Abstract
Biological developmental pathways require proper timing of gene expression. We investigated timing variations of defined steps along the lytic cascade of bacteriophage λ. Gene expression was followed in individual lysogenic cells, after induction with a pulse of UV irradiation. At low UV doses, some cells undergo partial induction and eventually divide, whereas others follow the lytic pathway. The timing of events in cells committed to lysis is independent of the level of activation of the SOS response, suggesting that the lambda network proceeds autonomously after induction. An increased loss of temporal coherence of specific events from prophage induction to lysis is observed, even though the coefficient of variation of timing fluctuations decreases. The observed temporal variations are not due to cell factors uniformly dilating the timing of execution of the cascade. This behavior is reproduced by a simple model composed of independent stages, which for a given mean duration predicts higher temporal precision, when a cascade consists of a large number of steps. Evidence for the independence of regulatory modules in the network is presented.
Keywords: bacteriophage λ, noise, precision, prophage induction, timing
Introduction
Cells respond to given stimuli by the activation of specific regulatory pathways, often characterized by a cascaded architecture of gene expression in which the product of one gene regulates the expression of others. Notable examples include MAP kinase pathways (Lamb 1996; Gustin et al, 1998), developmental programs (Davidson et al, 2002) and transcriptional cascades (Lee et al, 2002). A central issue in considering the proper functioning of genetic cascades is how noise in gene activity, due to sources such as the stochastic nature of gene expression (Berg, 1978; Rigney, 1979; McAdams and Arkin, 1997) and the fluctuating number of molecular components, propagates along a cascade (Thattai and van Oudenaarden, 2002; Becskei et al, 2005; Hooshangi et al, 2005; Pedraza and van Oudenaarden, 2005; Rosenfeld et al, 2005) and affects the fidelity of information transfer. This consideration has been the object of intense recent scrutiny, and has been tackled in depth in the context of synthetic, engineered transcriptional cascades (Hooshangi et al, 2005; Rosenfeld et al, 2005), natural cascades (Colman-Lerner et al, 2005) as well as in theoretical studies (Thattai and van Oudenaarden, 2002). These studies revealed that noise in the activity of one gene affects fluctuations in downstream gene expression and that noise amplification results in loss of synchrony among a cell population. Timing is an important aspect in the regulation of biological cascades (see e.g., Kalir et al, 2001). Recently, noise in the timing of cell-cycle start has been studied in single yeast cells (Bean et al, 2006).
The temporal variability in the orchestrated series of events that take place when Escherichia coli cells, lysogenic with bacteriophage λ, are induced allows the study of timing noise in a natural genetic cascade. The lysis–lysogeny decision of bacteriophage λ, a paradigm for the operation of developmental genetic networks, is composed of interlocked positive and negative feedback loops and is regulated by both phage and bacterial factors (Ptashne 2004; Dodd et al, 2005; Oppenheim et al, 2005). The lysogenic or off state of the cascade is maintained with high stability by multimers of the lambda repressor (CI), repressing both the phage pL and pR promoters (Dodd et al, 2001). A combination of negative and positive feedback mechanisms keeps the concentration of CI at about 150–200 copies per cell (Dodd et al, 2004). Prophage induction is triggered by DNA damage. Upon encountering DNA damage, replication forks stall and single-stranded DNA tracts form, activating the cell's SOS network (Little 1996; Friedman et al, 2005). This network, responsible for the repair/bypass of DNA lesions, consists of about 40 genes whose expression is downregulated by the LexA repressor. Polymerization of the RecA protein on the single-stranded DNA tracts endows RecA with a coprotease function that promotes the cleavage of both LexA and CI, activating both the SOS response and the lambda induction network. The lambda lytic cascade proceeds through three stages: early, delayed early and late. In the early stage, CI degradation results in the expression of Cro and N functions from the pR and pL promoters, respectively (Figure 1). In the delayed early stage, N assists in overriding terminators, allowing RNA polymerase (RNAP) to extend both transcripts beyond the cro and N genes, leading among others to the expression of Q. In the late stage, the Q protein modifies RNAP initiating transcription from the pR′ late promoter, to allow for transcription beyond the tR′ terminator, leading to expression of the late genes, which encode for phage morphogenesis functions and host cell lysis proteins (see Oppenheim et al, 2005 for details). Similar three-stage architectures are found in diverse phages, such as the virulent phage T4 (Endy et al, 2000).
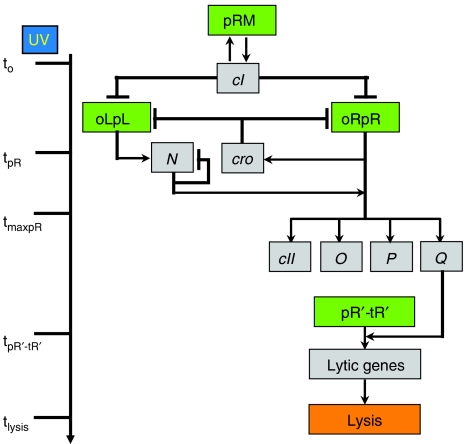
Figure 1.
Schematic model of lambda induction. Lambda promoters are colored in green and genes in gray. The lambda induction cascade is carried out in three stages. In the early stage, UV irradiation results in a decrease of CI levels, activating the pR and pL promoters at time tpR , leading to the expression of the cro and N genes. During the delayed early stage, RNAP is modified so as to override transcriptional terminators, allowing the continuation of both transcripts and the expression of CII, O, P and Q. During the late stage, the Q protein modifies RNAP, initiating transcription from the pR′ late promoter at time tpR′-tR′, to become resistant to transcription terminators present downstream, and allowing the expression of late genes that encode proteins for phage morphogenesis and host cell lysis. During the late stage of the cascade, the late gene products assemble phage virions and lyse the host at time tlysis (Oppenheim et al, 2005).
