Abstract
Efforts to construct therapeutically useful models of biological systems require large and diverse sets of data on functional connections between their components. Here we show that cellular responses to combinations of chemicals reveal how their biological targets are connected. Simulations of pathways with pairs of inhibitors at varying doses predict distinct response surface shapes that are reproduced in a yeast experiment, with further support from a larger screen using human tumour cells. The response morphology yields detailed connectivity constraints between nearby targets, and synergy profiles across many combinations show relatedness between targets in the whole network. Constraints from chemical combinations complement genetic studies, because they probe different cellular components and can be applied to disease models that are not amenable to mutagenesis. Chemical probes also offer increased flexibility, as they can be continuously dosed, temporally controlled, and readily combined. After extending this initial study to cover a wider range of combination effects and pathway topologies, chemical combinations may be used to refine network models or to identify novel targets. This response surface methodology may even apply to non-biological systems where responses to targeted perturbations can be measured.
Keywords: chemical genetics, combinations and synergy, metabolic and regulatory networks, simulation and data analysis
Introduction
Living organisms are built of interacting components, whose function and dysfunction can be described through dynamic network models (Davidson et al, 2002). Systems Biology involves the iterative construction of such models (Ideker et al, 2001), and may eventually improve the understanding of diseases using in silico simulations. Such simulations may eventually permit drugs to be prioritized for clinical trials, reducing potential risks and increasing the likelihood of successful outcomes.
Owing to the staggering complexity of biological systems, efforts to model them require large and diverse sets of data on connections between components and responses to system perturbations. Among the most advanced models are those developed for baker's yeast, Saccharomyces cerevisiae (Zhang et al, 2005), which are built upon protein–DNA (Lee et al, 2002) and protein–protein (Ho et al, 2002) associations, supplemented by correlated changes in gene expression (Hughes et al, 2000) or protein abundances (Gygi et al, 1999) under differing conditions. Information gleaned from targeted synergies, such as paired mutations (Tong et al, 2004) and gene–drug interactions (Parsons et al, 2004), have proven to be especially useful for revealing functional connections between components. Chemical combinations also show promise, and a proliferation experiment with yeast mutants in the presence of probe mixtures (Haggarty et al, 2003) has found that chemical profiles correlate with genetic similarity. This potential is confirmed by recent experiments using antibacterial combinations (Yeh et al, 2006) that show a relationship between synergy and chemical target relatedness.
Combination responses to varying concentrations of compounds provide a more detailed look at synergistic perturbations. Combination therapies have been used increasingly over the past century, and comprehensive reviews (Berenbaum, 1989; Greco et al, 1995) describe the experimental designs and combination analyses employed. Combinations of two or more agents can be tested using either exhaustive or efficient designs (Carter and Wampler, 1986), and the most widely used is the factorial design (also ‘checkerboard’ or ‘dose matrix’) where combinations are tested in all possible permutations of serially diluted single agent doses (Figure 1). A dose-matrix experiment comprehensively samples the underlying response surface with few assumptions about its shape. We have previously reported an approach for high-throughput dose-matrix screening of chemical combinations (Borisy et al, 2003; Keith et al, 2005; Zimmermann et al, 2007) in cell-based assays that preserve disease-relevant biological connections. Such screens yield a variety of response surfaces, with distinct shapes for combinations that work through different known mechanisms, suggesting that combination effects may contain information on the nature of functional connections between drug targets.
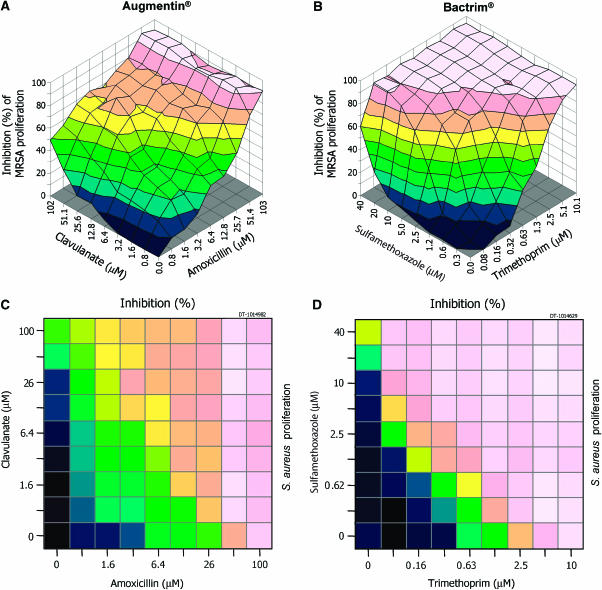
Figure 1.
The morphology of cell-based dose-matrix responses to chemical combinations differs between mechanisms. Data for two synergistic antibacterial combinations using a methicillin-resistant Staphylococcus aureus strain are shown in three-dimensional projections. In Augmentin® (A), clavulanate disables a bacterial defence against penicillin drugs, whereas both agents in Bactrim® (B) inhibit enzymes in folate metabolism. The mechanisms of action differ between the two combinations, as do the shapes of their response surfaces. Dose-matrix response surfaces show the inhibition of growth (1−treated/untreated) for each pairwise permutation of the serially diluted single agent doses. The 2D maps (C, D) show a top-down view of the surface, using the same colours for inhibition levels, to better display mathematical descriptions of the shape without obscuring any data, and permit many matrices to be shown together with clarity.
The past study of drug combinations has focused mainly on the question of whether a combination is more potent than equally effective doses of its constituents (Greco et al, 1995). Synergy over this level is especially important when justifying clinical uses, as it defines the point at which the combination can provide additional benefit over simply increasing the dose of either agent. This most widely used dose additivity model (Loewe, 1928) represents the expected response if both agents are actually the same compound. In that case, a slice through the response surface at any chosen iso-effect level (or ‘isobole’) should show a linear relationship between the doses of the two agents. For example, if 50% inhibition (I=T/U for treated and untreated samples) is achieved separately by 1 μM of drug A or 2 μM of drug B, a combination of 0.5 μM of A and 1 μM of B should also inhibit by 50%. Formally, the response at combined concentrations X,Y is the inhibition ILoewe that satisfies (X/XI)+(Y/YI)=1, where XI and YI are the single agent effective concentrations that produce ILoewe. Deviations from Loewe additivity are usually quantified using the combination index (Chou and Talalay, 1983) CI=(X/XI)+(Y/YI), which is essentially a ratio of total effective drug dose (combination versus single agents) required to achieve a given effect level. Loewe additivity is experimentally demanding because determining the effective concentrations requires well-sampled single agent curves. Moreover, the dose-additive constraint is not in closed form; so Loewe additive response surfaces generally must be computed using iterative root finding (Berenbaum, 1985). The other frequently used reference model is Bliss independence (Bliss, 1939), which is the expectation (multiplicative probabilities) for independent yet competing elimination agents, like bullets aimed at a limited set of targets. For inhibitions, IMult=IX+IY–IXIY, where IX and IY are the single agent inhibition levels at concentrations X and Y. Although this model is of questionable relevance to drugs in biological settings, Bliss independence has often been favoured because it can be directly calculated from minimally sampled experiments without single agent response curve interpolation or iterative root finding. A third useful reference is the highest single agent (HSA) model, or Gaddum's non-interaction (Berenbaum, 1989), where the expected combination effect is simply the maximum of the single agent responses at corresponding concentrations.
The relationship between combination effects and drug mechanism has been intuitively recognized for some time, but attempts to theoretically model this association (Ashford and Cobby, 1974; Harrap and Jackson, 1975; Jackson, 1993) have proven problematic. These studies simulated a branched metabolic pathway as a system of linked ordinary differential equations, and the final reaction velocity was calculated in response to varying concentrations of inhibitors targeting different modelled enzymes. Synergy over Loewe additivity was assessed for each pair of enzyme inhibitors by computing simulated dose matrices and reporting the smallest combination index at 50% inhibition. However, these efforts could not unambiguously predict synergy or antagonism, because it was possible to generate any combination index for most inhibitor pairs by varying one or more of the many kinetic parameters in the pathway model.
