively (Table 1). This finding was confirmed by immunoblotting experiments demonstrating an almost complete absence of Met phosphorylation in A549 cells (data not shown) as well as PC3 prostate carcinoma and BxPC3 pancreatic carcinoma cell lines (Fig. 3 C and D). Consistent with their lack of BxPC3 Met phosphorylation, V495G, D672N, and G498I mutants were completely inactive in stimulating BxPC3 cell proliferation at 0.1 and 2 μg/ml (Fig. 3E).
HGF Mutants as Met Antagonists.
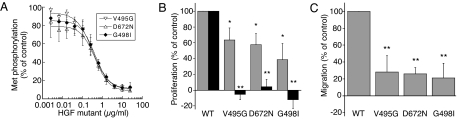
Because two-chain HGF mutants V495G, D672N, and G498I lacked agonist activity but maintained strong Met binding, we tested them as antagonists of HGF-dependent Met signaling. In KIRA assays, they showed concentration-dependent inhibition of Met phosphorylation in A549 cells and were equally potent, having IC50 values of 0.4 μg/ml (Fig. 4A). In addition, the HGF mutants inhibited BxPC3 cell proliferation by 38–67% at 2 μg/ml and showed complete inhibition at 20 μg/ml (Fig. 4B). Finally, all three mutants also inhibited MDA-MB435 cell migration at 20 μg/ml where >72% inhibition was measured (Fig. 4C).
Fig. 4.
Inhibition of Met phosphorylation, cell proliferation, and migration by HGF mutants. (A) HGF mutants (0.003–25 μg/ml) together with HGF (50 ng/ml) were added to A549 cell layer for 10 min at 37°C. Met phosphorylation by HGF mutants V495G (▿), D672N (▵), and G498I (♦) was quantified by using a KIRA assay and normalized to WT HGF activity (control). (B) Proliferation of BxPC3 cells was measured with 0.025 μg/ml HGF together with 2 μg/ml (gray bars) and 20 μg/ml (black bars) of HGF mutants. (C) MDA-MB435 cell migration was carried out in transwell plates in the presence of 0.1 μg/ml HGF together with HGF mutants at 20 μg/ml. HGF mutants were compared with WT HGF by using Student's t test: *, P < 0.05; **, P < 0.005.
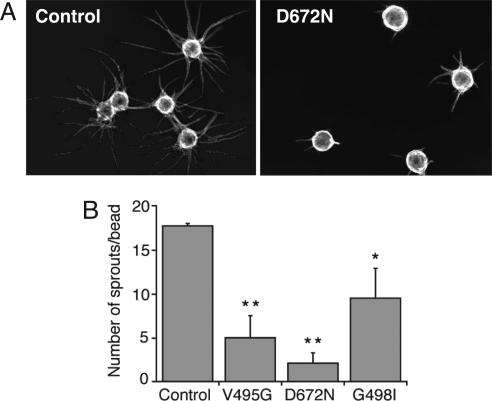
We also evaluated V495G, D672N, and G498I HGF mutants as antagonists for endogenously produced HGF in an endothelial tube formation assay, because HGF is proangiogenic (23, 24) and synergizes with VEGF to promote new blood vessel formation (25). In this assay, all three mutants reduced both the length and the number of endothelial sprouts emanating from the endothelial cell-coated beads. A representative image is shown for the most potent inhibitor, D672N (Fig. 5A). Quantification by the number of sprouts resulted in 72%, 88%, and 46% inhibition by V495G, D672N, and G498I, respectively (Fig. 5B).
Fig. 5.
Inhibition of in vitro endothelial tube formation by HGF mutants. Human umbilical vein endothelial cells attached to Cytodex 3 beads (average bead diameter was 170 μM) were embedded in a fibrin gel that was covered by Detroit 551 fibroblasts. HGF mutants in PBS or PBS alone (control) were added to the medium at a concentration of 10 μg/ml and tube formation was quantified after 6 days of culture. (A) Representative photographs of control and HGF mutant D672N. (B) Quantification of the number of endothelial sprouts per bead. HGF mutants were compared with a control by using Student's t test: *, P < 0.02; **, P < 0.005.