In the studies described below, we have activated the lytic pathway cascade by the induction of DNA damage in lysogenic cells with UV light, monitoring the timing of CI repressor inactivation and the onset of activity of the late gene activator Q as well as lysis in individual cells by time-lapse fluorescence microscopy. Our findings shed light on how different events along the lytic cascade are timed and organized, and how individual cell behavior depends on UV irradiation.
Results
Timing of the lytic cascade in individual cells following prophage induction
Lysogenic bacteria, harboring either pR-GFP or pR′-tR′-GFP reporter plasmids, were induced by UV irradiation. The first reporter fusion, pR-GFP, is under the direct repression of the CI repressor, and therefore monitors the inactivation of CI and reports on levels of expression from the earliest stage after induction of the cascade (see Figure 1). The second reporter fusion, pR′-tR′-GFP, monitors the expression of late lytic genes transcribed from the pR′ promoter, expression that is enabled by the antitermination activity of the Q protein at tR′. Typical snapshots of cells during derepression of pR-GFP and activation of pR′-tR′-GFP fusions after irradiation with 20 J/m2 are shown in Figure 2A, upper and lower panels, respectively. As the snapshots illustrate, in both cases the fluorescence increases monotonically with time, and pR-GFP is expressed earlier than pR′-tR′. Eventually, all cells undergo lysis at this level of irradiation, and disappear from the field of view. The fluorescence profiles of pR and pR′-tR′ as a function of time from individual cells in the snapshots, and the corresponding promoter activity profiles (see Materials and methods) are shown in Figures 2B and C, respectively (see Supplementary movies).
Figure 2.
Induction of individual lysogens following irradiation with 20 J/m2. (A) Snapshots of cells harboring the pR-GFP (top panels) and pR′-tR′-GFP (bottom panels) reporter plasmids undergoing induction taken at the times shown after irradiation. Some cells lyse and disappear from the field of view. Cell silhouettes were determined from dark-field microscopy images. (B) Fluorescence profiles of pR-GFP (blue) and pR′-tR′-GFP (red) of individual cells including those viewed in the top and bottom panels in (A), respectively, as a function of time. The sharp drop in every profile is due to lysis. (C) pR (blue) and pR′-tR′ (red) promoter activity profiles of individual cells derived from the fluorescence data in (B) as a function of time.
The pR and pR′-tR′ promoters exhibit different behaviors. Whereas fluorescence from pR-GFP commences at about 20 min following induction and saturates, that of pR′-tR′-GFP starts at ∼50 min and increases monotonically until lysis (Figure 2B). The time lag between the onset of expression from pR and pR′-tR′ as reported by the fusions, ∼35 min, cannot be accounted for by the time it takes RNAP to transcribe the 6 kbp tract between pR and the Q gene (<3 min) (Gotta et al, 1991). This lag is similar to the one observed after infection, and has been suggested to arise from a functional threshold effect for Q-dependent pR′-tR′ expression (Kobiler et al, 2005). The difference in the behavior of pR and pR′-tR′ can also be seen in their promoter activities (Figure 2C). The pR promoter fusion is active only within a well-defined time window: after the initial increase, it peaks at about 45 min, and shuts off at ∼80 min, before maximal activity of pR′-tR′ is attained. The molecular basis for this unexpected behavior is not understood. Promoter shutoff is not observed in control experiments in lysogenic bacteria harboring a pTac-GFP fusion reporter plasmid, and induced with IPTG before UV irradiation (data not shown). In contrast to the behavior of pR, the activity of pR′-tR′ increases until cell lysis. Note that in this analysis, we focused on the timing of specific stages, avoiding a detailed analysis of GFP expression levels that may be affected by variations in plasmid copy number.
Lysis, as measured by a sharp drop in the fluorescence of GFP and a much weaker image of a cell's silhouette compared to ordinary cells in our dark-field images, is an abrupt event that takes place with a timescale of seconds. The rapid disappearance from the field of view in the fluorescence images is most probably due to rapid diffusion of GFP out of cells following membrane lysis. Our experiments indicate that the time at which lysis occurs does not depend on the stage of the cell cycle at the time of irradiation, as measured by the cell length (data not shown). Control experiments in which plasmids are absent indicate that while the mean lysis time is affected by the presence of the reporter plasmids (an increase of 40 and 65% with the pR-GFP and pR′-tR′-GFP reporters, respectively, is observed), its relative noise is not. Relative noise, further discussed below, is measured by the coefficient of variation η, the unitless ratio of the standard deviation divided by the mean η=σ/〈τ〉.
Timing of expression of pR, pR′-tR′ and time of lysis are independent of UV dose
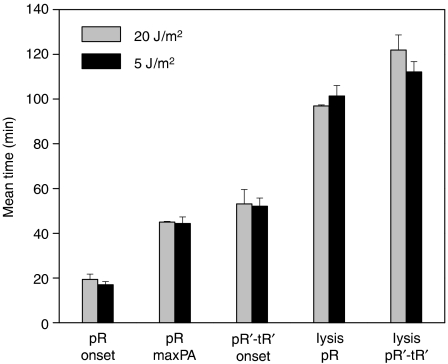
Recent experiments probing the SOS response in individual cells have revealed that the rate of SOS functions and cell growth decrease as the UV dose is increased (Friedman et al, 2005). This prompted us to ask whether timing of gene expression along the lytic cascade is also dependent on the amount of UV-induced DNA damage. To test whether the timing of execution of different cascade stages and cell–cell variability depend on UV dose, we also carried out experiments irradiating cells with 5 J/m2, and focused on four distinct events along the lytic cascade: (i) the onset time of induction of the cascade as monitored by the pR-GFP reporter fusion, (ii) the time at which maximum pR promoter activity is attained, (iii) the onset time of pR′-tR′ expression as determined by the pR′-tR′-GFP reporter plasmid and (iv) the time of lysis of individual cells in experiments using either reporter.