We approached the problem by focusing not just on the presence or absence of synergy relative to some reference level, but by using a set of shape models to characterize the morphology of full response surfaces. Here we show that there is a clear relationship between dose-matrix response shape for a combination and the connectivity of its targets (the type of connection between them), using simulated pathways and combination experiments. The simulations were used to define a reference set of shape models that represent connectivity-related responses, and the experiments demonstrated that the shapes predicted by the simulations do indeed occur in biological settings. We also use the data to show that the synergy profile produced by a drug across many combinations in a screen depends on the drug's mechanism, and can also be used to infer connectivity relationships between drug targets.
Results
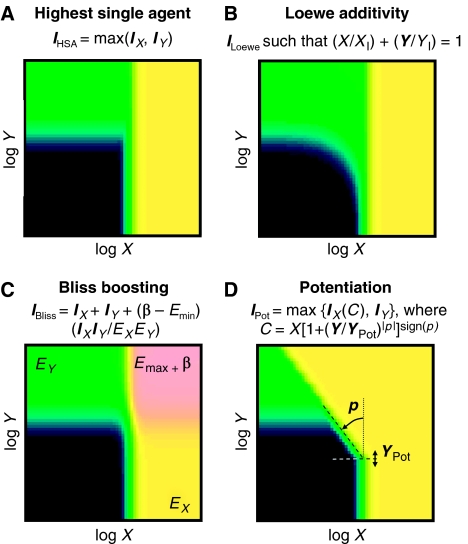
We considered four models for combination effect morphology (Figure 2 and Materials and methods) that reflect historical combination analyses and that represent many of the responses observed in our therapeutic combination screens. The HSA model describes a simple superposition of the single agent curves. Loewe additivity (Loewe, 1928) is the drug-with-itself reference to represent dose-additive pairings. The ‘Bliss boosting’ model extends Bliss Independence (Bliss, 1939) to allow variable boosts in effect at high concentrations. Finally, the ‘Potentiation’ model describes combinations like Bactrim® (Figure 1), where one agent's curve is shifted with a power-law slope and superposed on the enhancer's own activity. These models are not an optimally designed basis set of shape descriptors because they are not orthogonal (they produce degenerate, or indistinguishable, shapes for some parameter settings), and also are not complete, in that some observed surfaces do not fit any of them. Nevertheless, this set does cover many of the observed effects in our therapeutic screens, and permits a quantitative exploration of different synergy types. Each observed dose matrix can be analysed by fitting all the shape models to the data by means of least-squares minimization across any free parameters, and determining which shape models produce consistent fits. It is also useful to calculate a volume VHSA between the data and the HSA surface, normalized to the concentration sampling, to characterize the overall strength of combination effects.
Figure 2.
Combination response shape models that describe many of the observed response morphologies. Each model (shown using the same colour scale as Figures 1 and 3) is used to calculate an expected combination effect Imodel at any concentration X,Y, based on the single agent response curves. (A) HSA is a superposition of the X and Y single agent responses, calculated from the inhibitions IX at X and IY at Y. (B) Loewe additivity (Loewe, 1928) is the drug-with-itself reference for synergy, where ILoewe at X,Y yields additive doses relative to the components' effective concentrations XI,YI at ILoewe. (C) Bliss boosting describes combinations with a variable boost β above Emax (the greater of the single agent limiting efficacies EX,EY), at high combined concentrations. Useful reference levels for Bliss boosting are ‘cancelling’, ‘suppressing’, ‘masking’, ‘multiplicative’ corresponding to Bliss independence (Bliss, 1939), and ‘saturating’ (see Materials and methods). Finally, (D) potentiation can characterize responses similar to those of Bactrim® (Figure 1), where one single agent's curve IX(C) is shifted with a power-law slope p above an enhancer concentration Ypot, and superposed on the enhancer's own activity. These models can be extended to higher-order combinations, and used in the same form (with adjustments to Bliss boosting) for any type of measurement, provided that the activities of both agents vary monotonically with concentration. Of these models, only Loewe additivity has an a priori mechanistic basis.
Simulations of metabolic pathways
We used numerical simulations of metabolic pathways to show that pairs of targets with differing connectivity produce distinct combination effects (Figure 3). We constructed a branched, unregulated pathway of linked Michaelis–Menten reactions (Supplementary 1), and computed the inhibition of final reaction velocity after applying varying concentrations of paired competitive enzymatic inhibitors (affecting Km). Each pair of inhibitors was applied repeatedly over a dosing matrix, and we determined the response morphology by fitting our shape models to each simulated dose matrix, and finding which shape provided the best match over the whole surface (Figure 3). We found that when both inhibitors targeted the same enzyme, Loewe additive surfaces were produced as expected. When the inhibitors had separate targets within this pathway, the resulting surfaces were best fit by Bliss boosting, with differing levels depending on the target connectivity.
Figure 3.
Simulations of a multiply inhibited network yield distinct response shapes that depend on target connectivity. Substrates (black nodes) are metabolized through a series of Michaelis–Menten reactions (grey circles) from sources with constant reaction velocities to a limitless sink at the end-point. We calculated dose-matrix inhibitions of the end point velocity at RE to paired competitive inhibitors at various enzymes. The response surfaces for some combinations are shown, with the inhibitors indicated by joined markers. For Pathway A, same-target pairs produced Loewe additivity, and separated targets led to various Bliss boosts depending on where the targets were placed. Pathway B was added to investigate combination effects across pathways, with a bypass to ensure clear distinctions between cross-pathway boosting levels. Because both inputs are required for RAB, the junction is equivalent to a logical AND function. The negative feedback in Pathway B was introduced to represent processes like sterol biosynthesis in yeast. Inhibiting across pathways produced HSA-like masking effects, and inhibiting within the negative feedback loop yielded potentiation effects.
These differences in combination effect were robust to plausible variations of the simulated pathway that did not affect the network topology. Random perturbations of up to two orders of magnitude in all the reaction parameters recovered the original best-fit shapes (Supplementary Figure S2), suggesting that the response morphology is affected by only such extreme changes in kinetic parameters that alter the network topology. Similar Monte Carlo simulations perturbing specific parameter classes or individual enzymes showed no strong sensitivities to particular kinetic parameters or targets for this pathway (Supplementary 3). Converting the inhibitors to non-competitive (affecting Vm) kinetics likewise recovered the original best-fit shapes, altering mainly the single agent curves. An exception is that unbypassed serial combinations (e.g., R1xR6; Figure 3) yielded HSA-like responses for non-competitive inhibitors (Supplementary 2). Adjusting the end point to something more like proliferation by means of an exponential transform (see Materials and methods) distorted the resulting boost levels, but still did not change which shape model was preferred. All of these factors could strongly affect the single agent curves (e.g., potency, steepness, and limiting efficacy) but the best-fit shape model for each simulated combination effect was very stable to perturbation.
Introducing regulation by negative feedback did have strong effects on the combination responses. Pathway B was added to investigate synergy across pathways and to predict effects for sterol biosynthesis. Whereas inhibitors with serially placed targets in an unregulated linear pathway produced multiplicative boosts, enclosing such targets in a negative feedback loop yielded strong potentiation (although like unregulated serial inhibitors, these yield weaker effects for non-competitive inhibitors). The synergies under negative feedback are not immediately intuitive (Supplementary Figure S3) and an analytical derivation of the simulated power-law potentiation for this model would be challenging, underscoring that simulation is the most practical means for deriving even the simplest emergent properties of complex systems.
Another change affecting topology is to alter the type of junctions in a pathway. For the branches in Pathway A, the reaction could proceed down either side to the end point; so parallel pairs of inhibitors across these junctions (e.g., R3 with R5) are functionally analogous to logical OR operations. Inhibiting across such OR junctions produced saturating Bliss responses in our simulations. Replacing such junctions with functional AND operations (here implemented by pairing targets across Pathway A and Pathway B, both required for RAB) resulted in masking surfaces (close to HSA or Bliss masking) for combinations that straddled the junction. Such masking effects were produced even for cross-pathway combinations involving R11 and R12, which are bypassed in Pathway B to ensure clear distinctions between masking, multiplicative, and saturating boosts.