Discussion
The importance of proper N-terminal insertion of the protease-like HGF β domain into its activation pocket described herein provides previously unrecognized evidence for the structural and functional kinship of HGF with trypsin-like serine proteases. Not only does the activation cleavage of pro-HGF lead to the formation of a Met binding site in a region that corresponds to the serine protease active site (19, 20), but the integrity of the Met binding site depends on distinct electrostatic and hydrophobic interactions between the β-chain N terminus and residues in its activation pocket. Because the β-chain N terminus is not in direct contact with Met (Fig. 1) (20), we propose that impairment of N-terminal insertion influences the Met binding region allosterically, in a manner similar to N-terminal insertion in serine proteases and subsequent stabilization of their substrate/inhibitor interaction sites. Therefore, the HGF activation mechanism likely recapitulates the molecular activation mechanism that imparts functional competence to trypsin-like serine proteases.
It is intriguing that although the HGF β-chain N terminus is not part of the Met binding site, mutational changes disrupting the interaction of the N terminus with the activation pocket were generally more deleterious to HGF-dependent signaling than were changes within the actual Met binding site (19). Thus, just as found with serine proteases (16, 18, 21, 26–28), a buried N terminus is critical for ordering the activation domain. In the case of HGF, this results in optimal Met interaction and signaling, instead of proteolytic activity.
Hydrophobic and Electrostatic Interactions in the HGF Activation Pocket.
Changes introduced in the HGF β-chain were specifically designed to disrupt either electrostatic (D672N) or hydrophobic interactions (e.g., V495A, V495G) in the activation pocket. Of note, truncation of the V495 [c16] side chain by two methyl groups was sufficient to largely eliminate hydrophobic interactions with β-chain activation pocket residues V614 [c138], G616 [c140], and A632 [c158] as indicated by the loss of function by the V495A mutant. On the other hand, charge compensation for the N-terminal nitrogen by D672 [c194] was equally important, as suggested by results with the D672N [c194] mutant. Therefore, both hydrophobic and electrostatic interactions (i.e., the salt bridge) are important contributors for the stabilization of the buried N terminus, similar to findings of mutagenesis studies of trypsin (21).
The addition of a single methyl group to residue 498 (G498A) located at the entrance to the activation pocket was well tolerated, which agrees with the prediction from the van der Waals radii for this mutant. In contrast, bulkier side chains in G498I or G498V practically eliminated HGF function because of either an improperly buried N terminus or a displaced main chain at 498 having adverse interactions with a separate HGF β-chain (HGF β:HGF β dimer model) hypothesized in the active signaling complex (19, 20). The chemical modification experiments support the former, where Ile at 498 “kicks out” the N terminus from the activation pocket (Fig. 2).
Surprisingly, truncation of the N terminus by one residue (V495del) was not as deleterious as side-chain modifications made on the full-length N terminus (e.g., V495G). This finding suggested that the truncated N terminus was able to engage, albeit not optimally, in interactions with the activation pocket. This relatively mild effect contrasts with the >104 loss in BPTI binding and the >106 loss of substrate catalysis in N-terminally truncated trypsin (trypsin delI16) compared with trypsin (29). Comparing the HGF N-terminal segment with trypsin-like serine proteases does not reveal any amino acid insertion in HGF that might compensate for this truncation.
N-Terminal Insertion Equilibrium of HGF.
Previous studies with serine proteases have shown that the N terminus is in equilibrium between the buried state (active enzyme) and the “free” noninserted state (inactive enzyme). For highly active enzymes, the equilibrium favors the buried state. Furthermore, mutation or cofactor binding can affect a shift in the equilibrium concomitant with changes in enzymatic activity (30–32). For instance, in the absence of its cofactor tissue factor, N-terminal insertion of Factor VIIa is incomplete, consistent with a zymogen-like state having low catalytic activity (22). The addition of tissue factor stabilizes N-terminal insertion, reflected in the reduced availability of the N-terminal nitrogen for carbamylation, resulting in enhanced substrate interactions (22). Compared with WT, HGF β mutants V495G, D672N, and G498I exhibited significantly increased rates of chemical modification at the N terminus (Fig. 2). This finding implies that their N terminus was less buried in the activation pocket.