The behavior of cells undergoing the full lytic cascade after irradiation with 5 J/m2 is similar to the one described above for the higher 20 J/m2 dose. However, after irradiation with the lower dose, only a subpopulation of cells (40±15%) lyse, in contrast with the higher dose at which all cells lyse. This behavior is independent of the type of reporter plasmid present, and is similar to the observations in bulk cell experiments (Dutreix et al, 1985). Notably, for cells that completed the lytic cascade, the timing of the four events in the cascade did not show any significant dependence on the UV level within experimental error (Figure 3). The t-test P-values (for the null hypothesis that the times at high and low UV levels come from the same distribution) for pR onset, pR maximum promoter activity and pR′-tR′ onset times are 0.39, 0.94 and 0.27, respectively. A two-way ANOVA analysis of lysis (to take into account the effects of different reporter plasmids) yields a P-value of 0.96, indicating that the variability in timing is not due to the different UV doses received by each cell.
Figure 3.
Mean times of different events along the lytic cascade: onset time of expression from pR, the time corresponding to its maximum promoter activity, onset time of expression from pR′-tR′ and lysis time, following induction with UV doses of 5 J/m2 (black) and 20 J/m2 (gray). To exhibit the effect of plasmid type on the lysis time, the data were plotted separately for cells bearing either the pR-GFP or pR′-tR′-GFP reporter plasmids. Data were taken from three experiments at 5 J/m2 and two experiments at 20 J/m2. Error bars denote one standard error, reflecting variability between different experimental repeats.
Incomplete induction is observed at low UV doses
Cells that escape lysis after irradiation with the 5 J/m2 dose eventually divide, with the first division being delayed by ∼50 min beyond the typical division time of non-irradiated cells (doubling time of ∼60 min). The delay in cell division is inversely correlated with the cell length at the time of irradiation (data not shown). Escape from lysis is observed in experiments with either the pR or the pR′-tR′-GFP fusions, as well as in control experiments with cells bearing no plasmids. This indicates that the possible alteration of CI levels owing to the presence of the pR reporter plasmid does not have a significant effect on whether a cell will lyse or not.
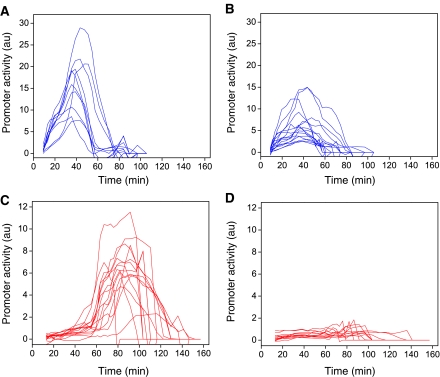
The promoter activity of pR reporters in individual cells undergoing incomplete induction is typically lower than that in cells that lyse in the same experiment (rank-sum P-value=2.4 × 10−5 for the null hypothesis of similar amplitudes), even though there are cases opposite to these (Figures 4A and B). As the measurement of promoter activity amplitude depends on the reporter plasmid copy number, which varies between cells, we cannot determine if there is a threshold value of pR promoter activity that could indicate whether cells will lyse or the cascade will be aborted. Note that the promoter activity of cells that lyse and those undergoing incomplete induction extends over a time window similar in extent. We cannot rule out that at the lower UV dose only a portion of the CI repressor molecules are inactivated, allowing for partial expression from the reporter plasmids without prophage induction. However, the similarity of the profiles of pR activity in all cells (lysing and non-lysing) indicates that the phage induction cascade may be activated also in non-lysing cells.
Figure 4.
Direct observation of incomplete induction in a subpopulation of cells, following irradiation with a UV dose of 5 J/m2. Whereas some cells harboring pR-GFP reporter plasmids (blue traces) complete the lytic cascade and exhibit promoter activity profiles (A) similar to those observed after irradiation with a high dose (see Figure 2C), a subpopulation of cells do not lyse, go through cell division after an ∼50 min delay and exhibit promoter activities (B) that are typically lower than cells that lyse. Similar experiments carried out with cells harboring pR′-tR′-GFP reporter plasmids (red traces) also show differences in promoter activity profiles between cells that lyse (C) and cells that undergo incomplete induction (D).
In contrast, cells undergoing incomplete induction do not exhibit a measurable Q function, as reported by the pR′-tR′-GFP fusion (compare Figures 4C and D), indicating that the commitment to lysis occurs before late gene expression. Incomplete induction, termed ‘subinduction’, has been previously inferred from measurements of repressor inactivation in cell ensembles (Bailone et al, 1979). The time window over which we observe pR to be active is consistent with the 30 min needed to carry out CI derepression by UV irradiation to completion (Bailone et al, 1979).
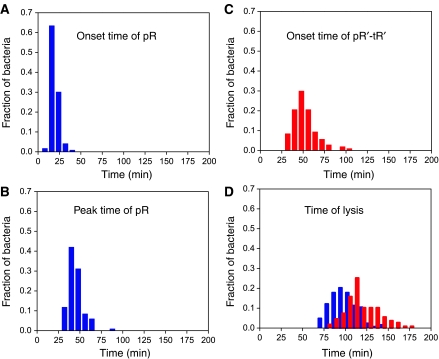
Analysis of timing noise along the cascade
In order to characterize the propagation of temporal fluctuations along the lytic cascade, we plot in Figure 5 histograms of the onset times of the derepression of pR and the activity of Q, as reported by the pR and pR′-tR′ reporter plasmids, the time at which the promoter activity of pR attains its maximum, as well as of the lysis times of individual cells after UV irradiation obtained in experiments using either of the reporter plasmids. The onset time of expression from pR, centered at ∼15 min after induction of DNA damage, is relatively narrow. In contrast, the time at which pR attains its maximum activity, centered around ∼40 min, the onset time of expression from pR′-tR′, centered at ∼50 min, as well as lysis times, centered at ∼95 min (determined using pR-GFP fusions) or ∼110 min (using the pR′-tR′-GFP fusions), are characterized by larger and larger timing fluctuations. The observed variability in pR expression timing is similar in extent to that observed for the pRecA promoter after induction of the SOS response in non-lysogens (Friedman et al, 2005).