The best-fit shapes for all the combinations in both simulated pathways are summarized in Figure 4, using symbols whose size reflects the HSA volume and where the colour and symbol denote the best-fit model. The ambiguity between the shape models for each combination is illustrated in Supplementary Figure S4. In these simulations, the fitted shapes clearly correspond to distinct mechanistic configurations. It is notable that all the cross-pathway combinations produced either HSA surfaces or other masking surfaces with weaker combination effects (with ∣VHSA∣∼1, between the measured effect and the HSA model surface) than are seen within each of the pathways (∣VHSA∣∼3–10).
Figure 4.
The best-fit shape models for our simulated pathway (Figure 3) consistently depend on target connectivity. Combination effects between competitive inhibitors are shown as symbols whose area is proportional to the HSA volume and whose colour indicates the best-fit model shape: black for HSA, blue for Loewe additivity, green for Bliss boosting, red for potentiation, and grey when two or more models could not be distinguished (see Supplementary Figure S4). The Bliss boosting model is subdivided using shape to indicate the boosting level (square for saturating, diamond for multiplicative, circle for masking, and open circle for suppressing). The bold lines separate Pathway A from Pathway B combinations, and the thin lines partition Pathway A into its component sections. All inhibitor-with-self pairings fit Loewe additivity, as expected. Other combinations within Pathway A consistently fit the Bliss model with distinct boosting levels. Cross-target combinations within the negative feedback in Pathway B consistently show potentiation, and cross-pathway combinations always have masking responses (close to Bliss masking, additivity, or HSA).
These simulations suggest specific hypotheses in terms of our shape models for experimental combinations of chemical inhibitors of metabolic pathways. Agents sharing a primary target should, of course, produce Loewe additive responses. Branched unregulated pathways should produce various levels of Bliss boosting, depending on the target connectivity, and linear pathways regulated by negative feedback should show power-law potentiation. Combinations with targets that cross between such pathways are likely to produce masking effects (similar to the HSA model or Bliss masking).
Yeast combinations experiment
To test some of the hypotheses from our simulations, we undertook a chemical combination screen focused on the sterol metabolism in yeast. This pathway is very well characterized with known inhibitors at a number of enzymes (Wills et al, 2000), and is regulated by negative feedback (Gardner et al, 2001). We selected 10 antifungal drugs (Table I), six with targets along a linear section of the pathway and four targeting other cellular functions. Because sterol biosynthesis is much the same across yeast species (Wills et al, 2000), we selected Candida glabrata, a frequent focus of antifungal treatment that we had used in past therapeutic screens, and measured proliferation using a metabolic assay (Materials and methods). After determining single agent activities for all the drugs, we designed dose-matrix experiments with concentration samples centred on each drug's effective concentration at 50% effect (EC50) when possible.
Table 1. Drugs used in the C. glabrata proliferation experiment.

The dose-matrix responses for each combination are presented in Figure 5 and Supplementary 4. The combination effects are visible towards the top right-hand corner of each response surface, especially between the sterol pathway inhibitors. Some of the single agents produced variable responses, and this is most obvious for those drug-with-itself dose matrices along the diagonal of the grid which show only one side appearing to be active. The self-combination of rapamycin illustrates how a spurious synergy can arise from instability in the measured potency of a drug, and the apparent synergy of miconazole and itraconazole shows evidence of single agent instability—its inhibition errors are exceptionally large, indicating inconsistency between the replicate blocks. Much more striking are the synergies between sterol inhibitors, which span a greater range of concentrations and which recapitulate the known antifungal synergy between statins and azoles (Lorenz and Parks, 1990).
Figure 5.
Validation of expected combination effects in a yeast experiment focused on sterol biosynthesis. In yeast, the sterol pathway (left) produces ergosterol, an essential cell wall component, through a series of metabolic enzymes (grey symbols) acting on intermediate substrates (black symbols), and regulated by negative feedback (dotted connections). The experiment tested C. glabrata proliferation responses to all combinations of 10 antifungal drugs (Table I), six with known targets on the sterol pathway (inhibitor markers). The results are shown (centre) with concentrations increasing from the bottom left of each drug pair's dose matrix. The combination effect symbols (right) summarize the observed response for each pair in terms of the models presented in Figure 2. Size shows synergy as measured by VHSA, the volume between the data and HSA surface. The best-fit shape model is indicated by colour: black for HSA, blue for Loewe additivity, green for Bliss boosting, red for potentiation, and grey when two or more models could not be distinguished (see Supplementary Figure S5). Drug pairs sharing a target mostly produced weak responses (VHSA<1) consistent with Loewe additivity, and cross-target sterol combinations produced potentiation, as expected for a linear pathway under negative feedback (∼70% prediction accuracy). Combinations involving non-sterol drugs showed fewer strong combination effects and their synergy profiles across all the drugs were more variable.
The shape and magnitude of each combination effect is summarized by symbols whose size represents the total synergy (as measured by VHSA) and whose colour denotes the best-fit shape model. Degeneracies between the models are measured by the comparative chi-squared (χ2) goodness-of-fit estimates (Supplementary 4 and Supplementary Figure S5). Although the data quality did not permit a clear distinction between the HSA model and Loewe additivity, the two most similar models in the set, most of the drug pairs sharing the same targets and agent-with-self combinations produced responses that were consistent with Loewe additivity. The exceptions all had relatively weak synergy, with only one, Itraconazole with Miconazole, exceeding the estimated systematic error level (VHSA>1; see Materials and methods). By contrast, all but one of the cross-target sterol combinations exceeded that level, and all produced surfaces that were best fit by potentiation. Across pathways, the combination effects were more variable, and strong synergies (VHSA>1; see Materials and methods) were relatively rare (5/28 versus 10/22 for same pathway), with an ∼8% Poisson probability (Press et al, 1997, §14.3.3).
The observed effects confirm predictions from our pathway simulations. We expected same-target combinations to produce Loewe additivity and cross-target sterol pairings to give rise to potentiation. Among the 11 same-target and 11 cross-target sterol combinations tested, the prediction accuracy is only 54% if we insist on unambiguously correct shape classifications (the predicted model has χ2min, and Δχ2>χ2min for the next best fit). However, almost all the failures are due to the degeneracy of the models for HSA-like combinations. If we count as a success any surface whose best fit was indistinguishable from the predicted shape, with Δχ2<χ2min, the prediction accuracy rises to 72% with an uncertainty of ∼22% due to the sample size, assuming Poisson statistics within each confusion matrix class. Across pathways, the relative diversity of combination effects makes sense, because their target connectivities are probably more varied, and the relative rarity of strong synergies accords with the predominantly masking effects in our simulated cross-pathway combinations.
Combination screen in human tumour cells
There is further support for our simulations from a prior combination screen aimed at discovering potential combination therapeutics. Human tumour cells (HCT116) were tested using a proliferation assay for responses to all pairwise combinations of 90 drugs and probes, most with known mechanism (Supplementary 5). This screen provides a broader sampling of possible target connectivities than had our C. glabrata experiment, and permits us to investigate the applicability of our shape models to cellular signalling networks. Because this screen had been designed for discovering potent synergies, a four-fold dilution factor had been used (compared to two-fold for our yeast experiment), and many compounds were either inactive alone or had not been sampled far above their EC50 concentrations, often obscuring distinctions between fitted shape models. Nevertheless, we expect strong potency shifts to be nearly always detectable, and other shape classes can be distinguished when the single agents have been appropriately sampled.
For each combination, we obtained an HSA volume VHSA and best-fit shapes as described above. We also examined ‘synergy profiles’ constructed from each agent's VHSA values across all the other probes, and recorded the correlation coefficient between the synergy profiles for each pair of drugs in the screen. Each pair of drugs was classified by target relatedness, using the known target annotations (Supplementary 5), as ‘Identical’ for drug-with-itself, ‘Same’ for drugs sharing a target, ‘Related’ for pairs with distinct targets in the same pathway or function, ‘Different’ for pairs targeting distinct functions, and ‘Unknown’ for pairs involving at least one uncharacterized drug. Inter-kinase combinations were classified separately as ‘Kinases’. Supplementary 6 presents the resulting scores and correlations for all the tested combinations, and Supplementary 7 shows their distributions grouped by target similarity.