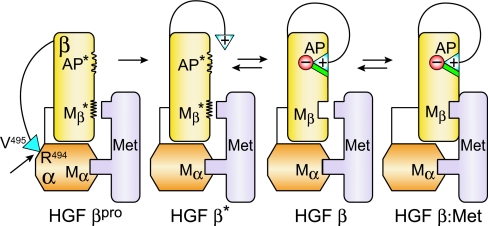
A proposed model of the allosteric linkage of N-terminal insertion with the Met binding site for the β-chain is shown in Fig. 6. Because pro-HGF binds Met with high affinity, activation may occur when the α-chain of pro-HGF is bound to Met at the high-affinity NK1 binding site (Mα in Fig. 6). However, pro-HGF activation and subsequent N-terminal insertion could also occur before any Met binding through the HGF α-chain (model not shown). The 1:1 binding complex of HGF:Met (HGF β:Met in Fig. 6) is consistent with 2:2 stoichiometry proposed for the quaternary signaling complex, comprising two HGF:Met complexes. Dimerization in the 2:2 complex may occur through HGF β:HGF β interactions (19, 20) and/or through NK1:NK1 interactions (33). A disfavored, yet additional possibility is a 1:2 signaling complex in which one bivalent HGF bridges two Met molecules. A structure of the elusive, intact signaling complex would be most enlightening.
Fig. 6.
Model of allosteric linkage of N-terminal insertion in the HGF β domain with its Met binding site. Labels refer to the state of the HGF β domain in each conformation. Single chain pro-HGF contains a disulfide bond (thin line) between the α-chain (orange) and β-chain (yellow). Pro-HGF first binds Met (light purple) at the NK1 region of the α-chain (Mα). The HGF β-chain is initially in the pro-HGF state (HGF βpro) where the β-chain activation pocket (AP*) and Met binding site (Mβ*) are in disordered conformational states and incompetent for Met signaling. Upon cleavage at R494–V495 (arrow), the positively charged HGF β-chain N terminus (blue triangle, +) is released, but not yet buried in its activation pocket (HGF β*). The N terminus can then insert into its activation pocket, which locks in the conformation (AP) observed in the HGF β crystal structure (HGF β) (19). A conformational change at its Met binding site (Mβ) results in Met binding at this site (HGF β:Met). In the HGF β and HGF β:Met states, the N-terminal V495 forms a salt bridge with the negatively charged D672 [c194] (red circle, −), and the V495 side chain engages in hydrophobic interactions (green). An equilibrium exists between the free N terminus (HGF β*) and the buried N terminus (HGF β), which favors the HGF β* state for the V495G, D672N, and G498I mutants. Pro-HGF activation, N-terminal insertion, and Mβ formation could also occur before any Met binding at Mα (model not shown).
How exactly does N-terminal insertion regulate the binding interaction of Met with HGF β-chain? This allosteric regulation likely involves two regions of the HGF activation domain, the 690s [c220s] loop (i.e., R695, G696) and the active site region (residues Y673 [c195] and E670 [c192]) situated close to D672, because they harbor residues that make direct structural contacts with Met (20). Although D672 does not directly contact Met, the likely movement of its side chain when forming the salt bridge with the N-terminal V495 will reorder the neighboring Y673 and E670, allowing for optimal interaction with Met. There are, however, limitations in comparing the β-chain with trypsin-like serine proteases. Even though the Met binding region largely corresponds to the substrate/inhibitor interaction region, there is no strict correspondence of the residues engaged in Met binding to those engaged in substrate/inhibitor binding by serine proteases (20). For instance, the P1 side chain of the substrate or inhibitor is completely buried in the S1 pocket of serine proteases, whereas Met residue E221 merely extends its side chain toward the pseudo S1 pocket in HGF β. Detailed insight of the allosteric linkage between N-terminal insertion and the Met binding site awaits the structure of “zymogen-like” uncleaved pro-HGF β-chain.
There is a formal possibility that the N terminus of one β-chain could interact with the activation pocket of another β-chain to form a dimer. Mechanistically, this hypothesis would explain the need for the proteolytic activation of pro-HGF to two-chain HGF required for Met signaling. In fact, there is precedence for intermolecular activation of serine proteases, where the N terminus of the bacterial cofactors staphylocoagulase and streptokinase insert into the activation pocket of prothrombin and plasminogen, respectively (34, 35). Although a β-chain dimer is indeed present in both the free and bound structures, the N terminus is buried intramolecularly in each monomer for both structures (19, 20). Thus, the likelihood that HGF might undergo an activation mechanism such as this is very remote.
Recently, Gherardi et al. (33) provided structural evidence to show that single-chain pro-HGF adopts a compact (closed) conformation, whereas two-chain HGF is elongated (open). Although the low-resolution structures do not provide a detailed view on the intramolecular contacts of the closed form, they appear to involve the HGF β-chain and kringle 2 domain (33). Because the closed-to-open transition is regulated by the activation cleavage and the liberation of the HGF β-chain N terminus, it is conceivable that the N-terminal insertion is essential to stabilize the elongated form and/or to destabilize the intramolecular interactions of the closed form.