Figure 5.
Histograms of event timing along the lytic cascade. Onset time of pR derepression (A), the time at which the activity of pR attains its maximum (B), the onset time of the Q-dependent pR′-tR′ expression (C) as well as lysis time (D) in individual cells completing the full lytic cascade after irradiation of lysogens with UV doses of either 5 or 20 J/m2. Histograms of cells harboring the pR-GFP or pR′-tR′-GFP plasmids are represented by blue and red bars, respectively. As observations of the expression of these promoters were carried out in different cells, each histogram has been normalized to unit area. Each observation was derived from at least 100 cells.
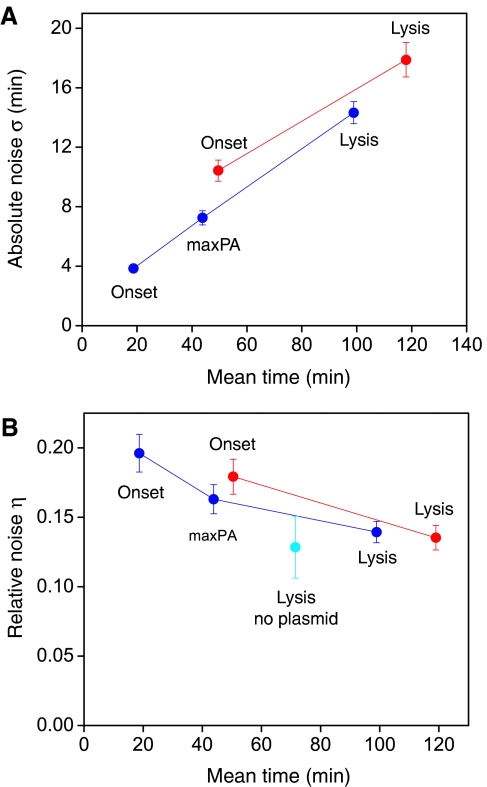
Figure 5 shows that there is an increase in the temporal variability of each successive stage of the cascade, as each stage contributes noise to those downstream. This is quantified in Figure 6A, where we plot the standard deviation 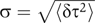 as a measure of the width of each histogram in Figure 5, and the average is taken over all cells except outliers (defined as cells for which σ>2.5; see Materials and methods). Here, τ is the time of occurrence of a particular event such as the onset time of promoter expression or the lysis time in a given cell. As downstream stages take longer to appear, differences in the precision of execution time between stages within a cascade are best compared by the coefficient of variation η=σ/〈τ〉. This ratio is analogous to the one used to quantify noise in protein and mRNA concentrations (Paulsson and Ehrenberg, 2001; Swain et al, 2002; Swain 2004). The values of η for the different stages in the cascade are plotted in Figure 6B. The salient feature of this plot is the decrease in η as the cascade progresses. Hence, the relative precision in timing increases, as the execution of the cascade progresses.
as a measure of the width of each histogram in Figure 5, and the average is taken over all cells except outliers (defined as cells for which σ>2.5; see Materials and methods). Here, τ is the time of occurrence of a particular event such as the onset time of promoter expression or the lysis time in a given cell. As downstream stages take longer to appear, differences in the precision of execution time between stages within a cascade are best compared by the coefficient of variation η=σ/〈τ〉. This ratio is analogous to the one used to quantify noise in protein and mRNA concentrations (Paulsson and Ehrenberg, 2001; Swain et al, 2002; Swain 2004). The values of η for the different stages in the cascade are plotted in Figure 6B. The salient feature of this plot is the decrease in η as the cascade progresses. Hence, the relative precision in timing increases, as the execution of the cascade progresses.
Figure 6.
Statistical analysis of fluctuations along the lytic cascade. (A) Standard deviation as a measure of width of histograms in Figure 5. Blue and red points represent data from cells harboring the pR-GFP or pR′-tR′-GFP reporter plasmids, respectively. Data were derived from at least four experimental repeats. Error bars represent one standard deviation (see Materials and methods). (B) Coefficient of variation η defined as the ratio between the standard deviation over the mean time of different stages along the lytic cascade. The value of η for lysis time is similar in experiments with pR-GFP or pR′-tR′-GFP plasmids (blue and red, respectively) and in experiments without reporter plasmids (cyan). The statistical significance for decrease in η between two adjacent points is less than 0.02 in all cases (see Materials and methods).
Correlation of lysis time with phage gene expression
The expression of late lytic genes such as S and R is obligatory for lysis to take place. Late lytic genes belong to the same operon, whose transcription starts at the pR′ promoter and requires Q activity for gene expression. We therefore inquired to what extent the lysis time tlysis and the onset time of pR′-tR′ expression tpR′-tR′ correlate. A statistical analysis (see Materials and methods) indicates that these two times are indeed correlated (Spearman correlation coefficient r=0.6137, P-value=2 × 10−12), indicating that cells in which Q activity is manifested early lyse earlier and vice versa.
One can imagine two sources for the observed cell–cell variations in lysis time: they could arise from random fluctuations of the various components of the induced lysis network, or alternatively, from cell factors uniformly changing the rate of execution of the cascade, speeding it up in some cells and slowing it down in others. Tapping the lytic cascade at an intermediate point, such as the onset of Q activity, offers the opportunity to discriminate between these two cases.