The shape models in Figure 2 provide reasonable coverage of the observed combination effects. Only ∼30% of 4092 drug pairs in the screen produced a formally acceptable fit (χ2min<2) to any of the models (Supplementary 7a). However, given the systematic errors produced by the experimental process (Materials and methods), a more fair assessment of the uncertainties can be obtained by examining the χ2 distribution of the Loewe additive model for the 87 drug-with-self combinations, as a negative control set. These ‘Identical’ combinations have a median additive χ2∼9, suggesting that the uncertainties are roughly three times larger than the formal estimates from replicate dose matrices, due to systematic errors that were not accounted for by the scatter between replicates. Adopting this χ2 level as more representative of the errors extends the coverage (χ2min<18) of our models to most of the screen, leaving only ∼10%, which are dominated by those combinations that are equally inconsistent with all four models.
For an analysis of the observed shape distributions, we selected a set of ‘optimally sampled’ combinations, where at least two concentration samples had been tested above the EC50 for both single agents, enabling distinctions between strong combination effect shapes. Among these 253 combinations, ∼60% fit masking models (HSA, additive, or Bliss masking), ∼30% were best fit by potentiation, with the remaining ∼10% accounted for by multiplicative or saturating Bliss boosts. These proportions hold even for the 91 optimal combinations with sub-saturated single agent activities (Emax<90%), for which masking and multiplicative effects would be clearly distinguishable. For all target connectivity groupings, the optimally sampled combinations were dominated by masking and potentiation effects.
Across the whole screen, combination effects and synergy profile correlations (Figure 6A and B) support the trends observed in our pathway simulations and the C. glabrata experiment. When we examined synergy score distributions (Supplementary 7), we found that drug pairs targeting different pathways or involving unknown targets had significantly more variable scores (Materials and methods) with a slight trend to lower synergy levels. The synergy profile correlations show a more striking trend. Pairs with the same or related targets ought to have similar connectivities with the rest of the set, because they are likely to have similar mechanistic relationships to the rest of the drug targets in the screen. Indeed there are significant differences between the profile correlation distributions with a stronger trend towards increasing correlation with target similarity.
Figure 6.
Combination effect statistics from the HCT116 screen show expected trends, and the exceptionally strong synergies between inhibitors of EGFR signalling (Table II) reflect known connections. The distributions of HSA volumes (A) and synergy profile correlations (B) are shown for drug pairs in the HCT116 tumour cell screen (Supplementary 6), grouped by target similarity (‘Identical’ for drug-with-self combinations; ‘Same’ for distinct drugs sharing a target; ‘Related’ for differing targets sharing a common functional group; ‘Different’ for pairs with different functions; and a separate group ‘Kinases’ for combinations between kinase inhibitors). There is a significant trend towards more variable synergy and lower profile correlations for more distantly related targets (Supplementary 7), and the most synergistic and highly correlated pairs within the ‘Kinases’ set are dominated by those targeting the EGFR signalling pathways, which are major drivers of HCT116 proliferation. (C) Combinations between MAPK/PI3K, MAPK/PKC, and PI3K/PKC targets show different levels of response that are consistent across mechanistic replicates, despite limited concentration sampling (only TUBC and ROTT were optimal) and evidence of off-target effects (synergy between PI3K inhibitors and discrepant ROTT effects compared to the other PKC inhibitors). Shape model degeneracies are shown in Supplementary Figure S6.
The responses to pairings of kinase inhibitors also provide support for an association between combination effects and connectivity. Whereas combinations between kinases in general had similar distributions to those for other related target pairs, combinations between drugs targeting the phosphatidylinositol-3 kinase (PI3K), protein kinase C (PKC), or mitogen-activated protein kinase (MAPK) signalling pathways (Table II) showed exceptionally strong synergies and correlations. HCT116 cells maintain an activating KRAS mutation that drives cellular growth and survival (Shirasawa et al, 1993), and are especially reliant upon epidermal growth factor receptor (EGFR)-mediated signalling for proliferation (Awwad et al, 2003). Because these pathways are central to EGFR signalling and interconnected (Schlessinger, 2004), we would expect HCT116 cells to be very sensitive to these combinations, and that inhibitors of those pathways should have similar synergy profiles. Moreover, the combination effects between the pathways show differences (Figure 6C) that are consistent across mechanistic replicates, despite limited concentration sampling and evidence of secondary target activity. Such differences may eventually provide novel insights into how these pathways interact in the context of HCT116 proliferation (e.g., that the MAPK/PKC connection may be less direct than the other target pairings).
Table 2. Drugs targeting EGFR signalling used in the HCT116 proliferation experiment.

Discussion
The simulations and experiments presented here add a new dimension to earlier studies investigating the effects of multiple perturbations to biological systems. Early studies of statistical epistatic associations between random mutagenesis and chemical stresses (Elena and Lenski, 1997; Kishony and Leibler 2003) were followed by systematic studies of mutant libraries in combination with chemical probes (Giaever et al, 2004; Lum et al, 2004; Parsons et al, 2004) and synergies between targeted chemical probes (Haggarty et al, 2003; Yeh et al, 2006). This study explores the additional information available from variable dose response matrices, and relates quantitative combination effects, as measured response shape classification or by HSA volumes, to target connectivity. We use the term ‘connectivity’ to capture both the distance and topology of the connection between the targets, rather than the more common ‘interaction’ (Parsons et al, 2004), denoting the presence of a functional connection. Our pathway simulations show that biological systems are expected to produce a useful variety of combination effects that are correlated with connectivity, and our experimental results demonstrate that the simulations are biologically relevant and predictive.
There was only partial consistency between specific chemical synergies in our yeast experiment and corresponding interactions observed in earlier genetic screens (Supplementary 8). The observed synergies between sterol inhibitors are reflected in homozygous synthetic lethalities (Stark et al, 2006) that are internal to the pathway, and rapamycin's synergy with terbinafine agrees with the reported interaction between TOR1 and ERG1. However, the mutant screen of caspofungin's target, FKS1, showed no correlation with our combination effects. Moreover, there is little correspondence between our results and the cross-activities identified in drug sensitivity screens of heterozygous yeast mutants (Giaever et al, 2004; Lum et al, 2004; Parsons et al, 2004). The mixed results of these comparisons are not very surprising. Aside from experimental errors (see Materials and methods), some of the disagreement can be ascribed to the quantitative nature of dose-matrix response data, because the responses to varying doses (Hartman and Tippery, 2004), or even time-resolved experiments (Warringer et al, 2003), can detect the effects of non-essential genes that would have been missed in lethality screens. Another contribution may come from likely differences between genetic and protein networks (Ozier et al, 2003). Chemical combinations can interfere directly with protein interactions without mediation through gene expression, and although the protein and gene networks are intertwined, connectivity differences between the two types of targets should be expected.
There was more agreement between our simulation results and a prior flux balance analysis (FBA) of the yeast metabolic network (Segrè et al, 2005) in response to paired mutations. The simulated mutations should correspond roughly to the high-concentration limit of our dose matrices. Both approaches predict strong synergies (synthetic lethality) when the system is inhibited across parallel alternative pathways, and find masking effects (buffering) when unrelated pathways are impaired, provided that the two pathways are not competing for a common supply of precursors (Supplementary Figure S2 in Segrè et al). The main difference is that the FBA simulations produced more multiplicative than masking pairings, especially for cross-pathway combinations. This may be due to the partial view of combination effects provided by the epistasis scores (Supplementary Figure S1), or to an implicit assumption, in the FBA simulations, that competition for the same metabolic ingredients is widespread. The masking responses found in our cross-pathway simulations are supported experimentally, by the weaker synergy levels seen in our C. glabrata experiment, and by the large fraction of masking effects (∼60%) in both the HCT116 screen and prior antibacterial combination experiments (Yeh et al, 2006).