Therapeutic Potential for HGF Variants as Met Antagonists.
A very recent and comprehensive domain binding analysis has shown Met binding affinities of 0.15 μM for the NK1 domain in the α-chain and 1.4 μM for the HGF β domain (36). Because the high-affinity binding site of HGF resides in the α-chain, full-length mutants D672N, V495G, and G498I efficiently competed with WT HGF for Met binding and were potent inhibitors of HGF-mediated cell migration, proliferation, tubulogenesis, and Met phosphorylation. Therefore, by introducing a single amino acid change, two-chain HGF was converted from an agonist into an antagonist. Recently, strong antitumorigenic activity using an uncleavable form of pro-HGF (scHGF) has been observed in murine models (37). For HGF mutants D672N and V495G, the complete absence of agonistic activity paired with WT Met binding is comparable to scHGF. As such, these new Met antagonists hold promise for new therapeutic approaches that target the HGF/Met pathway in cancer (5).
Materials and Methods
Construction, Expression, and Purification of HGF Mutants.
N-terminal insertion mutations in full-length HGF were constructed, expressed in Chinese hamster ovary cells, and purified by HiTrap Sepharose SP cation exchange chromatography (GE Healthcare, Piscataway, NJ) as described in ref. 19. Mutant forms of the HGF β-chain itself were made using C-terminal His6-tagged HGF β constructs in the C604S [c128] background, expressed in insect cells, and purified as described in ref. 19. Examination by SDS/PAGE showed that all HGF mutants were >95% pure; full-length mutants were completely converted into the two-chain form. Sequencing of HGF β mutants confirmed the amino acid changes at the N terminus.
HGF-Dependent Phosphorylation of Met.
Cell lines A549, BxPC3, and PC3 (American Type Culture Collection, Manassas, VA) were used for Met phosphorylation studies. Cells were serum-starved for 20 h and then incubated with 100 ng/ml HGF or HGF mutant for 10 min. Samples were prepared by washing cells twice with cold PBS followed by addition of gel sample buffer. Samples containing 25 μg of protein were analyzed by SDS/PAGE on 4–12% Tris-glycine gels (Invitrogen, Carlsbad, CA). Proteins were transferred onto nitrocellulose membranes and subsequently treated with anti-Met antibody DL-21 (Upstate, Lake Placid, NY) or anti-phospho-Met antibody Tyr-1234/1235 (Cell Signaling Technology, Danvers, MA) overnight at 4°C. After washing, membranes were incubated with IRDye800-conjugated goat anti-mouse IgG (Rockland, Gilbertsville, PA) and AlexaFluor 680 goat anti-rabbit IgG (Invitrogen) for 1 h. Proteins were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
An electrochemiluminescence-based KIRA assay was used to quantify Met phosphorylation on A549 cells as described in ref. 19. HGF mutants (1–500 ng/ml) were added to A549 cell layers for 10 min at 37°C, and Met phosphorylation was determined. For inhibition studies, HGF mutants (0.003–25 μg/ml) were mixed with HGF (50 ng/ml) and added to the A549 cell layer for 10 min at 37°C. For each experiment, the Met phosphorylation induced by HGF mutants was expressed as a percentage of the maximal signal obtained with HGF. Results are the average ± SD of at least three assays.
Chemical Modification of the N Terminus.
The relative extent of the N terminus buried in the HGF β mutants was determined by chemical modification of the N-terminal primary amine, chemically accessible only when “free,” and subsequent N-terminal sequencing by Edman degradation. Fifty-microliter solutions of HGF β-chain and mutants at 0.2 mg/ml in PBS were mixed with 3 μl of a freshly prepared 10 mM EZ-link Sulfo-NHS-LC-Biotin (Pierce, Rockford, IL) stock solution and immediately incubated on ice. After 0, 30, 60, and 120 min, 3-μl aliquots were taken and applied to a PVDF membrane; for initial sequence data at t0, 3 μl of water was added instead of the NHS reagent. The protein spots were excised from the PVDF membrane and subjected to N-terminal sequence analysis with the Applied Biosystems (Foster City, CA) Procise Sequencer, Model 494. The peak height of the second valine residue was chosen for data analysis to eliminate noise from the first sequencing cycle. Values are the average ± SD (n = 3).
Binding of HGF and HGF β-Chain Mutants to Met.