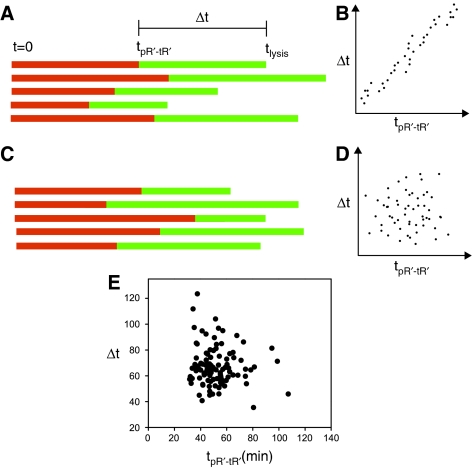
Lysis time can be looked at as a sum of two intervals, namely, the time elapsed until the onset of Q activity tpR′-tR′, and the interval Δt=tlysis−tpR′-tR′ in which late gene activation takes place, culminating in lysis. If indeed a major source of variability is the different rates at which the lytic cascade progresses in different cells, one would expect uniform dilation of timelines, which is different between cells, as depicted schematically in Figures 7A and B. If on the other hand, variations in tlysis are due to random fluctuations in the cascade components, one would expect the behavior depicted in Figures 7C and D. The actual measured values of Δt versus tpR′-tR′, plotted in Figure 7E show more similarity to Figure 7D, suggesting that fluctuations in lysis time are not due to the existence of factors uniformly affecting cascade rates. Furthermore, a correlation analysis yields a Spearman correlation coefficient r=−0.0522 (P-value of 0.73), confirming that both quantities are not correlated. Hence, the time elapsed from induction to the onset of Q activity and the time it takes cells to lyse once the expression of late genes is activated are independent intervals. Probing the cascade at the time at which maximal pR promoter activity is attained instead of at tpR′-tR′ yields a similar behavior albeit less pronounced (Supplementary Figure S1).
Figure 7.
Correlation of events along the lytic cascade. (A) Schematic timelines of individual cells along the lytic cascade from the time of induction at t0=0, through the onset time of pR′-tR′ expression, tpR′-tR′, and ending in the time of lysis, tlysis, assuming uniform time dilation from cell to cell, yielding the schematic correlation between the intervals Δt=tlysis−tpR′-tR′ (green) and tpR′-tR′−t0 (red) shown in (B). (C) Individual cell timelines assuming the execution of the cascade is affected by random fluctuations in timing of the various stages, in which case Δt and tpR′-tR′ vary independently, yielding the schematic correlation shown in (D). (E) Measured intervals Δt in individual cells completing the lytic cascade as a function of tpR′-tR′. A correlation analysis yields a P-value equal to 0.59, suggesting that both quantities are not correlated.
A similar analysis of the onset time of expression from the pR promoter is not possible, as we detect no correlation between tpR and tlysis. This may be due to the narrow spread of the onset time of pR expression, and the large variability in lysis timing. In addition, no correlation was found between the maximum fluorescence levels from the pR-GFP reporter and tlysis. This indicates that although these levels can discriminate between cells undergoing incomplete induction and those lysing, they are not good predictors of when lysis will occur.
Discussion
The induction of bacteriophage λ provides an interesting model for the study of timing fluctuations in a natural biological cascade. The molecular circuitry underlying the induction cascade is known to a large extent, and the initiation of the cascade can be synchronously induced in all cells by UV irradiation. We accessed directly the cell–cell timing variability during the execution of different stages along the cascade by following expression from two reporter fusions and the lysis of individual cells.
UV irradiation of lysogenic cells leads to the activation of two interrelated networks: that of the host SOS and that of prophage lytic development. The fact that prophage induction can proceed in cells in which the SOS network remains inactive (deduced from experiments with a non-inducible lexA mutation; Quillardet et al, 1982) suggests that the key event in the interaction between these two networks is mainly at the initial repressor cleavage stage. However, these two networks show different responses to the UV dose, one of the reasons being the different affinities of LexA and CI for active RecA (Mustard and Little, 2000). In contrast to the SOS network (Friedman et al, 2005), UV dose governs the proportion of cells in which the lytic cascade is turned on, but does not affect timing. Thus, after the initial induction step, the lambda network proceeds autonomously of the SOS response.
Analysis of the timing of different events as the phage cascade progresses reveals that while the absolute variation in timing of each stage increases, the coefficient of variation η decreases. Hence, the relative precision in the execution time of each successive stage of the cascade increases. A general theoretical consideration determines when such a behavior can be observed. In a cascade composed of independently distributed interval lengths, the absolute standard deviation will always increase, as it is the sum of the non-negative variances of the individual stages. For such a cascade of independently distributed interval lengths, the coefficient of variation η may increase or decrease, depending on the distributions of the various stages. A necessary and sufficient condition for a monotonic decrease in η, stemming from the additivity of the mean and the variance, is 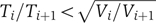 for all i<n, where Ti and Vi denote the cumulative mean and variance up to stage i of the cascade, respectively, and n is the number of stages in the cascade. For example, in a cascade of exponentially distributed stages, in which there is a rate-limiting step, all precision built up until that point may be lost and the process returns to approximately an exponential.
for all i<n, where Ti and Vi denote the cumulative mean and variance up to stage i of the cascade, respectively, and n is the number of stages in the cascade. For example, in a cascade of exponentially distributed stages, in which there is a rate-limiting step, all precision built up until that point may be lost and the process returns to approximately an exponential.