Thus the dose-matrix response shapes and synergy profiles from systematic combination screens can provide valuable new insights into network connectivity. A possible application in chemical genetics would be to extend phenotypic profiling for mechanism inference (Perlman et al, 2004; Macdonald et al, 2006), using combinations effectively as extra assays. Query drugs with unknown targets could have their synergy profiles compared to a previously assembled profile library for a diverse set of chemical probes. The known targets for those probes whose profiles correlate most closely with a query drug's are likely candidates for the drug's mechanism. For constructing biological network models, observed combination effects can be compared to expectations from target connectivity through existing protein interaction networks, and inconsistencies can be used to improve the models by means of a prediction-validation procedure (Ideker et al, 2001). Combining chemicals is much simpler than the process of constructing double mutant strains, and chemical combinations can yield detailed information about connections between proteins that are not accessible to single agents or mutagenesis, enabling the study of disease networks like those involved in cancer (Schoeberl et al, 2002) and inflammation (Bouwmeester et al, 2004) signalling.
Of course, there are limitations to this approach for probing biological networks. The collection of full dose matrices is expensive, and will necessitate a trade-off between the added information obtained from response shapes and the loss of assay fidelity from the use of high-throughput measurements (e.g., single time points for growth rather than time-resolved growth curves). The mechanistic information that can be extracted will depend strongly on the generality and specificity of the set of shape models being used. The four models presented here represent only a subset of the observed responses, and the pathway simulations sampled only a small minority of possible target connectivities. Even with more extensive simulations, there will certainly be mechanistic degeneracies associated with each distinct response shape. For example, it will probably be possible to produce potentiation like that seen in Bactrim using several distinct target configurations. Thus, response shapes cannot be used to uniquely define combination mechanisms, but merely to exclude inconsistent connections. Finally, chemical probes often have poorly characterized mechanisms or multiple targets, and combination screens will have to account for this by covering biological targets with several chemically distinct probes. Despite all these limitations, the present study has shown that even with a limited set of shape models, dose-matrix data can provide useful connectivity information.
The results of this initial exploration suggest several directions for expanding the response surface approach to combination analysis. First, it is essential to extend the current shape model set by using more comprehensive simulations of larger networks. The most obvious way to do this would be to define a basis set that represents simple observed network connectivities, including serial and parallel arrangements, bifurcations, both AND and OR junctions, and some regulation mechanisms (Milo et al, 2002). Combination responses from pathway simulations covering the connectivity basis could then be used to define shape models for comparison with simulated responses for larger networks, as well as with experimental data. Another approach would be to systematically explore perturbations of an existing biological network, such as S. cerevisiae metabolism (Famili et al, 2003), and to model drug inhibition by partially restricting the reaction rates at metabolic enzymes, essentially the Segrè et al's FBA for partial inhibitors. The challenge with this approach would be to identify meaningful connectivity measures that correlate with simulated synergy types. A third very promising area for future exploration is higher order combinations. The emergence of resistance to treatment for afflictions like cancer (Fojo and Bates, 2003) can be effectively addressed by combinations of three or more drugs (Komarova and Wodarz, 2005). Combination approaches can also offer improved control by perturbing multiple components of the disease network (Csermely et al, 2004), and higher order combinations should provide yet more details on mechanistic connections between their targets. The response surface approach presented here can be readily adapted to three or more drugs, as can the specific shape models shown in Figure 2, and a metabolic FBA of higher order combinations should provide important insights into the effectiveness of multi-target treatments. Finally, because the simulated combination responses were insensitive to the reaction details, this methodology may even be extendable to non-biological network problems where responses to targeted perturbations can be measured (Milo et al, 2002).
Materials and methods
Combination response shape models
Experimental dose-matrix responses include both potency shifts, where a combination alters the apparent potency of the single agents, and efficacy boosts, where the joint effect exceeds levels possible for either of the constituents. Many of these responses can be described by simple models that use the single agent curves to predict the combination effect (Figure 2), and whose parameters provide quantitative measures. Strong potency shifts relative to a simple HSA surface can be described using a power-law potentiation model, and efficacy boosts are quantifiable using a model that extends Bliss Independence (Bliss, 1939) to variable combination effect levels at high concentration. These models are expressed in terms of inhibitions, but can be adapted to other types of measurement, or for combinations involving more than two perturbing agents, without altering their qualitative properties. The shapes presented are not a complete orthogonal set covering all expected combination responses, but were chosen to represent some of the variety seen in our therapeutic screens, as well as to reflect historical reference models used in the literature.
The HSA model, or Gaddum's non-interaction (Berenbaum, 1989), is based simply on the intuition that if a combination's effect exceeds those of its constituents, there must be some interaction. Mathematically, the HSA model is a superposition of the single agent curves where, at any combined concentration (X,Y), the inhibition IHSA=max(IX,IY), where IX and IY are effects produced by the single agents at (X,0) and (0,Y). The model values are calculated at each dose-matrix point, where IX and IY were determined using sigmoidal fits to the single agent response data. The volume VHSA=ΣX,Yln fX ln fY(Idata−IHSA) between the data and the HSA surface, adjusted for variable dilution factors fX, fY, can be readily calculated and provides a practical overall measure of synergy (in units of inhibition), summed over dimensionless log-concentration space.
We also considered Loewe additivity (Loewe, 1928), the drug-with-itself standard. At each combined concentration (X,Y), we used an iterative approach (Berenbaum, 1985) to find the inhibition ILoewe that satisfies (X/XI)+(Y/YI)=1, where XI and YI are the single agent effective concentrations. Starting with a guess that I=IHSA, we interpolated the single agent curves to find XI,YI that produce I, calculated the corresponding combination index, and used bisection (Press et al, 1997) to converge on a value of I with combination index CI=1. Loewe additivity reduces to HSA at concentrations that are very different from XI,YI, as well as for I above an agent's limiting activity (effectively XI,YI → ∞).
To model boosts in efficacy at high concentrations different from what the single agents can achieve, we used a ‘Bliss boosting’ model, adapting the Bliss independence model (Bliss, 1939) for boost levels other than multiplicative. Mathematically, IBliss=IX+IY+(β−Emin)(IX IY/EX EY), where Emin is the lesser of EX and EY, the limiting single agent efficacies. The one free parameter β, in units of effect, determines the amount of boosting above Emax, the greater of the single agent efficacies. For inhibition effects, there are a number of useful reference levels: β=−Emax produces ‘cancelling’, with zero effect at high concentration; β=Emin−Emax is ‘suppressing’, where the less effective agent prevails; β=0 yields ‘masking’, where the more effective agent prevails; β=Emin(1−Emax) is multiplicative (Bliss independence); and β=(1−Emax) produces ‘saturating’ at 100% inhibition. At the high concentration limit, the suppressing, masking, multiplicative, and saturating levels correspond to ‘suppression’, ‘buffering’, ‘no epistasis’, and ‘synthetic lethal’ in recent classifications for epistasis (Elena and Lenski, 1997; Segrè et al, 2005; Yeh et al, 2006). Bliss boosting can be used in this form for any type of effect measure provided that the single agent responses increase monotonically with concentration. Only the saturating and multiplicative reference levels need to be adjusted according to the measurement scale. For example, if we consider fitness ratios (treated over untreated growth ratios) with f=ln(T/U) in place of inhibitions, there is no saturating level and β=Emin indicates multiplicative. Note that Bliss masking is similar to both HSA and Loewe additivity for very high or very low concentrations where the single agent effects are almost flat.
Finally, we used a power-law ‘potentiation’ model to describe strong shifts in potency similar to that seen for Bactrim® (Figure 1). For such combinations, an active single agent's response curve shows an increase in potency as the enhancer is titrated in, which is seen as linear iso-effect contours in serially diluted dose matrices (logarithmic concentration space). The inhibition IPotent=max(IX(C), IY), where IX(C) is the single agent response curve of the potentiated compound, at a shifted concentration C=X[1+(Y/Ypot)∣p∣]sign(p), where sign(p) is a unit sign function evaluated at (−1, 0, +1) corresponding to the sign of its argument. Although the functional form presented here is awkward, it was the simplest way we could find to achieve power-law potentiation and depotentiation above a threshold enhancer concentration, with the enhancer's own activity superimposed. The two free parameters for this model are the threshold concentration Ypot above which potentiation takes effect, and the potentiation slope p (synergy for positive and antagonism for negative p). There is no potentiation for p=0, where this model reduces to an HSA surface. Just as for HSA and Loewe additivity, the form of this model is identical for any type of effect measure.