Binding of HGF β-chain mutants or full-length HGF mutants was measured in a competition binding assay using Met–IgG fusion protein, as described in ref. 19. Relative binding affinities were expressed as the IC50(mutant)/IC50(WT) and are the average ± SD of at least three assays.
Cell Migration and Proliferation Assays.
The promigratory effects of HGF full-length mutants (100 ng/ml) on MDA-MB435 cells (HTB-129; American Type Culture Collection) were measured by use of a collagen-coated transwell system as described in ref. 19. For inhibition assays, HGF mutants (20 μg/ml) in serum-free medium were mixed with 100 ng/ml WT HGF. Results are the average ± SD (n = 5–8).
The activity of the full-length HGF mutants V495G, D672N, and G498I on BxPC3 cell proliferation was determined essentially as described in ref. 11. Each mutant was assayed 2 to 4 times; values are expressed as percent of HGF activity (0.1 μg/ml) after subtracting basal proliferation in the absence of HGF. For inhibition studies, cells were incubated with HGF mutants (2 μg/ml and 20 μg/ml) and 25 ng/ml HGF in serum-free medium for 3 days, after which cells were quantified. Results are expressed as percent of HGF activity as a control ± SD (n = 3–4).
Endothelial Tube Formation Assay.
The assay was carried out essentially as described in ref. 38. Human umbilical vein endothelial cells (Cambrex Bio Science, Walkersville, MD) were seeded onto Cytodex 3 beads (GE Healthcare, Piscataway, NJ) at a ratio of 400 cells per bead in 1–2 ml of EGM2 medium (Cambrex Bio Science). After overnight incubation, the endothelial cell-coated beads were washed and resuspended in EGM2 medium containing 2.5 mg/ml fibrinogen (Sigma, St. Louis, MO), and ≈200 beads per milliliter were added to 12-well tissue culture plates (BD Biosciences, Franklin Lakes, NJ) containing 20 μl of 0.06 units/ml thrombin (Sigma). After the fibrin gel had solidified, 4–8 × 104 Detroit 551 fibroblasts (American Type Culture Collection) in 2 ml of EGM2 were added. HGF mutants were added at 10 μg/ml, and medium was changed every 2 days. Endothelial sprouts were photographed and quantified after 6 days. Results are the average ± SD (n = 4).
Abbreviations
- BPTI
bovine pancreatic trypsin inhibitor
- HGF
hepatocyte growth factor
- HGF β
β-chain of HGF
- KIRA
kinase receptor activation.
Footnotes
Conflict of interest statement: The authors of this work are employed by Genentech, Inc.
References
- 1.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 2.Jiang WG, Martin TA, Parr C, Davies G, Matsumoto K, Nakamura T. Crit Rev Oncol Hematol. 2005;53:35–69. doi: 10.1016/j.critrevonc.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R. Cytokine Growth Factor Rev. 2002;13:41–59. doi: 10.1016/s1359-6101(01)00029-6. [DOI] [PubMed] [Google Scholar]
- 4.Trusolino L, Comoglio PM. Nat Rev Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 5.Christensen JG, Burrows J, Salgia R. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 6.Donate LE, Gherardi E, Srinivasan N, Sowdhamini R, Aparicio S, Blundell TL. Protein Sci. 1994;3:2378–2394. doi: 10.1002/pro.5560031222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tordai H, Banyai L, Patthy L. FEBS Lett. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 8.Shimomura T, Miyazawa K, Komiyama Y, Hiraoka H, Naka D, Morimoto Y, Kitamura N. Eur J Biochem. 1995;229:257–261. doi: 10.1111/j.1432-1033.1995.tb20463.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee SL, Dickson RB, Lin CY. J Biol Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 10.Herter S, Piper DE, Aaron W, Gabriele T, Cutler G, Cao P, Bhatt AS, Choe Y, Craik CS, Walker N, et al. Biochem J. 2005;390:125–136. doi: 10.1042/BJ20041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchhofer D, Peek M, Lipari MT, Billeci K, Fan B, Moran P. FEBS Lett. 2005;579:1945–1950. doi: 10.1016/j.febslet.2005.01.085. [DOI] [PubMed] [Google Scholar]