The observed decrease in the coefficient of variation can be reproduced by a simple model of independent, identically distributed cascaded events, in which the time elapsed between an event and the next one is distributed with an exponential distribution 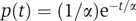 , the single rate parameter α determining both the mean time and the standard deviation (see e.g., Golding et al, 2005). The time of execution of such a cascade composed of k events is therefore a sum of k exponentially distributed random variables. In the case in which all rates are equal, known as the Erlang distribution (which is a special case of the gamma distribution with integer k), the average time of execution is kα, whereas the standard deviation is
, the single rate parameter α determining both the mean time and the standard deviation (see e.g., Golding et al, 2005). The time of execution of such a cascade composed of k events is therefore a sum of k exponentially distributed random variables. In the case in which all rates are equal, known as the Erlang distribution (which is a special case of the gamma distribution with integer k), the average time of execution is kα, whereas the standard deviation is 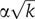 , leading to η=1/
, leading to η=1/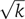 . As the two parameters α and k can be changed independently, a higher temporal accuracy can be achieved by using a large number of fast stages rather than a small number of slow steps. Note that the
. As the two parameters α and k can be changed independently, a higher temporal accuracy can be achieved by using a large number of fast stages rather than a small number of slow steps. Note that the 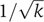 decrease of η with k can be obtained without assuming an exponential distribution, requiring only independent, identically distributed interval lengths between successive stages in the cascade.
decrease of η with k can be obtained without assuming an exponential distribution, requiring only independent, identically distributed interval lengths between successive stages in the cascade.
There exist many intermediate steps between any two points at which the lambda cascade was tapped, each step being subjected to fluctuations. Even the synthesis of a single protein consists of many stochastic events (Paulsson, 2005). Furthermore, in a genetic cascade, expression of a gene depends on the amount of the protein at the previous stage, and therefore the assumption of independence between stage interval lengths is no longer valid. In spite of these caveats, when we calculated η from published stochastic simulation data of a seven-stage transcriptional cascade with identical rate constants (Hooshangi et al, 2005), we still obtained a decrease in the coefficient of variation as expected from the simple model described above. Note that a different choice of rate constants may possibly lead to an increase.
Our experiments provide evidence for a correlation between tpR′-tR′, the onset time of Q activity monitored by pR′-tR′-GFP expression, and lysis time tlysis. A correlation is also observed between the time at which pR attains its maximal activity and tlysis. These correlations are expected, as the events we study are arranged in a cascade. The higher coefficient between tpR′-tR′ and tlysis may stem from the fact that lysis is ultimately caused by the expression of the late lytic genes, all of which reside on the single pR′-tR′ operon, whereas pR shutoff may originate from an earlier stage in the lytic cascade.
Sources of variability have been classically divided into intrinsic and extrinsic noise (Swain et al, 2002; Raser and O'Shea, 2005). Intrinsic noise sources such as transcription initiation (Golding et al, 2005) and mRNA translation are stochastic in nature, and create differences in the expression of two equivalent, independent gene reporters within the same cell. Global extrinsic sources such as number variations in a factor affecting gene expression—for example, ribosomes—affect the two reporters and the expression of all genes in the cell similarly, but induce cell–cell differences. The existence of ‘fast’ and ‘slow’ cells may be viewed as the simplest manifestation of global extrinsic noise, in which all stages in a cascade are affected similarly by the metabolic status of the cell (which is assumed to remain constant throughout the cascade progression). In such a case, one would expect a correlation between the time interval from UV irradiation to the onset of pR′-tR′ expression (tpR′-tR′) and the interval from the onset of pR′-tR′ expression to lysis (tlysis−tpR′-tR′). The lack of such a correlation in our data indicates that timing in the lambda cascade is not determined uniformly for each cell, and that the sources of noise in the two parts of the cascade are distributed independently. Therefore, variability in lysis time is not due to differences in the global rate of cascade progression, but probably to random fluctuations in the execution time of the various cascade stages.
In addition to the study of temporal fluctuations in gene expression, the present study has revealed a couple of important features specific to the phage lambda induction network: first, it has provided direct evidence for the dose-dependent existence of two phenotypic outcomes, that is, cells displaying either incomplete induction or lytic fates; and second, the shutoff of the pR promoter. A subpopulation of cells that did not go through the lytic cascade, but went into cell division (after some delay), was only observed after irradiation with the low UV dose. These cells were further characterized by a low activity from the pR reporter and no activity from the pR′-tR′ reporter. Low levels of UV damage typically lead to an earlier shutoff of the SOS response than high levels (Friedman et al, 2005). Furthermore, the rate of cleavage of CI by the activated RecA filament is much lower than that of the LexA repressor (Mustard and Little, 2000). Under these conditions, it is conceivable that CI concentrations are not sufficiently depleted to activate the lytic pathway, and positive feedback through pRM may restore CI concentration to achieve repression. Our pR-GFP reporter plasmids do not possess the full operator architecture that enables the formation of the CI octamer in the prophage (see Materials and methods). Hence, it is conceivable that derepression of the pR-GFP fusion may occur under conditions in which the lytic cascade in the prophage is not activated. We note, however, that incomplete induction is observed in the presence or absence of reporter plasmids. We suspect that the variability in numbers of CI molecules among cells (Baek et al, 2003) is such that it may lead to the phenotypic variation observed in our experiments. The increase in the proportion of cells undergoing full induction as the UV dose is increased is consistent with the increase in infective centers observed in ensemble experiments (Baluch and Sussman, 1978), and with theoretical models probing the basins of attraction of different fixed points in the network (Chung and Stephanopoulos, 1996). The present kinetic analysis has also revealed a delay in Q activity similar to the one found after infection, as well as an unexpected shutoff in the promoter activity of pR. Whereas the delay in Q activity was attributed to a threshold effect (Kobiler et al, 2005), the cause for the pR promoter shutoff remains elusive. A potential candidate is the Cro protein, which is known to downregulate the expression from pR. However, recent experiments indicate that Cro reduces pR expression by only about two-fold (Svenningsen et al, 2005; Atsumi and Little, 2006). In addition, our preliminary results suggest that the shutoff of pR activity does not depend on Cro alone, as it is also observed in induction experiments with Cro mutant prophages (Oppenheim et al, 2004).