These models are not orthogonal; so we defined a fitting order to ensure an unambiguous choice. The shape models have degeneracies where two or more models can describe similar response shapes. Loewe additivity and HSA, for example, are largely degenerate, in that only when the single agents increase slowly over the sampled concentrations can a distinction between the two models be made. Nevertheless, both can be readily distinguished from Bliss boosting and potentiation, which themselves are distinct over a large range of their parameters, and many observed surfaces fit only one model effectively (Supplementary 2). To ensure an unambiguous choice that applies the principle of Occam's razor when fitting several models to an observed surface, we ordered them by increasing complexity (HSA, Loewe, Bliss, potentiation) and declared as best fit the first model that was consistent with the observed surface. Within Bliss boosting, we classified combinations as cancelling, suppressing, masking, multiplicative, or saturating, depending on which fell closest to the best-fit β for the observed response shape. Note that for some pairings, the boost levels cannot be separated. For example, if one single agent reaches 100% inhibition at high concentration, it will be impossible to distinguish saturating from multiplicative or masking levels. If both single agents reach 100%, only cancelling effects will be distinguishable from the others. For such boost level degeneracies, the first consistent level in the sequence above was chosen.
We note that these models were used primarily to characterize response surface shapes. Indeed, of the four shape models discussed here, only Loewe additivity has an a priori physical foundation as a predictive combination model. Although the original Bliss independence model (corresponding to the multiplicative level of Bliss boosting) is based on statistical independence, that foundation has no clear relevance to biological systems, where widely separated targets are neither statistically independent nor cleanly competing for the same end point. The Bliss boosting and potentiation models were included in our analysis to describe distinct shapes observed in our screens, and for well-sampled dose matrices neither is very good at fitting the effects that the other was designed to describe.
Pathway simulations
Simulations of metabolic pathways were performed using the XPPAUT ordinary differential equation simulation software (Ermentrout, 2002), on Boston University's Biowulf computing cluster. The network shown in Figure 3 was modelled as a system of Michaelis–Menten enzymatic reactions, each with three kinetic parameters: the limiting reaction rate Vm, the rate constant Km, and an exponential degradation rate Dg. The two pathways were driven by supplying substrates S1 and S11 from limitless sources with constant reaction velocities, and converting them through the enzymatic reactions before passing through the end-point reaction RE into a limitless sink. For symmetry and simplicity, we set all Vm=1 and Km=1, and the degradations Dg to a lower level of 0.01 in most cases. We used Dg=1 for substrates at bifurcations and for the final product of Pathway A to ensure illustrative differences in the single agent inhibitions (reducing response shape degeneracies) and to balance the flux through both pathways (for stable simulations). The system was initially started with all substrate levels set to 0.1 and run to steady state using an RK4 integration algorithm calculated at time intervals of 0.001 over 1 000 000 iterations, and the resulting substrate levels and parameter settings are shown in Supplementary 1. The combination response for each pair of inhibitors was determined using 100 simulations over a 10 × 10 dose matrix, where for each dosing point the targeted reactions were restricted by competitive inhibitors (that affect Km) and the simulation was run for a further 600 000 iterations using a step size of 0.005, before the inhibition I=1–T/U was calculated on the treated end-point reaction velocity T, relative to the untreated velocity U. Optimal 9-point inhibitor concentration samples were chosen for each single agent by using a bisection search to find the single agent's EC10 and EC90 concentrations, defining six samples with a fixed dilution factor f=(EC90/EC10)0.2 to cover that range, and extending the total range with two additional concentration steps above the EC90 and one more below the EC10 at the same dilution factor f.
The resulting combination response data are tabulated in Supplementary 2. Each surface was compared to the models presented in Figure 2 (assuming 1% inhibition errors), and the best fit was determined to be HSA, Loewe additivity, Bliss boosting, or potentiation, in that order, depending on which was first consistent with the best-fitting model's χ2 (where consistency meant that its χ2 was less than twice the best fit's). Bliss boosting models were further classified as cancelling, suppressing, masking, multiplicative, or saturating, where it could be uniquely specified given the single agent limiting efficacies. The shape classifications, chi-squared, and best-fit parameter values are listed in Supplementary 2, and the shapes are also summarized in Figure 3. Supplementary Figure S1 compares the shape classifications and target connectivities from this simulation to epistasis calculations (Segrè et al, 2005; Yeh et al, 2006) based on single concentration points taken from the matrices.
We varied the model parameters to explore the sensitivity of our results to the kinetic parameters. First, we performed Monte Carlo tests with 100 simulations, each time allowing all of the parameters to randomly vary with uniform probability density within a multiplicative range 10±Δ, and counted how often the best-fit model surface agreed with the original fit before perturbation (Supplementary Figure S2). The Monte Carlo tests were repeated with steadily increasing Δ, to determine the level at which the unperturbed simulations failed to predict the outcome, both with all Vm, Km, and Dg varied simultaneously, and with each parameter class perturbed separately. Finally, we explored sensitivity to individual nodes by perturbing each enzyme in Pathway A separately.
We also explored some simulation assumptions to test the robustness of our results. First, we replaced the competitive inhibitor kinetics with non-competitive (affecting Vm) inhibition, performed the simulations, and repeated the model fitting to determine the best-fit response shapes. Second, we addressed the concern that a metabolic reaction velocity might provide a poor comparison to proliferation, the most accessible experimental end point. As proliferating cells undergo exponential doubling, a linear reduction of velocity by a factor T/U for a growth-limiting metabolic reaction should slow the doubling time by a proportional factor. Over a time interval corresponding to n uninhibited doublings, the resulting inhibition of cell population becomes Igrowth∼1–2nIv, in terms of the velocity-based inhibition IV. We applied this transformation to our simulated dose-matrix inhibitions for Pathway A, assuming n∼10 doublings, and repeated the surface fitting procedure on the transformed dose matrices to determine the best-fit response shapes.
Combination screening experiments
The chemical library was archived in robotically accessible vials, to which diluent (dimethyl sulphoxide or water) was added in preparation for addition to 384-well master plates by a Tecan Freedom liquid-dispensing robot. Liquid transfers to dilution and assay plates were handled using a Perkin-Elmer MiniTrak station adapted for the combination high-throughput procedure. Each 384-well assay plate contained six 6 × 6 dose-matrix blocks, with four serial dilutions (two-fold for yeast and four-fold for HCT116) of the top concentration for each agent. Additional wells were reserved for transfer and untreated control wells. The compound mixtures were then added to the biological component of the assay as appropriate.
For the yeast experiment, C. glabrata (ATC #90030) cultures were cultured overnight in flasks with RPMI media in a 32°C shaker at 250 r.p.m. The next day, the cultures were diluted into media until they had an absorbance equivalent to 50% of McFarland standard, corresponding to a density of ∼8000 cells/ml, and dispensed with automated dispensers into 384-well assay plates (35 μl wells containing media and test compounds) to yield ∼160 cells/well. The plates were incubated in the presence of drugs and media at 32°C for 18 h (∼10 doublings). Upon completion, cell populations were measured at a single time point, with a metabolic Alamar Blue fluorescence reagent (at 590 nm emission) using a Perkin-Elmer Fusion plate reader after excitation at 535 nm.
For the tumour cell line experiment, the HCT116 (ATC #CCL-247) cells were cultured in flasks with RPMI-1640 media for 2–20 passages, drawing test populations along the way. Extracts were trypsinized, counted, and seeded into 384-well plates at a density of 1500 cells/well in 35 μl media using automated dispensers. To ensure adherence, the plated cells were incubated at 37°C, with 5% CO2 overnight, after which the compounds were added and the plates were incubated again at 37°C with 5% CO2 for 72 h (or about 2–3 doublings). After incubation, 10.5% Alamar Blue fluorescence dye (in 40 μl media) was added and plates were incubated for 6 h. Cell population measurements were made at a single time point upon completion, using a Perkin-Elmer Victor II plate reader (excitation at 535/590 nm emission).