- 12.Peek M, Moran P, Mendoza N, Wickramasinghe D, Kirchhofer D. J Biol Chem. 2002;277:47804–47809. doi: 10.1074/jbc.M209778200. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann G, Naldini L, Weidner KM, Sachs M, Vigna E, Comoglio PM, Birchmeier W. Proc Natl Acad Sci USA. 1992;89:11574–11578. doi: 10.1073/pnas.89.23.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lokker NA, Mark MR, Luis EA, Bennett GL, Robbins KA, Baker JB, Godowski PJ. EMBO J. 1992;11:2503–2510. doi: 10.1002/j.1460-2075.1992.tb05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naldini L, Tamagnone L, Vigna E, Sachs M, Hartmann G, Birchmeier W, Daikuhara Y, Tsubouchi H, Blasi F, Comoglio PM. EMBO J. 1992;11:4825–4833. doi: 10.1002/j.1460-2075.1992.tb05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedstrom L. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 17.Khan AR, James MN. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber R, Bode W. Acc Chem Res. 1978;11:114–122. [Google Scholar]
- 19.Kirchhofer D, Yao X, Peek M, Eigenbrot C, Lipari MT, Billeci KL, Maun HR, Moran P, Santell L, Wiesmann C, Lazarus RA. J Biol Chem. 2004;279:39915–39924. doi: 10.1074/jbc.M404795200. [DOI] [PubMed] [Google Scholar]
- 20.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. EMBO J. 2004;23:2325–2335. doi: 10.1038/sj.emboj.7600243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedstrom L, Lin TY, Fast W. Biochemistry. 1996;35:4515–4523. doi: 10.1021/bi951928k. [DOI] [PubMed] [Google Scholar]
- 22.Higashi S, Nishimura H, Aita K, Iwanaga S. J Biol Chem. 1994;269:18891–18898. [PubMed] [Google Scholar]
- 23.Bussolino F, Di Renzo MF, Ziche M, Bocchietto E, Olivero M, Naldini L, Gaudino G, Tamagnone L, Coffer A, Comoglio PM. J Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant DS, Kleinman HK, Goldberg ID, Bhargava MM, Nickoloff BJ, Kinsella JL, Polverini P, Rosen EM. Proc Natl Acad Sci USA. 1993;90:1937–1941. doi: 10.1073/pnas.90.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin X, Yang S, Ingle G, Zlot C, Rangell L, Kowalski J, Schwall R, Ferrara N, Gerritsen ME. Am J Pathol. 2001;158:1111–1120. doi: 10.1016/S0002-9440(10)64058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bode W, Schwager P, Huber R. J Mol Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]
- 27.Bode W, Huber R. FEBS Lett. 1976;68:231–236. doi: 10.1016/0014-5793(76)80443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bode W. J Mol Biol. 1979;127:357–374. doi: 10.1016/0022-2836(79)90227-4. [DOI] [PubMed] [Google Scholar]
- 29.Pasternak A, Liu X, Lin TY, Hedstrom L. Biochemistry. 1998;37:16201–16210. doi: 10.1021/bi980951d. [DOI] [PubMed] [Google Scholar]
- 30.Persson E, Kjalke M, Olsen OH. Proc Natl Acad Sci USA. 2001;98:13583–13588. doi: 10.1073/pnas.241339498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovan RJ, Ruf W. Biochemistry. 2002;41:9302–9309. doi: 10.1021/bi0202169. [DOI] [PubMed] [Google Scholar]
- 32.Camire RM. J Biol Chem. 2002;277:37863–37870. doi: 10.1074/jbc.M203692200. [DOI] [PubMed] [Google Scholar]
- 33.Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, Miguel RN, Blundell TL, Vande Woude GF, Skoglund U, Svergun DI. Proc Natl Acad Sci USA. 2006;103:4046–4051. doi: 10.1073/pnas.0509040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Nature. 2003;425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Reed GL, Hedstrom L. Eur J Biochem. 2000;267:3994–4001. doi: 10.1046/j.1432-1327.2000.01434.x. [DOI] [PubMed] [Google Scholar]
- 36.Holmes O, Pillozzi S, Deakin JA, Carafoli F, Kemp L, Butler PJG, Lyon M, Gherardi E. J Mol Biol. 2007;367:395–408. doi: 10.1016/j.jmb.2006.12.061. [DOI] [PubMed] [Google Scholar]
- 37.Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, Comoglio PM, Michieli P. J Clin Invest. 2004;114:1418–1432. doi: 10.1172/JCI22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatsu MN, Sainson RC, Aoto JN, Taylor KL, Aitkenhead M, Perez-del-Pulgar S, Carpenter PM, Hughes CC. Microvasc Res. 2003;66:102–112. doi: 10.1016/s0026-2862(03)00045-1. [DOI] [PubMed] [Google Scholar]