A number of recent studies have analyzed noise propagation in synthetic transcriptional cascades (Hooshangi et al, 2005; Pedraza and van Oudenaarden, 2005; Rosenfeld et al, 2005). In these studies, cellular individuality in a clonal population arises from variability in protein and RNA numbers. Our study, conducted at the single-cell level, addressed a different aspect of individuality, namely, that arising from variability in the times of execution of different stages of a complex, natural biological cascade. Our findings reveal that the relative noise η in the timing of successive stages of the cascade decreases as the cascade progresses. This behavior can be accounted for by simple but very general theoretical considerations as those presented here and is probably due, in part, to the fact that bacteriophages such as lambda are made of a number of relatively independent modules (Hendrix, 2003). Interestingly, lambda utilizes transcription termination and antitermination extensively as a control motif in its polycistronic modules, allowing the phage to sense the host's physiological state. Theoretical evidence for the reduction of noise due to a polycistronic architecture has been provided (Swain, 2004). Transcription antitermination may therefore enable the phage to differentially express genes on the same operon, while maintaining a lower level of noise between expressed genes. A future challenge is to develop experimental systems for the continuous monitoring of phage DNA replication and transcriptional and post-transcriptional steps of the phage genetic network. As recently reviewed, the organization of the lambda genome may yet teach us much more about how information and function are encoded in the gene order of eukaryotic genomes (Kosak and Groudine, 2004).
Materials and methods
Plasmid, bacterial and phage strains
Promoter-GFP fusion reporter plasmids were constructed as described by Kobiler et al (2005). Briefly, the lambda promoters were inserted upstream of the promoterless fast-folding gfpmut3 gene (Cormack et al, 1996) of the medium-copy plasmid pSA11 (Schlosser-Silverman et al, 2000), which carries a pBR322 origin of replication and appears at approximately 20 copies per cell. Response time of GFP fluorescence following IPTG induction of pSA11-bearing bacteria was ∼15 min. Promoters used were pR (between lambda coordinates 5′-37905 and 38034-3′) and pR′-tR′ (between lambda coordinates 5′-44300 and 44831-3′). Monolysogenic bacteria were created by infecting E. Coli W3110 with λ c+ knR (Kobiler et al, 2002). Bacteria containing a single prophage were selected as knR immune lysogens that were sensitive to λ c17.
Experimental setup
Experiments were carried out in a home-built inverted microscope, as described previously (Friedman et al, 2005). Samples were illuminated with an argon laser (488 nm) and images were collected using an intensified CCD and stored on a computer for later analysis. In all experiments, bacteria were humidified and the temperature was controlled and set to 37°C.
Sample preparation
Cells were grown overnight in an LB medium and diluted into a fresh medium used in the experiments (M9+20 amino acids except tryptophan, 50 mg/l; thiamine 20 mg/l; thymine 20 mg/l; biotin 1 mg/l; glucose 0.4% v/v). After reaching midlog phase (OD600=0.25–0.4), cells were placed on a preheated agarose slab (experimental medium+2% agarose) and incubated for 5 min at 37°C. For lambda prophage induction, cells were irradiated in situ with UV light (wavelength 254 nm), using a low-pressure mercury germicidal lamp at levels between 5 and 20 J/m2 (20 J/m2 corresponds to an irradiation time of 14 s). After UV irradiation, bacteria were covered with a coverslip and monitored using the fluorescence microscope. The thin layer of agar allowed for an efficient supply of oxygen and nutrients to reach the cells during the experiment. Our irradiation procedure avoids any inhomogeneities due to the considerable UV absorption by the liquid media.
Determination of bacterial fluorescence intensity
Images were recorded every 3 min. Each bacterial cell was manually marked on each frame, and represented as a piecewise continuous line, describing the long axis of the cell. The total intensity of pixels along this line was then calculated as a measure of the total fluorescence emanating from each cell. The total intensity corresponding to each cell was then corrected for photobleaching using the recursive equation 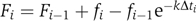 , where fi and Fi are the measured and corrected total intensities, respectively, per cell in image i, Δti is the exposure time of the image and k is the measured bleaching rate of GFP in cells.
, where fi and Fi are the measured and corrected total intensities, respectively, per cell in image i, Δti is the exposure time of the image and k is the measured bleaching rate of GFP in cells.
Determination of promoter activity
The promoter activity of individual cells was defined and calculated according to PA(t)=(dI(t)/dt)/L(t), where I(t) and L(t) are the intensity and the length of the cell at time t. The division by L(t) normalizes the data to concentration units (as bacteria grow in one dimension) and is analogous to the normalization by the optical density in measurements of cell populations. To reduce noise in the PA calculation, which stems mainly from focus changes between consecutive images, the derivative was calculated by a least squares linear fit to the intensity using a 15 min sliding window centered at time t.
Determination of gene expression times
The initial rise in the fluorescence was fit to a piecewise continuous function consisting of a line stitched to a parabolic growth function (which corresponds to a linear increase in promoter activity):
![]()
The onset time t0, which corresponds to the stitching point, was determined by a nonlinear least square fit using Origin 7.0 (OriginLab). The time at which pR attained its maximum promoter activity was determined by a Gaussian fit to 10 data points around the maximum.
Analysis of noise
Data from replicate experiments for each plasmid were normalized to a similar mean, to exclude the effect of differences between experimental runs. Data corresponding to each plasmid were filtered to remove outliers whose times deviated by more than 2.5 s.d. from the mean (resulting in the removal of <3% of cells for each plasmid). We suspect that outliers represent the behavior of cells in which genes within the lambda cascade have been damaged by the UV irradiation. Errors in noise calculation were estimated by using the bootstrap method. The P-values for the decrease in the relative noise were calculated by counting the number of times a decrease in the relative noise occurs in 10 000 bootstrap runs. An analysis of correlations was carried out by calculating the Spearman rank-sum correlation to reduce the effect of outliers. For all bacteria, each event was given a value according to its index in the sorted times list for that event. Correlations were then calculated on the resulting indices rather than the original times. The null hypothesis probability was calculated by a two-sided comparison of this C-value to the C-values of random permutations of these event times. In all cases, the rank-sum correlation P-values were very similar to results obtained by a normal Pearson correlation test.