This experimental procedure has a variety of sources for error in final proliferation measurements. Each compound is dissolved once in a single stock tube, and delivered into X- and Y-diluted master plates. Aliquots are then transferred from the master plates to each assay plate to produce the combinations. While this approach permits orthogonal serial dilutions and aids uniformity across instances of each master plate, there can be systematic errors in compound plating between the X and Y masters for less stable compounds. Cells are seeded in media at high densities, to reduce the well-to-well variability, but occasional clogs in a pipette can lead to non-uniform distribution. During incubation, slight variations in temperature or humidity can lead to varying growth rates across wells on a plate and between plates. Because the single cell population measurement is made during the exponential growth phase, variations can be amplified in the final readout, and this variation can be exaggerated when cells are sensitized by test compounds. The result of these various factors is that cell population measurements vary by ∼1–3% on each plate, across untreated wells, and the variations do not fit a normal distribution. Moreover, some 1–2% of wells on each plate show occasional spikes that are very different from their neighbours.
The contents of each plate were tracked in an automated laboratory information management system, using integrated barcode scanners in the liquid handling equipment, and stored in an Oracle database. Plates lacking compound transfers or with insufficient dynamic range (signal-to-noise ratio <5 between untreated controls and a cell-free background) were rejected and repeated. The remaining plates were inspected using custom quality control software. Individual wells that fell outside the expected range for normal assay readouts, or which were discontinuous with their neighbours, were marked for exclusion. Finally, the single agent wells in each combination block were visually inspected for consistency across the experiment, and combination blocks containing the most discrepant single agents were marked for exclusion.
Data analysis
Dose matrices were assembled from replicate combination blocks on experimental plates. Some of the data showed systematic variations across the plate, most likely owing to temperature or humidity gradients during incubation. In order to correct these effects, our assay plates had 40 untreated wells arranged around and between the blocks. The systematic variations were removed by fitting a smooth function of plate location to the untreated well data values, and dividing out the modelled spatial variation. After plate effect correction, fluorescence counts T from each treated well were converted to inhibitions Idata=(U−T)/U relative to the median U of 20 untreated wells. Replicate blocks for each dose matrix were merged, using the median inhibition at each dosing point. Standard error estimates σdata for each median inhibition were also calculated, based on the quadrature sum of a minimum acceptable 3% error, the median absolute deviation (MAD) of the corrected untreated data on each plate normalized to their median, and the MAD between replicate inhibition data between plates, using an empirical conversion from MAD to standard deviations (Filliben, 2005).
Each dose matrix was scored for synergy, and combination effects were compared to our response morphology models (Figure 2). Sigmoidal dose responses I=ECa/(Sa+Ca) as a function of concentration C were fitted to the single agent data, where E is the limiting efficacy at high concentration, S is the effective concentration, and a is the sigmoidicity or steepness of transition. These fitted curves were used for combination effect models. The HSA, Bliss boosting, and potentiation models were calculated, and the Loewe additivity model was solved at each dosing point by iterative root finding (Berenbaum, 1985). As a quantitative estimate of the combination activity, we computed HSA volume excess scores VHSA=∑X,Y ln fX ln fY(Idata–IHSA), summed over all CX,CY, along with an associated error estimate σHSA=sqrt[∑X,Y ln2 fX ln2 fYσ2data], both expressions using adjustments to account for differing dilution factors fX, fY. This volume measures the total excess inhibition, summed over dimensionless log-concentration space. Data surfaces were compared to the shapes presented in Figure 2 by varying the model parameters, using a Nelder-Mead simplex algorithm (Press et al, 1997, §10.4) to minimize χ2=Σdata[(Idata–Imodel)/σdata]2/Ndata, the reduced chi-squared. All four models were explored, and the overall best fit with the least chi-square χ2min was chosen to represent the shape of the combination. A best fit was chosen from HSA, additivity, boosting, or potentiation, in that order, depending on which was first consistent with the χ2min (Δχ2<χ2min).
Finally, systematic errors that were not captured by our error estimates from replicates can be estimated by examining the drug-with-self combinations as a negative control for combination effect. For both screens, the median Loewe additivity χ2 was ∼9, suggesting that the true reproducibility of drug response was roughly three times the estimated levels from the scatter between replicates. This suggests systematic errors of ∼10% at each dose-matrix point, leading to integrated volume error estimates of σHSA∼0.4. Thus, only combination effects with VHSA>1 are strong enough to be reliably distinguishable from drug-with-itself combinations.
For the HCT116 combination screen, synergy profiles were constructed for each drug by collecting into a vector all the VHSA scores involving that chemical. These profiles were then compared for all pairs of drugs in the screen by means of the Pearson correlation coefficient. The synergy profile correlations and synergy scores were grouped by the target similarity (Supplementary 5) of the two drugs in each combination, and classified as follows: ‘Identical’ for drug-with-itself; ‘Same’ for distinct drugs inhibiting the same target; ‘Related’ for differing targets sharing a common functional group; ‘Kinases’ for combinations of kinase inhibitors; ‘Different’ for unrelated pairs of drugs; and ‘Unknown’ for pairs involving one or more unknown mechanism. The target similarity group distributions were compared for significant differences using a chi-square test that assumes only Poisson counting statistics within each bin in the histograms (Press et al, 1997, §14.3.3).
Supplementary Material
Supplementary figures
XPPAUT ODE file specifying the reaction network shown in Fig. 3
Drugs used in the HCT116 tumour cell proliferation screen Sup_5
Response surfaces and shape model fits for the pathway simulations
Monte Carlo simulation results for the multiply-inhibited branched pathway
Combination response data and shape fitting results for the C. glabrata proliferation experiment
Combination scores from the HCT116 tumour cell proliferation screen
Statistical results for the HCT116 tumour cell proliferation screen
S. cerrevisiae drug sensitivity and genetic interaction data relevant to the C. glabrata experiment
Acknowledgments
This work was enabled and assisted by many people at CombinatoRx. We are grateful to Boston University's Department of Bioengineering for providing access to the Biowulf computing cluster, and to Tim Gardner for co-advising Andrew Krueger's thesis. Herbert Sauro, Baltz Aguda, Mike Foley, Trey Ideker, and Leroy Hood gave encouragement and advice at key points. We also thank the referees for very helpful comments. Brent Stockwell is supported in part by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Competing financial interests statementThe authors declare competing financial interests.