Supplementary Material
Supplementary Figure and Movie captions
Supplementary Figure S1
Supplementary Movie 1
Supplementary Movie 2
Acknowledgments
This work is dedicated to the memory of AB Oppenheim (1934–2006), who passed away during the revision of the manuscript. We thank R Weiss and S Hooshangi for supplying the data from the cascade simulation, S Vardi for valuable assistance and one of the reviewers for valuable comments. This research was supported in part by The Israel Science Foundation (grant # 489/01-1 and 340/04).
References
- Atsumi S, Little JW (2006) Role of the lytic repressor in prophage induction of phage lambda as analyzed by a module-replacement approach. Proc Natl Acad Sci USA 103: 4558–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Svenningsen S, Eisen H, Sneppen K, Brown S (2003) Single-cell analysis of lambda immunity regulation. J Mol Biol 334: 363–372 [DOI] [PubMed] [Google Scholar]
- Bailone A, Levine AJ, Devoret R (1979) Inactivation of prophage λ repressor in vivo. J Mol Biol 131: 553–572 [DOI] [PubMed] [Google Scholar]
- Baluch J, Sussman R (1978) Correlation between UV dose requirement for lambda bacteriophage induction and lambda repressor concentration. J Virol 26: 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR (2006) Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell 21: 3–14 [DOI] [PubMed] [Google Scholar]
- Becskei A, Kaufmann BB, van Oudenaarden A (2005) Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Gen 37: 937–944 [DOI] [PubMed] [Google Scholar]
- Berg OG (1978) A model for the statistical fluctuations of protein numbers in a microbial population. J Theor Biol 71: 587–603 [DOI] [PubMed] [Google Scholar]
- Chung JD, Stephanopoulos G (1996) On physiological multiplicity and population heterogeneity of biological systems. Chem Eng Sci 51: 1509–1521 [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Renekov O, Endy D, Pesce G, Brent R (2005) Regulated cell-to-cell variation in a cell-fate decision system. Nature 437: 699–706 [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173: 33–38 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan ZJ, Schilstra MJ, Clarke PJC, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H (2002) A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB (2001) Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev 15: 3013–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Shearwin KE, Egan JB (2005) Revisited gene regulation in bacteriophage lambda. Curr Opin Genet Dev 15: 145–152 [DOI] [PubMed] [Google Scholar]
- Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB (2004) Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev 18: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M, Bailone A, Devoret R (1985) Efficiency of induction of prophage lambda mutants as a function of recA alleles. J Bacteriol 161: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D, You LC, Yin J, Molineux IJ (2000) Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc Natl Acad Sci USA 97: 5375–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J (2005) Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PloS Biol 3: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Gotta SL, Miller OL, French SL (1991) Ribosomal-Rna transcription rate in Escherichia-coli. J Bacteriol 173: 6647–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW (2003) Bacteriophage genomics. Curr Opin Microbiol 6: 506–511 [DOI] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R (2005) Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci USA 102: 3581–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U (2001) Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Kobiler O, Koby S, Teff D, Court D, Oppenheim AB (2002) The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci USA 99: 14964–14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O, Rokney A, Friedman N, Court DL, Stavans J, Oppenheim AB (2005) Quantitative kinetic analysis of the bacteriophage lambda genetic network. Proc Natl Acad Sci USA 102: 4470–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Groudine M (2004) Gene order and dynamic domains. Science 306: 644–647 [DOI] [PubMed] [Google Scholar]
- Lamb TD (1996) Gain and kinetics of activation in the G-protein cascade of phototransduction. Proc Natl Acad Sci USA 93: 566–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Little JW (1996) The SOS regulatory system. In Regulation of Gene Expression in E. coli, Lin ECC, Simon A (eds) pp 453–479. New York: RG Landes Company [Google Scholar]
- McAdams HH, Arkin A (1997) Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA 94: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustard JA, Little JW (2000) Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of recA. J Bacteriol 182: 1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S (2005) Switches in bacteriophage lambda development. Annu Rev Genet 39: 409–429 [DOI] [PubMed] [Google Scholar]
- Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL (2004) In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology 319: 185–189 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2005) Models of stochastic gene expression. Phys Life Rev 2: 157–175 [Google Scholar]
- Paulsson J, Ehrenberg M (2001) Noise in a minimal regulatory network: plasmid copy number control. Q Rev Biophys 34: 1–59 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Ptashne M (2004) Genetic Switch: Phage Lambda Revisited. Woodbury, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Quillardet P, Moreau PL, Ginsburg H, Mount DW, Devoret R (1982) Cell survival, UV-reactivation and induction of prophage lambda in Escherichia coli K12 overproducing RecA protein. Mol Gen Genet 188: 37–43 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigney DR (1979) Stochastic model of constitutive protein levels in growing and dividing bacterial cells. J Theor Biol 76: 453–480 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Schlosser-Silverman E, Elgrably-Weiss M, Rosenshine I, Kohen R, Altuvia S (2000) Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J Bacteriol 182: 5225–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Costantino N, Court DL, Adhya S (2005) On the role of Cro in lambda prophage induction. Proc Natl Acad Sci USA 102: 4465–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PS (2004) Efficient attenuation of stochasticity in gene expression through post-transcriptional control. J Mol Biol 344: 965–976 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A (2002) Attenuation of noise in ultrasensitive signaling cascades. Biophys J 82: 2943–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure and Movie captions
Supplementary Figure S1
Supplementary Movie 1
Supplementary Movie 2