References
- Ashford JR, Cobby JM (1974) A system of models for the action of drugs applied singly or jointly to biological organisms. Biometrics 30: 11–31 [PubMed] [Google Scholar]
- Awwad RA, Sergina N, Yang H, Ziober B, Willson JK, Zborowska E, Humphrey LE, Fan R, Ko TC, Brattain MG, Howell GM (2003) The role of transforming growth factor alpha in determining growth factor independence. Cancer Res 63: 4731–4738 [PubMed] [Google Scholar]
- Berenbaum MC (1985) The expected effect of a combination of agents: the general solution. J Theor Biol 114: 413–431 [DOI] [PubMed] [Google Scholar]
- Berenbaum MC (1989) What is synergy? Pharmacol Rev 41: 93–141 [PubMed] [Google Scholar]
- Bergstrom JD, Bostedor RG, Masarachia PJ, Reszka AA, Rodan G (2000) Alendronate is a specific, nanomolar inhibitor of farnesyl diphosphate synthase. Arch Biochem Biophys 373: 231–241 [DOI] [PubMed] [Google Scholar]
- Bliss CI (1939) The toxicity of poisons applied jointly. Ann Appl Biol 26: 585–615 [Google Scholar]
- Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehár J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, Keith CT (2003) Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA 100: 7977–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (2004) A physical and functional map of the human tnf-alpha/nf-kappa b signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Carter WH Jr, Wampler GL (1986) Review of the application of response surface methodology in the combination therapy of cancer. Cancer Treat Rep 70: 133–140 [PubMed] [Google Scholar]
- Chou T-C, Talalay P (1983) Analysis of combined drug effects: a new look at a very old problem. Trends Pharmacol Sci 4: 450–454 [Google Scholar]
- Csermely P, Ágoston V, Pongor S (2004) The efficiency of multi-target drugs: the network approach might help drug design. arXiv 1–9. http://arxiv.org/abs/q-bio.MN/041204 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H (2002) A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Davis PD, Elliott LH, Harris W, Hill CH, Hurst SA, Keech E, Kumar MK, Lawton G, Nixon JS, Wilkinson SE (1992) Inhibitors of protein kinase C. 2. Substituted bisindolylmaleimides with improved potency and selectivity. J Med Chem 35: 994–1001 [DOI] [PubMed] [Google Scholar]
- Douglas CM, D'Ippolito JA, Shei GJ, Meinz M, Onishi J, Marrinan JA, Li W, Abruzzo GK, Flattery A, Bartizal K, Mitchell A, Kurtz MB (1997) Identification of the fks1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob Agents Chemother 41: 2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE (1997) Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–398 [DOI] [PubMed] [Google Scholar]
- Ermentrout GB (2002) Simulating, Analyzing, and Animating Dynamical Systems: A Guide to xppaut for Researchers and Students. Philadelphia, PA: Society for Industrial & Applied Mathematics [Google Scholar]
- Famili I, Forster J, Nielsen J, Palsson BO (2003) Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA 100: 13134–13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM (1998) Idenfification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 273: 18623–18632 [DOI] [PubMed] [Google Scholar]
- Filliben JJ (2005) Exploratory data analysis. Engineering Statistics Handbook, Internet, National Institute of Standards and Technology http://www.itl.nist.gov/div898/handbook/eda/section3/eda356.htm [Google Scholar]
- Fojo T, Bates S (2003) Strategies for reversing drug resistance. Oncogene 22: 7512–7523 [DOI] [PubMed] [Google Scholar]
- Fox T, Coll JT, Xie X, Ford PJ, Germann UA, Porter MD, Pazhanisamy S, Fleming MA, Galullo V, Su MSS, Wilson KP (1998) A single amino acid substitution makes ERK2 susceptible to pyridinyl imidazole inhibitors of p38 MAP kinase. Protein Sci 7: 2249–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Shan H, Matsuda SP, Hampton RY (2001) An oxysterol-derived positive signal for 3-hydroxy- 3-methylglutaryl-CoA reductase degradation in yeast. J Biol Chem 276: 8681–8694 [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW (2004) Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci USA 101: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Bravo G, Parsons JC (1995) The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47: 331–385 [PubMed] [Google Scholar]
- Gschwendt M, Muller HJ, Kielbassa K, Zang R, Kittstein W, Rincke G, Marks F (1994) Rottlerin, a novel protein kinase inhibitor. Biochem Biophys Res Commun 199: 93–98 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999 [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Clemons PA, Schreiber SL (2003) Chemical genomic profiling of biological networks using graph theory and combinations of small molecule perturbations. J Am Chem Soc 125: 10543–10545 [DOI] [PubMed] [Google Scholar]
- Harrap KR, Jackson RC (1975) Enzyme kinetics and combination chemotherapy: an appraisal of current concepts. Adv Enzyme Regul 13: 77–96 [DOI] [PubMed] [Google Scholar]
- Hartman IV JL, Tippery NP (2004) Systematic quantification of gene interactions by phenotypic array analysis. Genome Biol 5: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Lang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L (2001) A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2: 343–372 [DOI] [PubMed] [Google Scholar]
- Jackson RC (1993) Amphibolic drug combinations: the design of selective antimetabolite protocols based upon the kinetic properties of multienzyme systems. Cancer Res 53: 3998–4003 [PubMed] [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4: 1–8 [DOI] [PubMed] [Google Scholar]
- Kishony R, Leibler S (2003) Environmental stresses can alleviate the average deleterious effect of mutations. J Biol 2: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova NL, Wodarz D (2005) Drug resistance in cancer: principles of emergence and prevention. Proc Natl Acad Sci USA 102: 9714–9719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Loewe S (1928) Die quantitativen probleme der pharmakologie. Ergebn Physiol 27: 47–187 [Google Scholar]
- Lorenz RT, Parks LW (1990) Effects of lovastatin (mevinolin) on sterol levels and on activity of azoles in Saccharomyces cerevisiae. Antimicrob Agents Chemother 34: 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD (2004) Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116: 121–137 [DOI] [PubMed] [Google Scholar]
- MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang Z, Huang Z, Yu H, Dias J, Minami T, Michnick SW, Westwick JK (2006) Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Bio 2: 329–337 [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Okada T, Sakuma L, Fukui Y, Hazeki O, Ui M (1994) Blockage of chemotactic peptide-induced stimulation of neutrophils by wortmannin as a result of selective inhibition of phospatidylinositol 3-kinase. J Biol Chem 269: 3563–3567 [PubMed] [Google Scholar]
- Ozier O, Amin N, Ideker T (2003) Global architecture of genetic interactions on the protein network. Nat Biotechnol 21: 490–491 [DOI] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C (2004) Integration of chemical–genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22: 62–69 [DOI] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ (2004) Multidimensional drug profiling by automated microscopy. Science 306: 1194–1198 [DOI] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP (1997) Numerical Recipes in c: The Art of Scientific Computing, 2nd edn. Cambridge: CUP [Google Scholar]
- Rine J, Hansen W, Hardeman E, Davis RW (1983) Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci USA 80: 6750–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J (2004) Common and distinct elements in cellular signaling via EGF and FGF receptors. Science 306: 1506–1507 [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G (2002) Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol 20: 370–375 [DOI] [PubMed] [Google Scholar]
- Segrè D, Deluna A, Church GM, Kishony R (2005) Modular epistasis in yeast metabolism. Nat Genet 37: 77–83 [DOI] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, Sasazuki T (1993) Altered growth of human colon cancer cell lines disrupted at activated ki-ras. Science 260: 85–88 [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) Biogrid: a general repository for interaction datasets. Nucleic Acids Res 34 (Database issue): D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell CL, Hughes CE (1992) Antifungal agents used for deep-seated mycotic infections. Mayo Clin Proc 67: 69–91 [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F (1991) The bisindolylmaleimide gf 109203x is a potent and selective inhibitor of protein kinase C. J Biol Chem 266: 15771–15781 [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A (2003) High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci USA 100: 15724–15729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills EA, Redinbo MR, Perfect JR, Del Poeta M (eds) (2000) New Potential Targets for Antifungal Development. Expert opinion on therapeutic targets. London: Ashley [Google Scholar]
- Yano H, Agatsuma T, Nakanishi S, Saitoh Y, Fukui Y, Nonomura Y, Matsuda Y (1995) Biochemical and pharmacological studies with KT7692 and LY294002 on the role of phosphatidylinositol 3-kinase in Fc epsilon RI-mediated signal transduction. Biochem J 312: 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P, Tschumi AI, Kishony R (2006) Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38: 489–494 [DOI] [PubMed] [Google Scholar]
- Zhang LV, King OD, Wong SL, Goldberg DS, Tong AH, Lesage G, Andrews B, Bussey H, Boone C, Roth FP (2005) Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J Biol 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann GR, Lehár J, Keith CT (2007) Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today 12: 34–42. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17198971 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
XPPAUT ODE file specifying the reaction network shown in Fig. 3
Drugs used in the HCT116 tumour cell proliferation screen Sup_5
Response surfaces and shape model fits for the pathway simulations
Monte Carlo simulation results for the multiply-inhibited branched pathway
Combination response data and shape fitting results for the C. glabrata proliferation experiment
Combination scores from the HCT116 tumour cell proliferation screen
Statistical results for the HCT116 tumour cell proliferation screen
S. cerrevisiae drug sensitivity and genetic interaction data relevant to the C. glabrata experiment